Browse DAXX in pancancer
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF03344 Daxx N-terminal Rassf1C-interacting domain |
||||||||||
| Function |
Transcription corepressor known to repress transcriptional potential of several sumoylated transcription factors. Down-regulates basal and activated transcription. Its transcription repressor activity is modulated by recruiting it to subnuclear compartments like the nucleolus or PML/POD/ND10 nuclear bodies through interactions with MCSR1 and PML, respectively. Seems to regulate transcription in PML/POD/ND10 nuclear bodies together with PML and may influence TNFRSF6-dependent apoptosis thereby. Inhibits transcriptional activation of PAX3 and ETS1 through direct protein-protein interactions. Modulates PAX5 activity; the function seems to involve CREBBP. Acts as an adapter protein in a MDM2-DAXX-USP7 complex by regulating the RING-finger E3 ligase MDM2 ubiquitination activity. Under non-stress condition, in association with the deubiquitinating USP7, prevents MDM2 self-ubiquitination and enhances the intrinsic E3 ligase activity of MDM2 towards TP53, thereby promoting TP53 ubiquitination and subsequent proteasomal degradation. Upon DNA damage, its association with MDM2 and USP7 is disrupted, resulting in increased MDM2 autoubiquitination and consequently, MDM2 degradation, which leads to TP53 stabilization. Acts as histone chaperone that facilitates deposition of histone H3.3. Acts as targeting component of the chromatin remodeling complex ATRX:DAXX which has ATP-dependent DNA translocase activity and catalyzes the replication-independent deposition of histone H3.3 in pericentric DNA repeats outside S-phase and telomeres, and the in vitro remodeling of H3.3-containing nucleosomes. Does not affect the ATPase activity of ATRX but alleviates its transcription repression activity. Upon neuronal activation associates with regulatory elements of selected immediate early genes where it promotes deposition of histone H3.3 which may be linked to transcriptional induction of these genes. Required for the recruitment of histone H3.3:H4 dimers to PML-nuclear bodies (PML-NBs); the process is independent of ATRX and facilitated by ASF1A; PML-NBs are suggested to function as regulatory sites for the incorporation of newly synthesized histone H3.3 into chromatin. In case of overexpression of centromeric histone variant CENPA (as found in various tumors) is involved in its mislocalization to chromosomes; the ectopic localization involves a heterotypic tetramer containing CENPA, and histones H3.3 and H4 and decreases binding of CTCF to chromatin. Proposed to mediate activation of the JNK pathway and apoptosis via MAP3K5 in response to signaling from TNFRSF6 and TGFBR2. Interaction with HSPB1/HSP27 may prevent interaction with TNFRSF6 and MAP3K5 and block DAXX-mediated apoptosis. In contrast, in lymphoid cells JNC activation and TNFRSF6-mediated apoptosis may not involve DAXX. Shows restriction activity towards human cytomegalovirus (HCMV). Plays a role as a positive regulator of the heat shock transcription factor HSF1 activity during the stress protein response (PubMed:15016915). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000187 activation of MAPK activity GO:0006323 DNA packaging GO:0006333 chromatin assembly or disassembly GO:0006334 nucleosome assembly GO:0006338 chromatin remodeling GO:0007254 JNK cascade GO:0007257 activation of JUN kinase activity GO:0008625 extrinsic apoptotic signaling pathway via death domain receptors GO:0009755 hormone-mediated signaling pathway GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0031098 stress-activated protein kinase signaling cascade GO:0031396 regulation of protein ubiquitination GO:0031497 chromatin assembly GO:0032147 activation of protein kinase activity GO:0032872 regulation of stress-activated MAPK cascade GO:0032874 positive regulation of stress-activated MAPK cascade GO:0033674 positive regulation of kinase activity GO:0034728 nucleosome organization GO:0043401 steroid hormone mediated signaling pathway GO:0043405 regulation of MAP kinase activity GO:0043406 positive regulation of MAP kinase activity GO:0043410 positive regulation of MAPK cascade GO:0043506 regulation of JUN kinase activity GO:0043507 positive regulation of JUN kinase activity GO:0045860 positive regulation of protein kinase activity GO:0046328 regulation of JNK cascade GO:0046330 positive regulation of JNK cascade GO:0048545 response to steroid hormone GO:0051403 stress-activated MAPK cascade GO:0065004 protein-DNA complex assembly GO:0070302 regulation of stress-activated protein kinase signaling cascade GO:0070304 positive regulation of stress-activated protein kinase signaling cascade GO:0070997 neuron death GO:0071103 DNA conformation change GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071824 protein-DNA complex subunit organization GO:0071900 regulation of protein serine/threonine kinase activity GO:0071902 positive regulation of protein serine/threonine kinase activity GO:0097191 extrinsic apoptotic signaling pathway GO:1901214 regulation of neuron death GO:1901216 positive regulation of neuron death GO:1903320 regulation of protein modification by small protein conjugation or removal |
| Molecular Function |
GO:0002039 p53 binding GO:0003714 transcription corepressor activity GO:0005057 receptor signaling protein activity GO:0008047 enzyme activator activity GO:0019207 kinase regulator activity GO:0019209 kinase activator activity GO:0019887 protein kinase regulator activity GO:0030295 protein kinase activator activity GO:0031072 heat shock protein binding GO:0031625 ubiquitin protein ligase binding GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0042393 histone binding GO:0044389 ubiquitin-like protein ligase binding GO:0047485 protein N-terminus binding GO:0050681 androgen receptor binding GO:0051427 hormone receptor binding |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0016604 nuclear body GO:0016605 PML body GO:0070603 SWI/SNF superfamily-type complex GO:0098687 chromosomal region |
| KEGG |
hsa04010 MAPK signaling pathway hsa04210 Apoptosis |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804757: Regulation of TP53 Degradation R-HSA-6806003: Regulation of TP53 Expression and Degradation R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
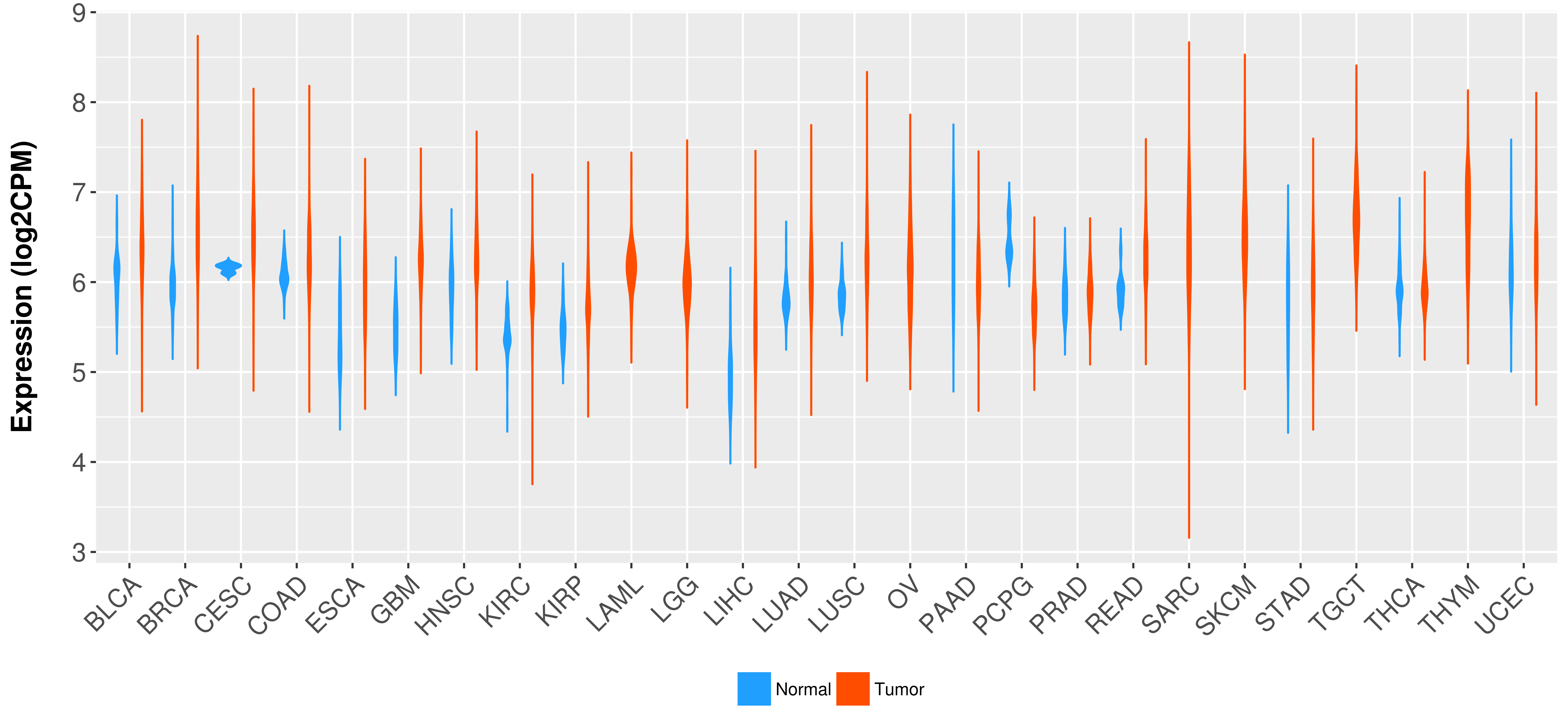
Differential expression analysis for cancers with more than 10 normal samples
|
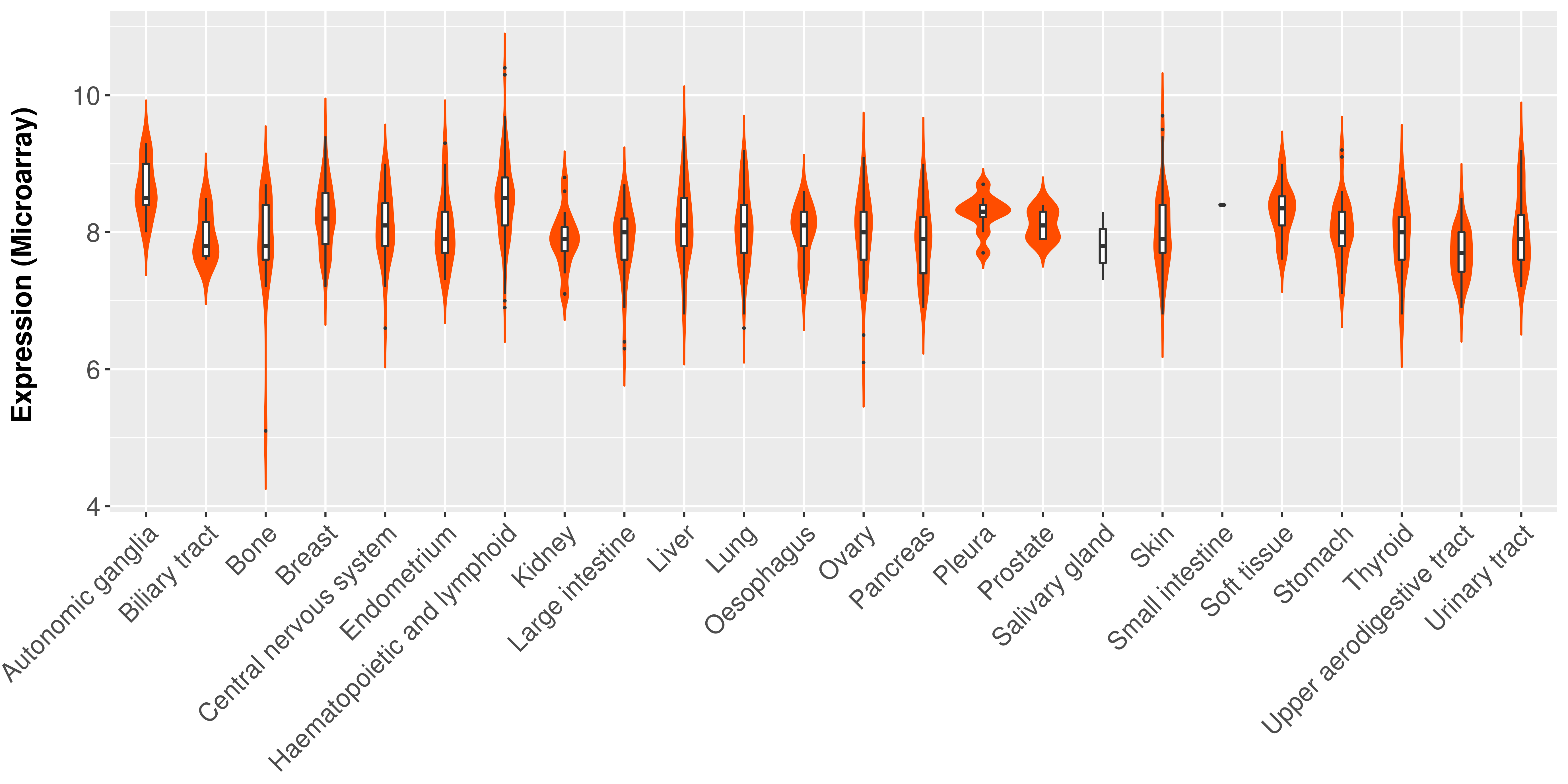
|
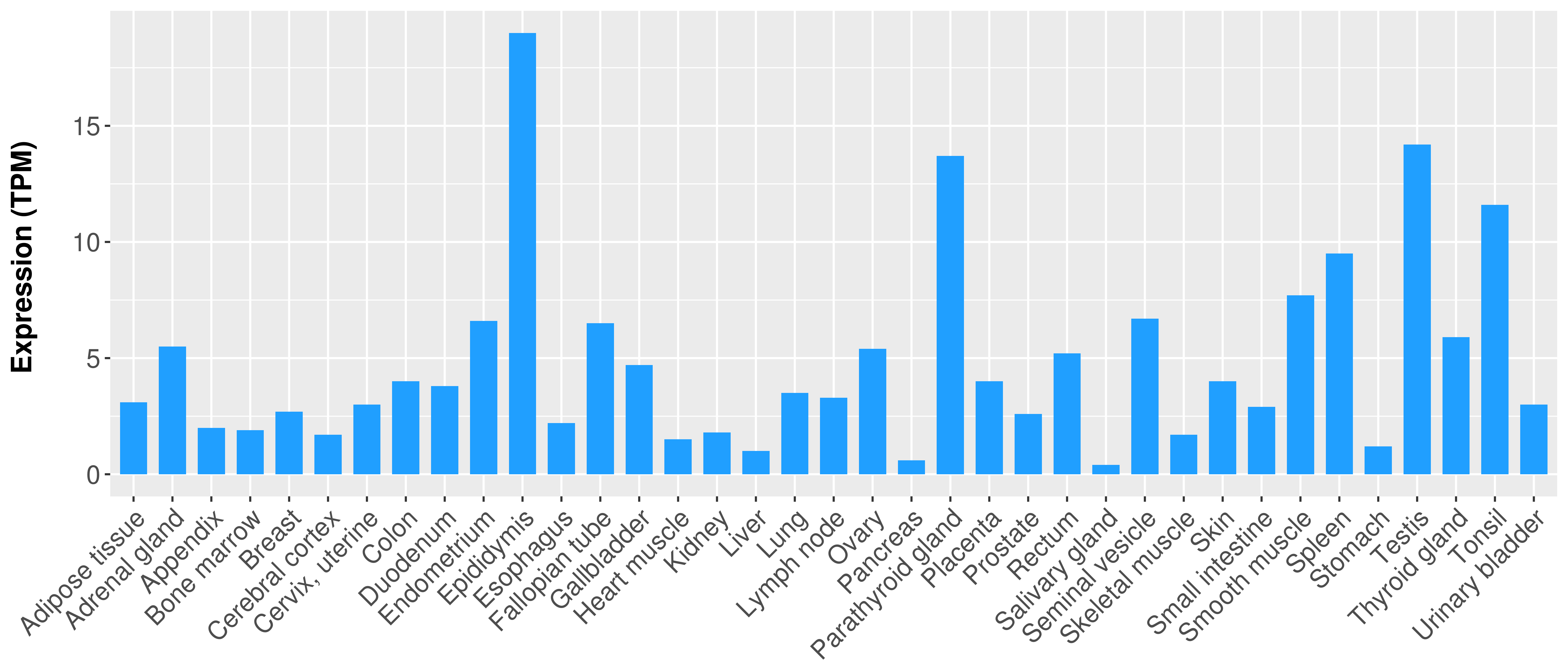
|
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
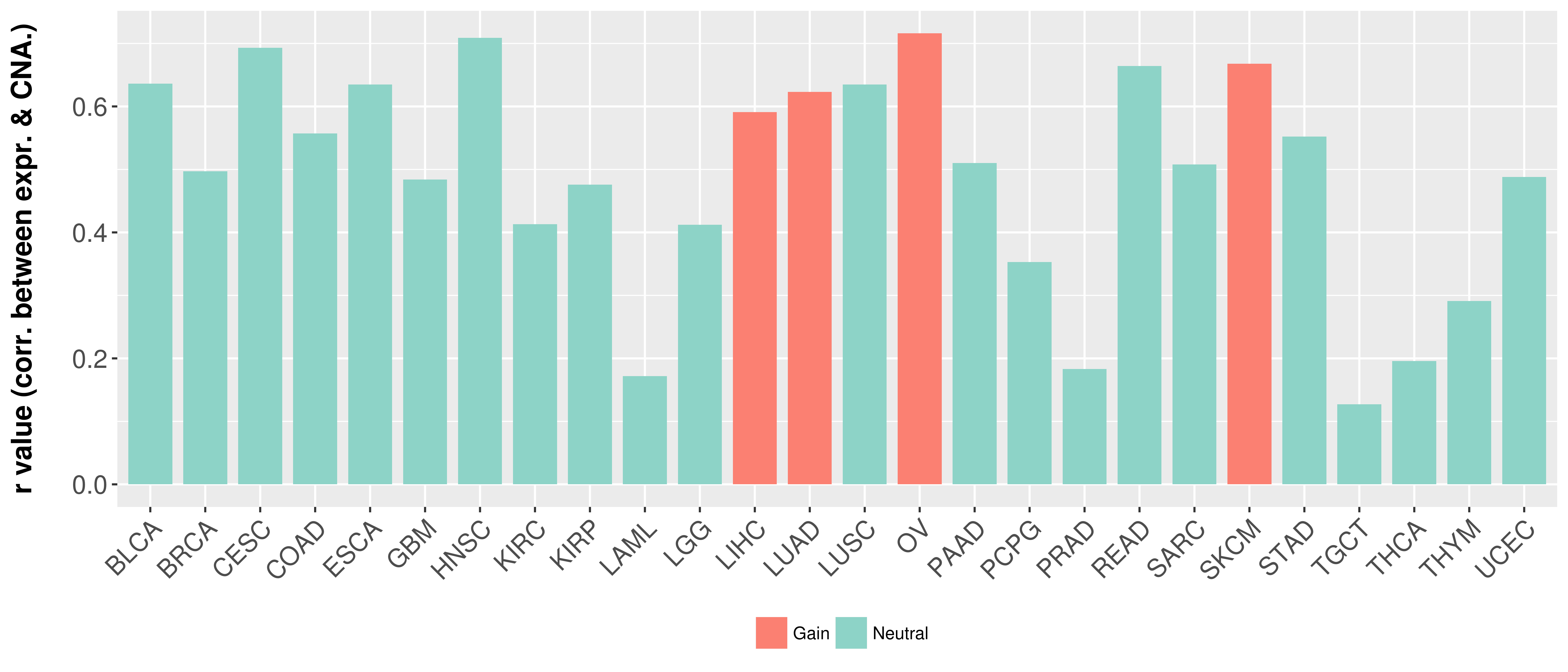
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
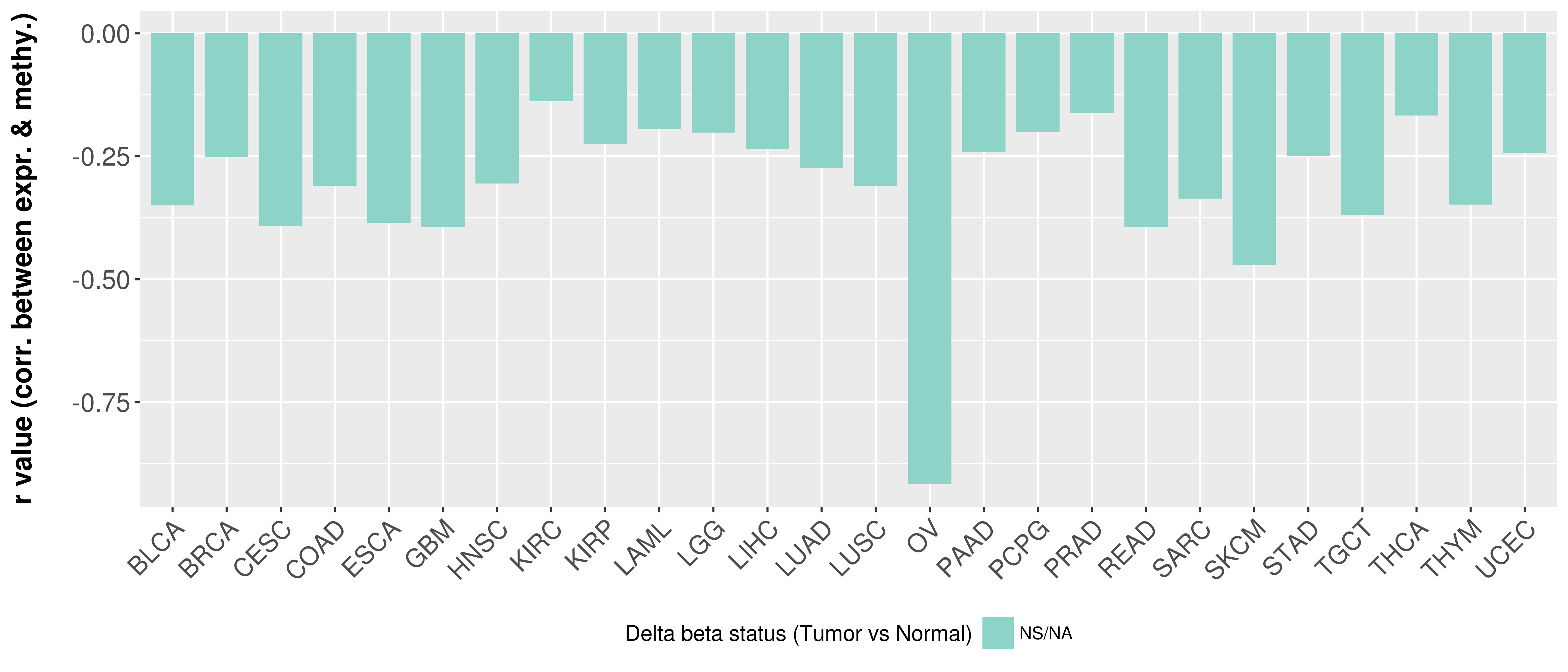
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
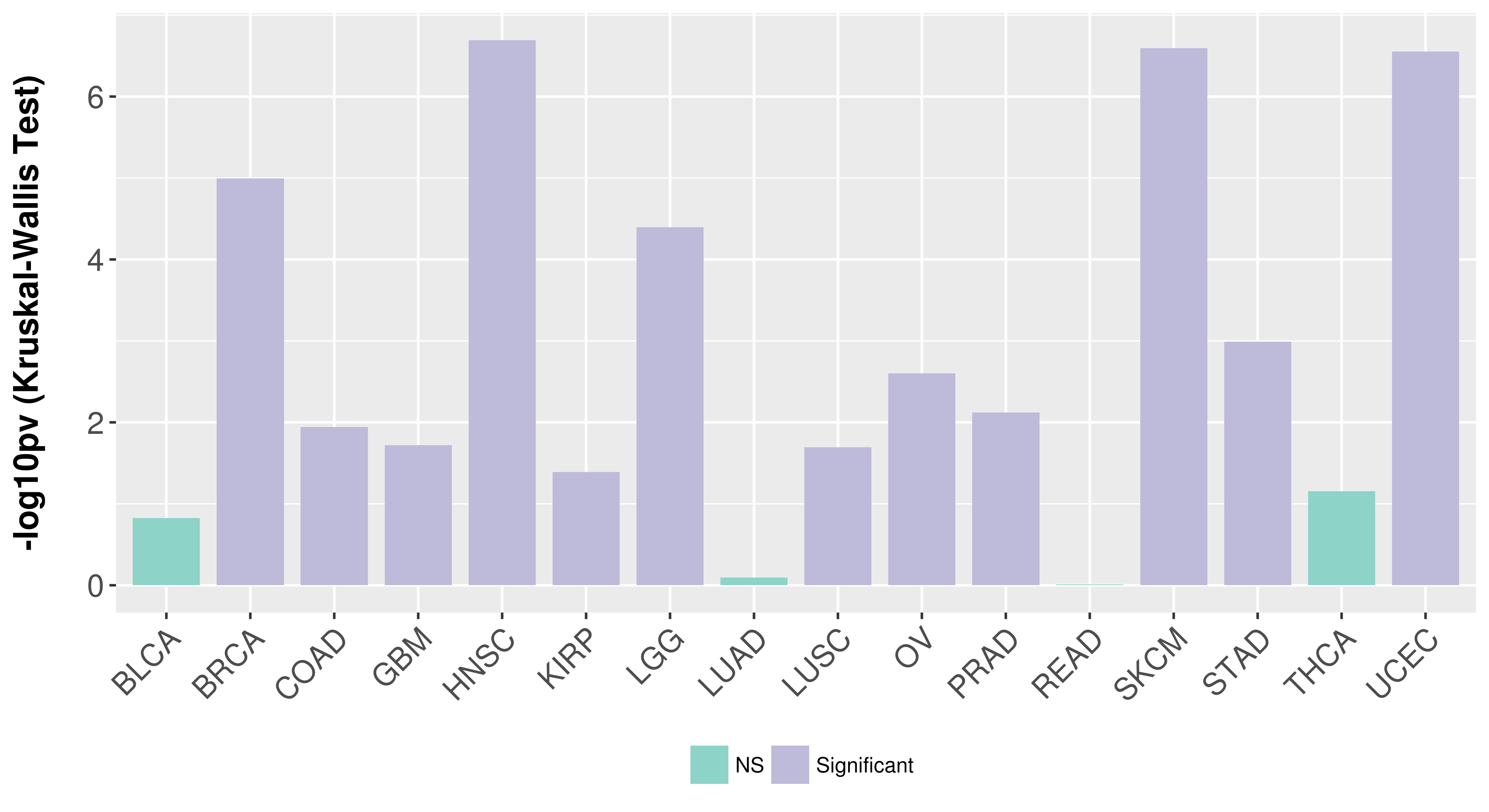
Association between expresson and subtype.
|
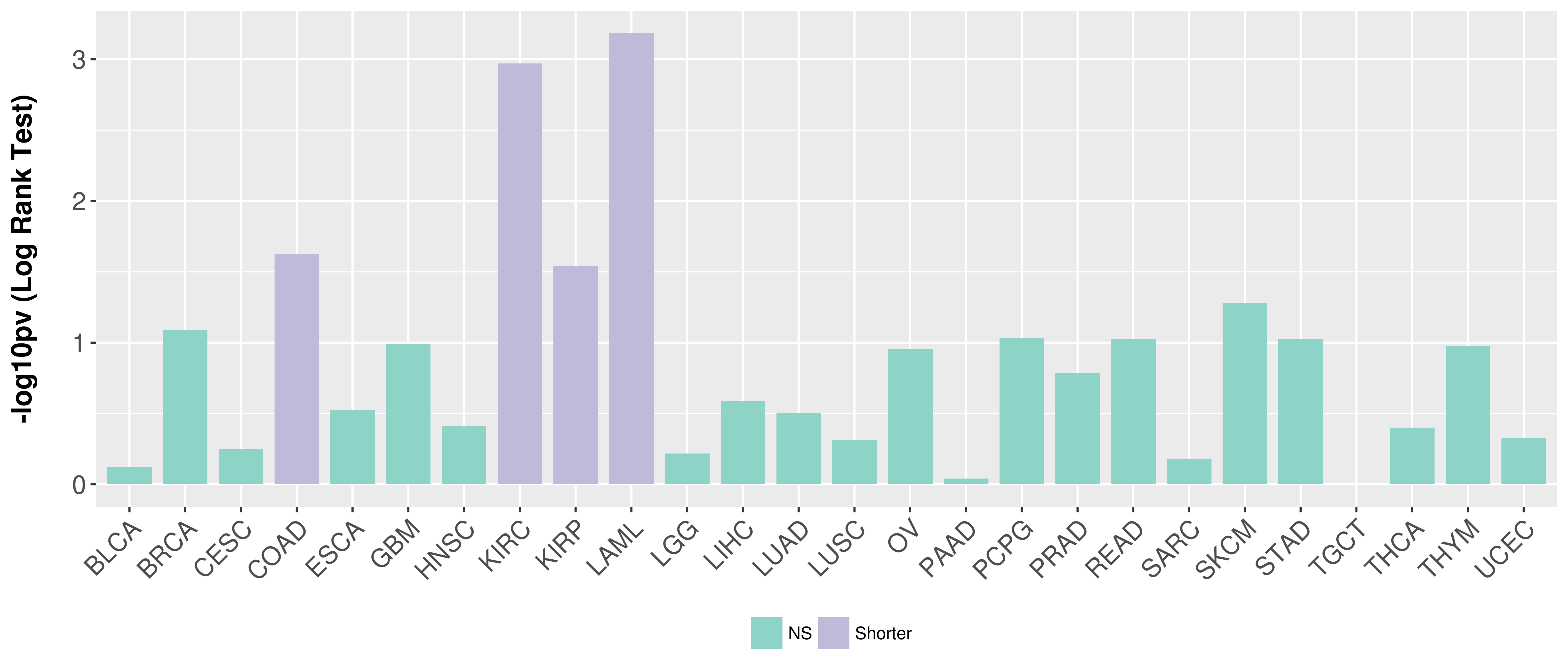
Overall survival analysis based on expression.
|
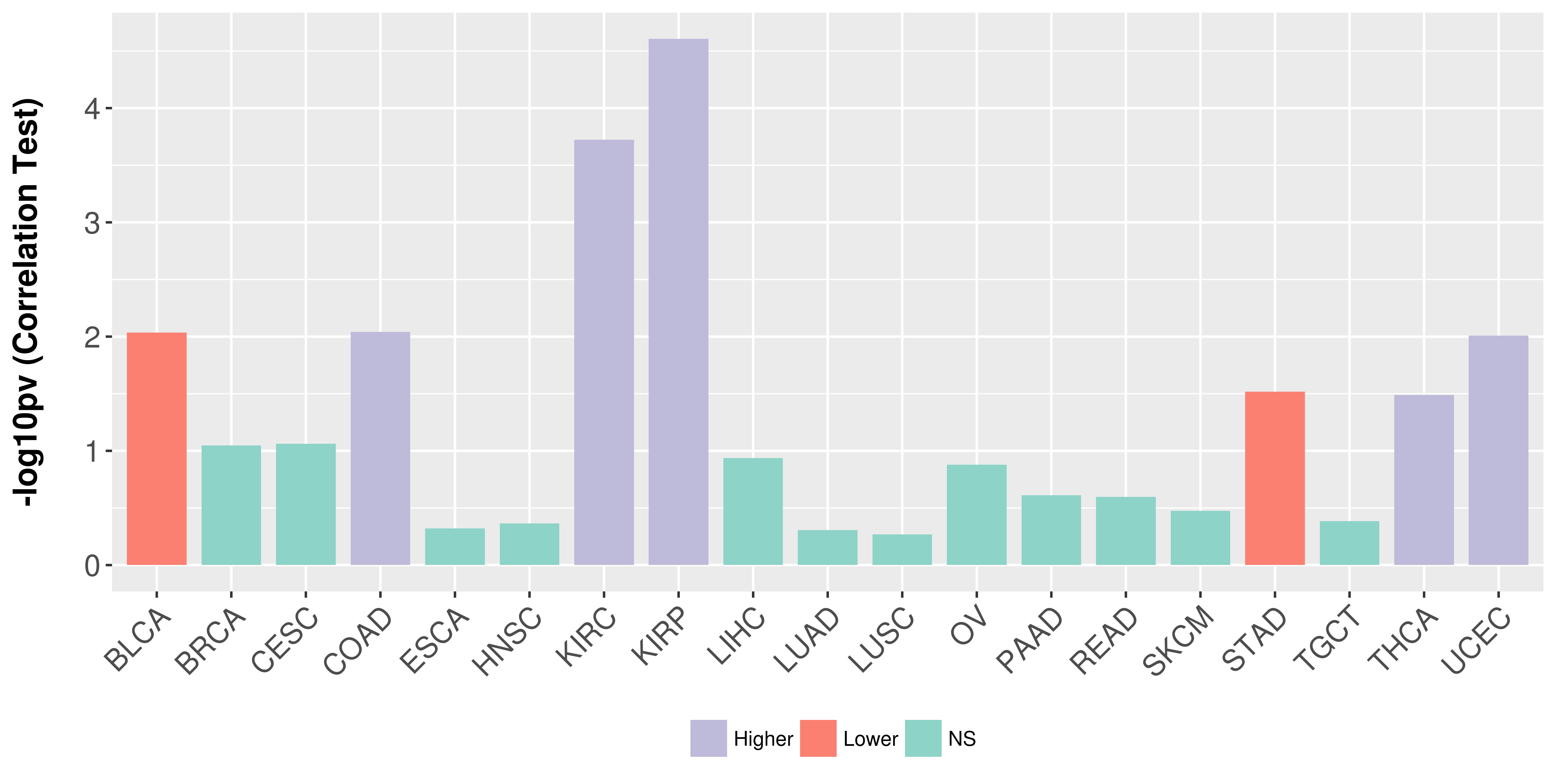
Association between expresson and stage.
|
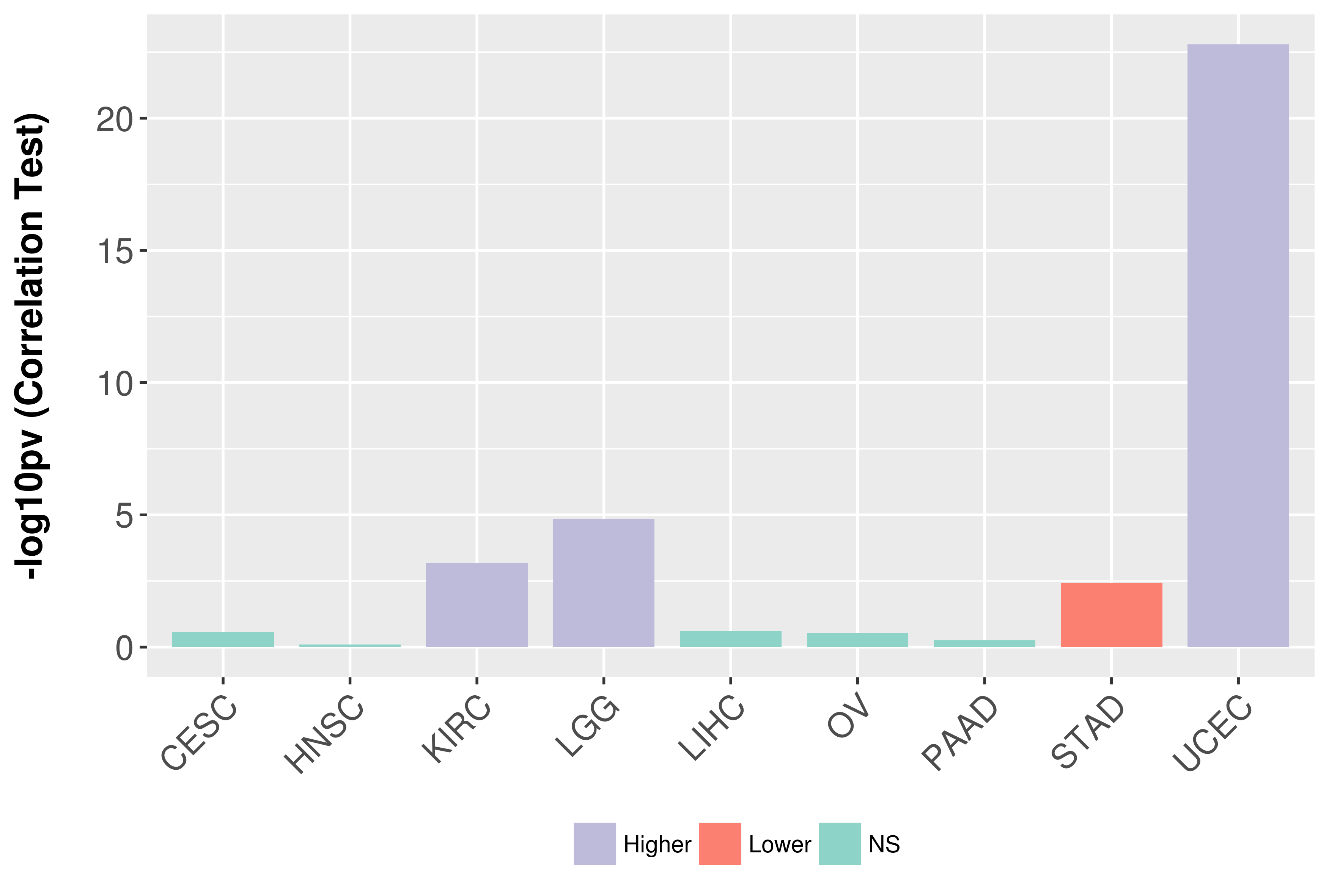
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for DAXX. |
| Summary | |
|---|---|
| Symbol | DAXX |
| Name | death domain associated protein |
| Aliases | DAP6; death-associated protein 6; BING2; CENP-C binding protein; ETS1-associated protein 1; Fas-binding prot ...... |
| Location | 6p21.32 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|