Browse EYA1 in pancancer
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain | - | ||||||||||
| Function |
Functions both as protein phosphatase and as transcriptional coactivator for SIX1, and probably also for SIX2, SIX4 and SIX5 (By similarity). Tyrosine phosphatase that dephosphorylates 'Tyr-142' of histone H2AX (H2AXY142ph) and promotes efficient DNA repair via the recruitment of DNA repair complexes containing MDC1. 'Tyr-142' phosphorylation of histone H2AX plays a central role in DNA repair and acts as a mark that distinguishes between apoptotic and repair responses to genotoxic stress (PubMed:19234442). Its function as histone phosphatase may contribute to its function in transcription regulation during organogenesis (By similarity). Has also phosphatase activity with proteins phosphorylated on Ser and Thr residues (in vitro) (By similarity). Required for normal embryonic development of the craniofacial and trunk skeleton, kidneys and ears (By similarity). Together with SIX1, it plays an important role in hypaxial muscle development; in this it is functionally redundant with EYA2 (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000132 establishment of mitotic spindle orientation GO:0000226 microtubule cytoskeleton organization GO:0001501 skeletal system development GO:0001655 urogenital system development GO:0001656 metanephros development GO:0001657 ureteric bud development GO:0001658 branching involved in ureteric bud morphogenesis GO:0001704 formation of primary germ layer GO:0001707 mesoderm formation GO:0001708 cell fate specification GO:0001710 mesodermal cell fate commitment GO:0001763 morphogenesis of a branching structure GO:0001822 kidney development GO:0001823 mesonephros development GO:0003007 heart morphogenesis GO:0003151 outflow tract morphogenesis GO:0003156 regulation of animal organ formation GO:0003263 cardioblast proliferation GO:0003264 regulation of cardioblast proliferation GO:0003266 regulation of secondary heart field cardioblast proliferation GO:0006282 regulation of DNA repair GO:0006302 double-strand break repair GO:0006470 protein dephosphorylation GO:0007163 establishment or maintenance of cell polarity GO:0007219 Notch signaling pathway GO:0007369 gastrulation GO:0007389 pattern specification process GO:0007423 sensory organ development GO:0007498 mesoderm development GO:0007501 mesodermal cell fate specification GO:0007507 heart development GO:0007605 sensory perception of sound GO:0008593 regulation of Notch signaling pathway GO:0009314 response to radiation GO:0010212 response to ionizing radiation GO:0014706 striated muscle tissue development GO:0016311 dephosphorylation GO:0016570 histone modification GO:0016576 histone dephosphorylation GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0030010 establishment of cell polarity GO:0030323 respiratory tube development GO:0030324 lung development GO:0035088 establishment or maintenance of apical/basal cell polarity GO:0035239 tube morphogenesis GO:0035335 peptidyl-tyrosine dephosphorylation GO:0035904 aorta development GO:0035909 aorta morphogenesis GO:0038034 signal transduction in absence of ligand GO:0040001 establishment of mitotic spindle localization GO:0042471 ear morphogenesis GO:0042472 inner ear morphogenesis GO:0042473 outer ear morphogenesis GO:0042474 middle ear morphogenesis GO:0043583 ear development GO:0045165 cell fate commitment GO:0045739 positive regulation of DNA repair GO:0045747 positive regulation of Notch signaling pathway GO:0048332 mesoderm morphogenesis GO:0048333 mesodermal cell differentiation GO:0048514 blood vessel morphogenesis GO:0048562 embryonic organ morphogenesis GO:0048568 embryonic organ development GO:0048645 animal organ formation GO:0048663 neuron fate commitment GO:0048665 neuron fate specification GO:0048704 embryonic skeletal system morphogenesis GO:0048705 skeletal system morphogenesis GO:0048706 embryonic skeletal system development GO:0048752 semicircular canal morphogenesis GO:0048754 branching morphogenesis of an epithelial tube GO:0048839 inner ear development GO:0048844 artery morphogenesis GO:0050673 epithelial cell proliferation GO:0050678 regulation of epithelial cell proliferation GO:0050679 positive regulation of epithelial cell proliferation GO:0050954 sensory perception of mechanical stimulus GO:0051052 regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051293 establishment of spindle localization GO:0051294 establishment of spindle orientation GO:0051640 organelle localization GO:0051653 spindle localization GO:0051656 establishment of organelle localization GO:0060037 pharyngeal system development GO:0060428 lung epithelium development GO:0060479 lung cell differentiation GO:0060487 lung epithelial cell differentiation GO:0060537 muscle tissue development GO:0060541 respiratory system development GO:0060562 epithelial tube morphogenesis GO:0060675 ureteric bud morphogenesis GO:0060795 cell fate commitment involved in formation of primary germ layer GO:0060840 artery development GO:0060872 semicircular canal development GO:0060914 heart formation GO:0060993 kidney morphogenesis GO:0061138 morphogenesis of a branching epithelium GO:0061245 establishment or maintenance of bipolar cell polarity GO:0061323 cell proliferation involved in heart morphogenesis GO:0061326 renal tubule development GO:0061333 renal tubule morphogenesis GO:0071599 otic vesicle development GO:0071600 otic vesicle morphogenesis GO:0072001 renal system development GO:0072006 nephron development GO:0072009 nephron epithelium development GO:0072028 nephron morphogenesis GO:0072073 kidney epithelium development GO:0072078 nephron tubule morphogenesis GO:0072080 nephron tubule development GO:0072088 nephron epithelium morphogenesis GO:0072163 mesonephric epithelium development GO:0072164 mesonephric tubule development GO:0072171 mesonephric tubule morphogenesis GO:0072513 positive regulation of secondary heart field cardioblast proliferation GO:0090102 cochlea development GO:0090103 cochlea morphogenesis GO:0090596 sensory organ morphogenesis GO:0097191 extrinsic apoptotic signaling pathway GO:0097192 extrinsic apoptotic signaling pathway in absence of ligand GO:1901099 negative regulation of signal transduction in absence of ligand GO:2000027 regulation of organ morphogenesis GO:2000136 regulation of cell proliferation involved in heart morphogenesis GO:2000826 regulation of heart morphogenesis GO:2001020 regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus GO:2001233 regulation of apoptotic signaling pathway GO:2001234 negative regulation of apoptotic signaling pathway GO:2001236 regulation of extrinsic apoptotic signaling pathway GO:2001237 negative regulation of extrinsic apoptotic signaling pathway GO:2001239 regulation of extrinsic apoptotic signaling pathway in absence of ligand GO:2001240 negative regulation of extrinsic apoptotic signaling pathway in absence of ligand |
| Molecular Function |
GO:0004721 phosphoprotein phosphatase activity GO:0004725 protein tyrosine phosphatase activity GO:0016791 phosphatase activity GO:0042578 phosphoric ester hydrolase activity |
| Cellular Component |
GO:0032993 protein-DNA complex |
| KEGG | - |
| Reactome |
R-HSA-5693606: DNA Double Strand Break Response R-HSA-5693532: DNA Double-Strand Break Repair R-HSA-73894: DNA Repair R-HSA-5693565: Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for EYA1. |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
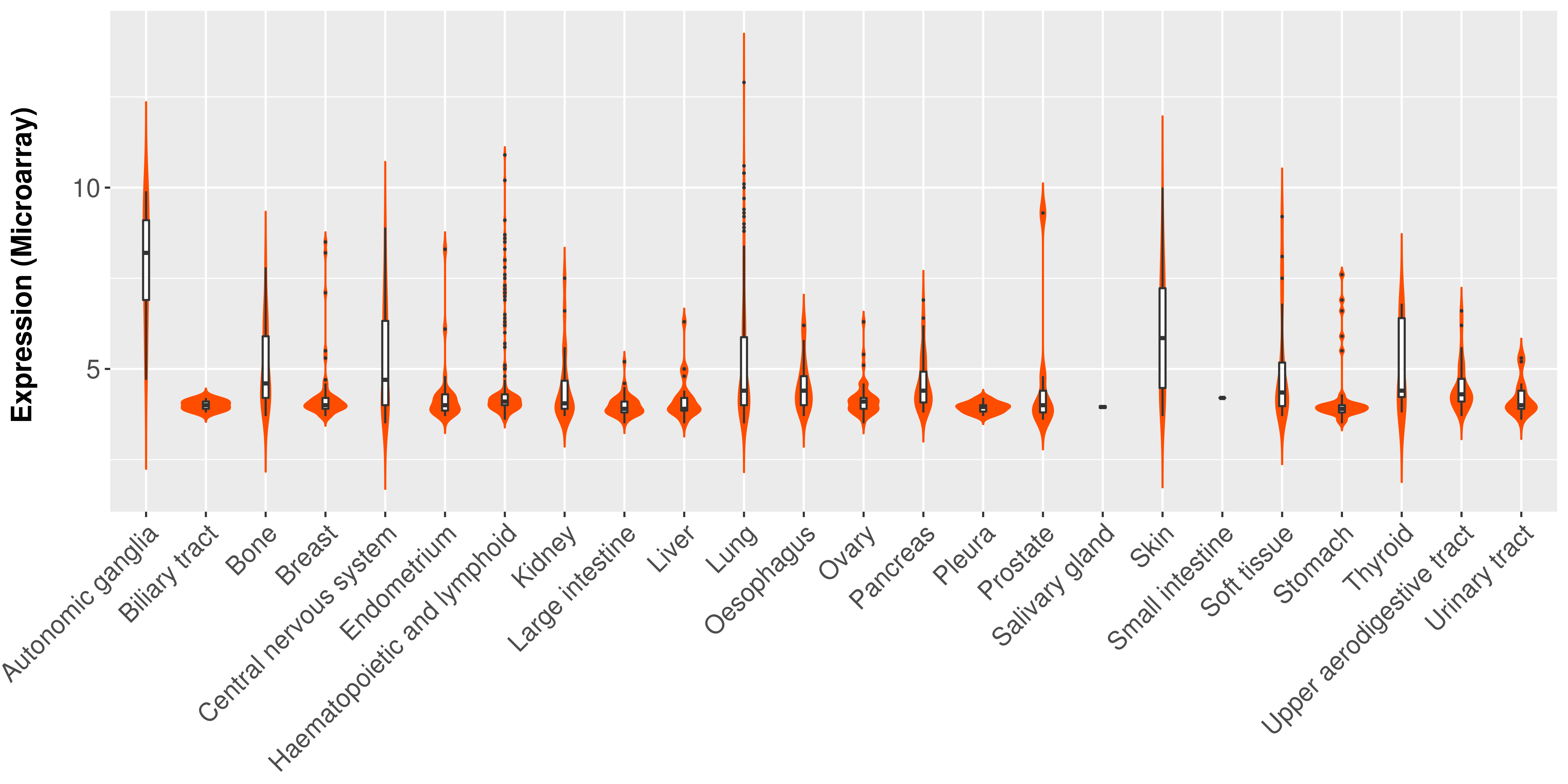
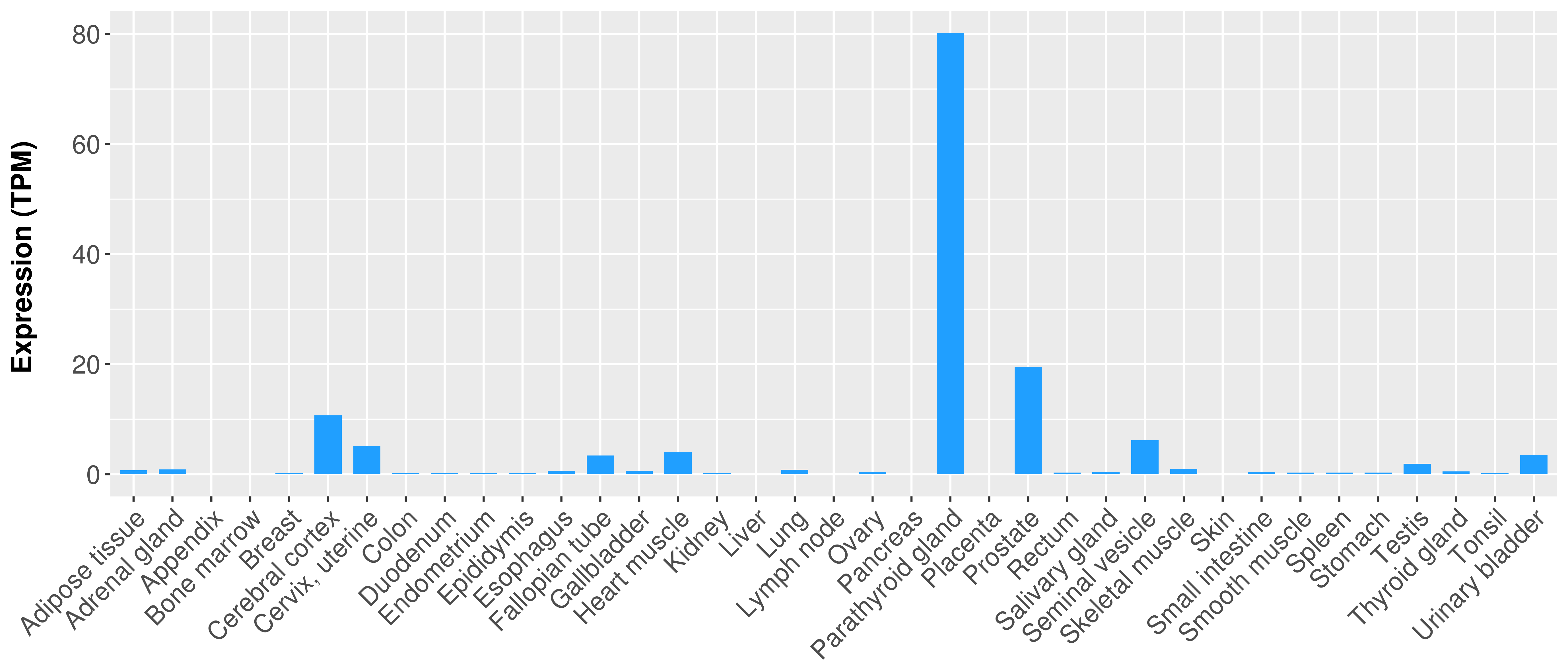
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
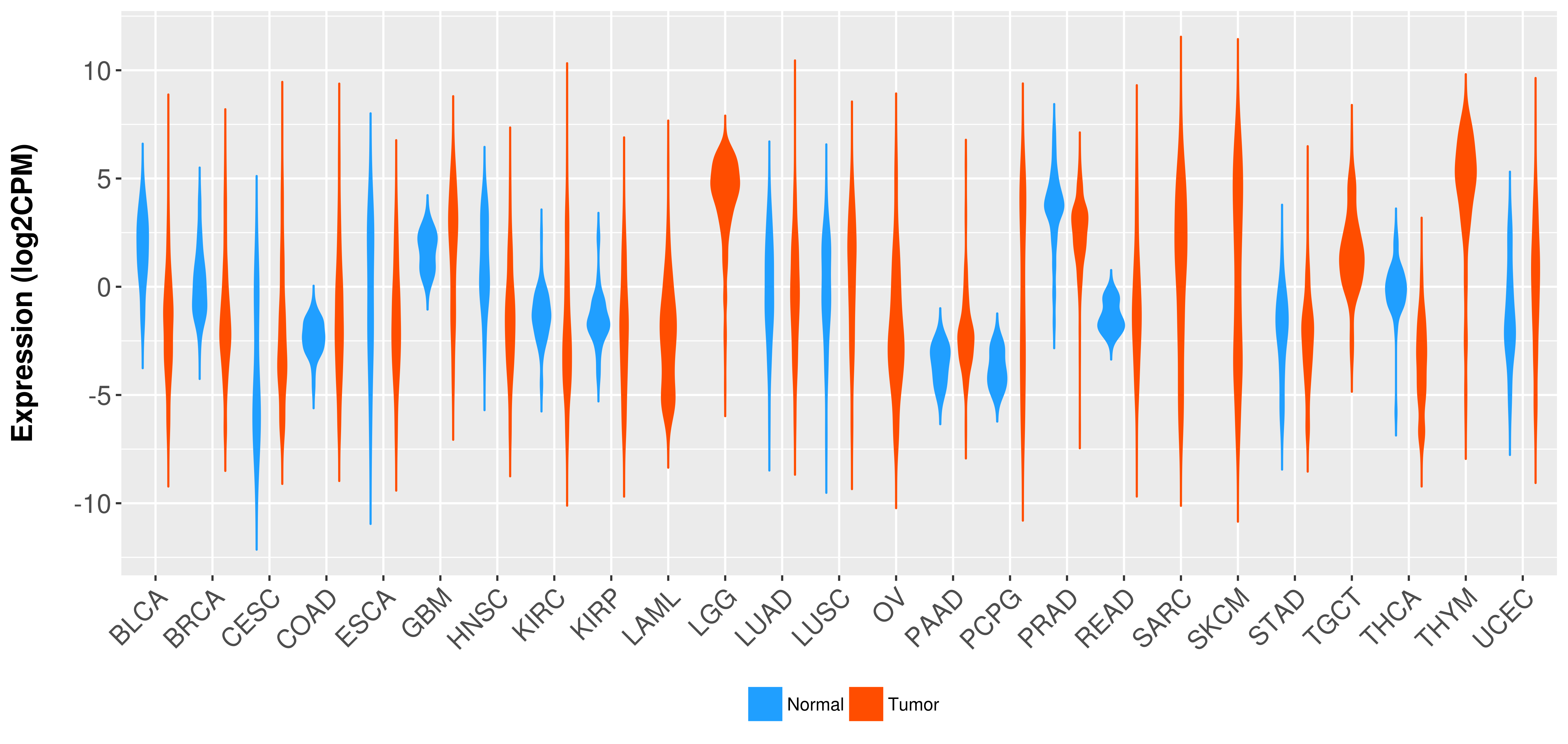
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
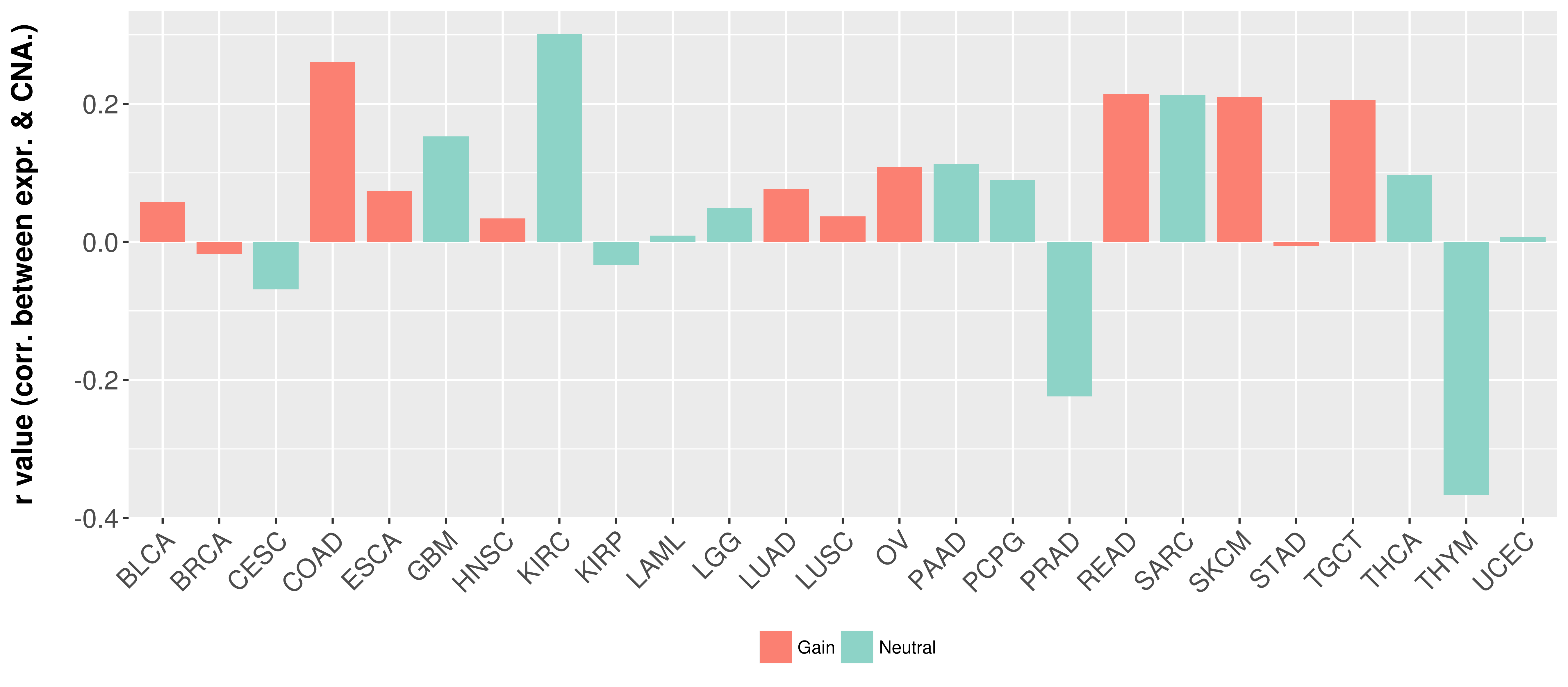
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
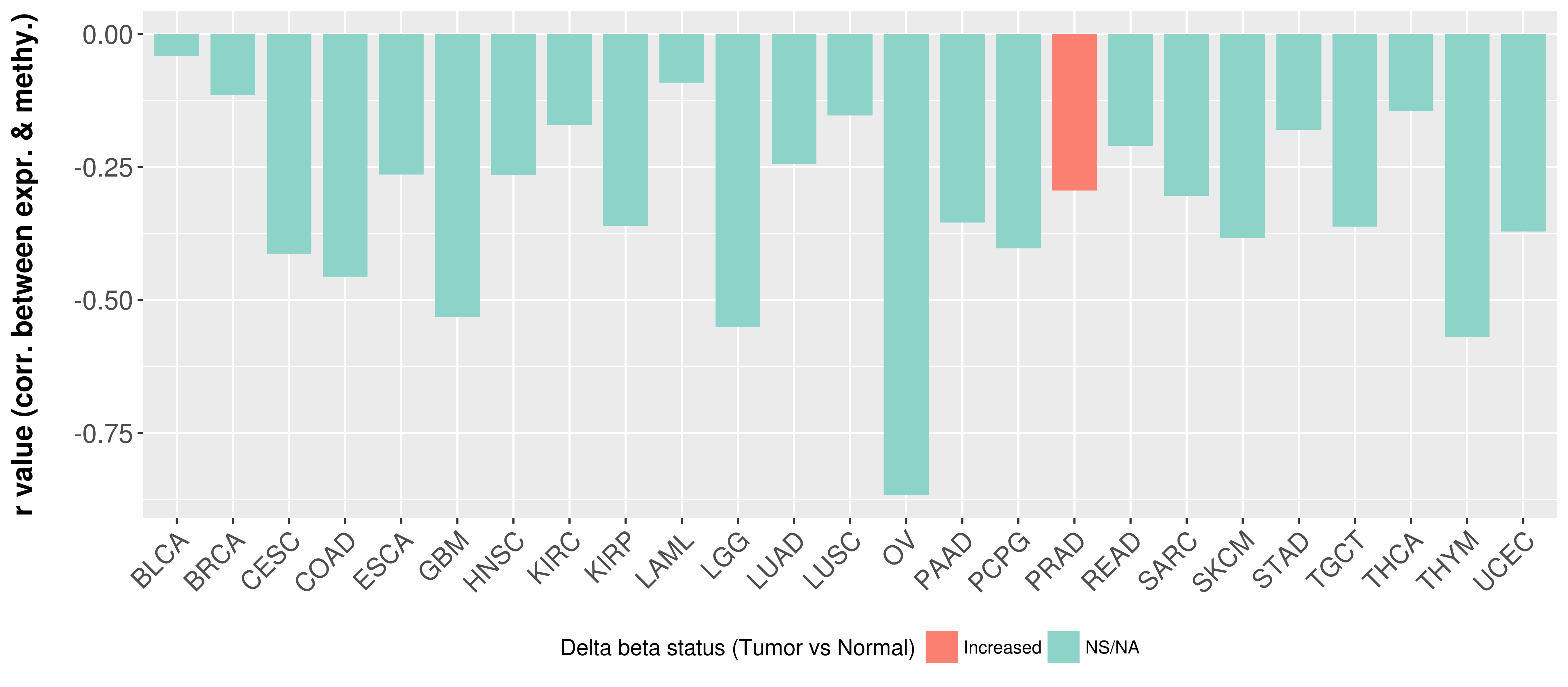
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
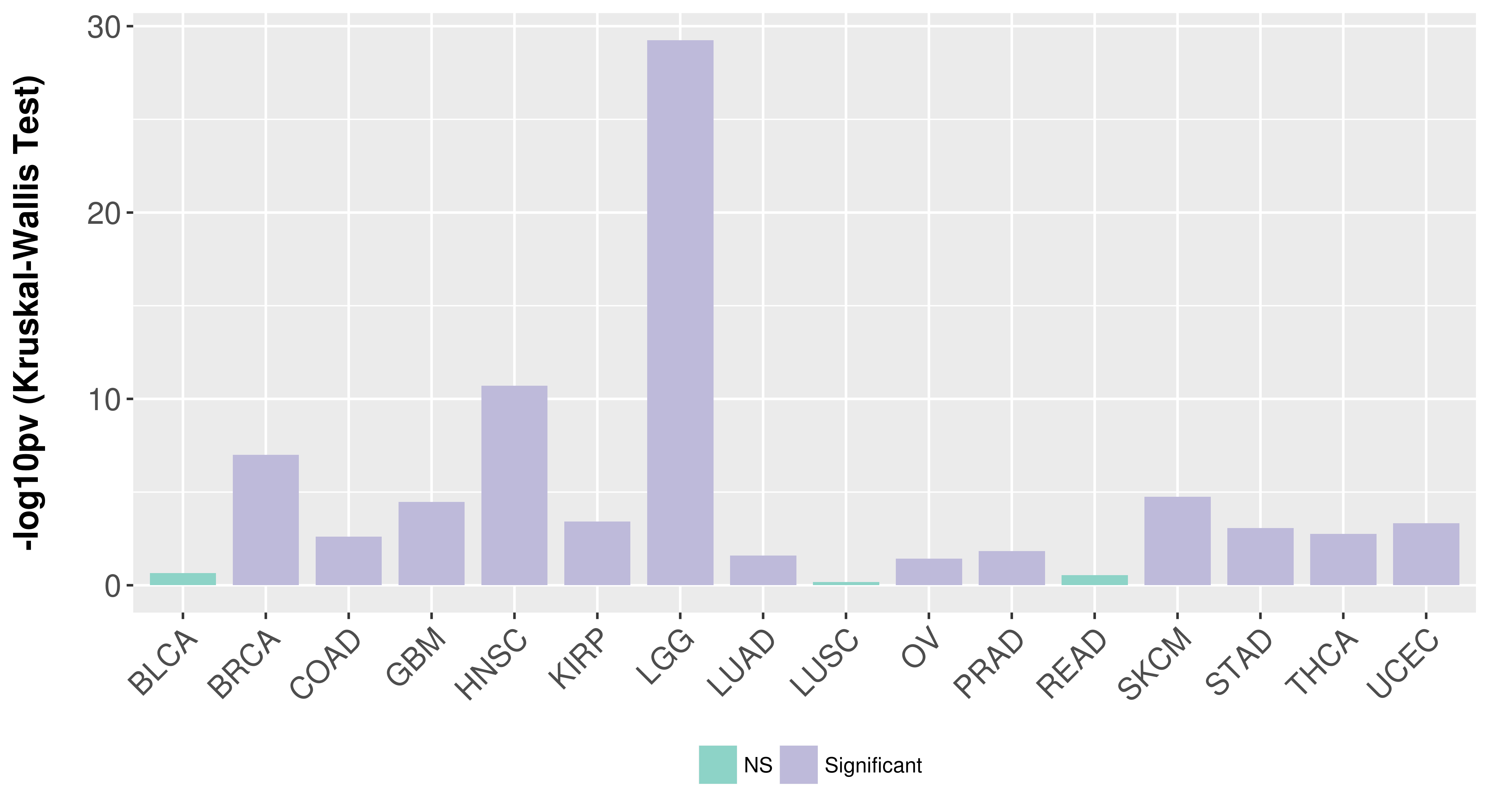
Association between expresson and subtype.
|

Overall survival analysis based on expression.
|
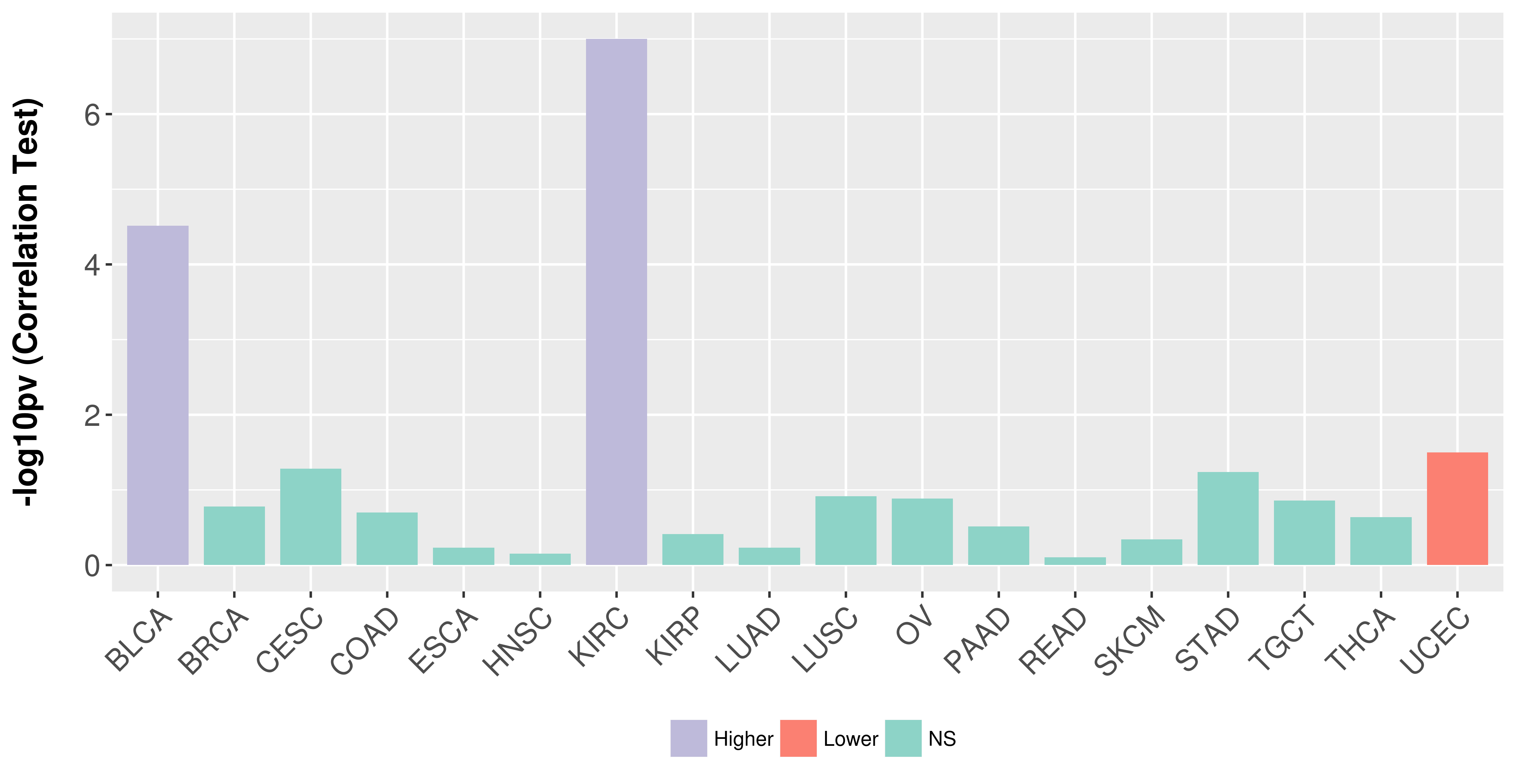
Association between expresson and stage.
|
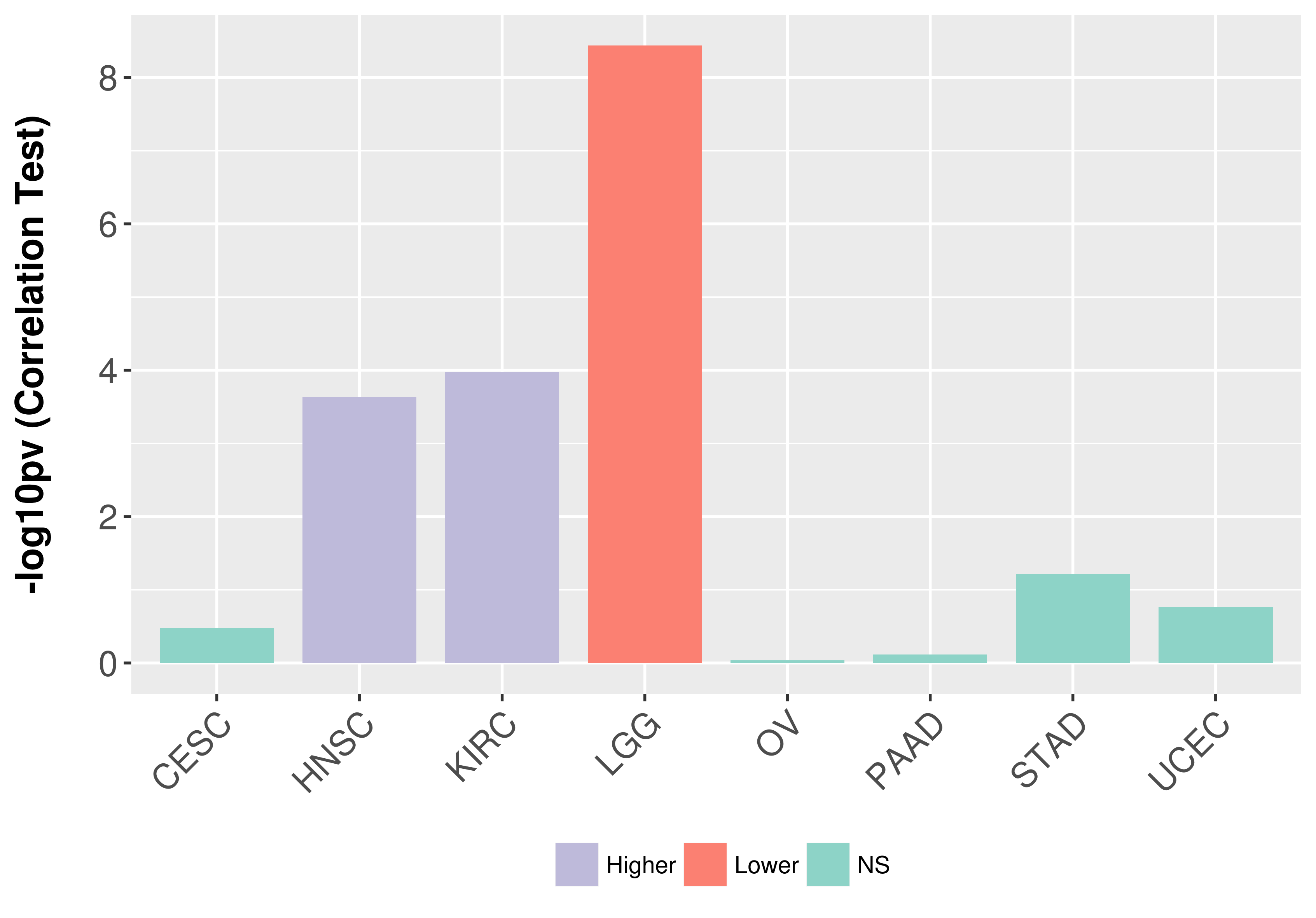
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for EYA1. |
| Summary | |
|---|---|
| Symbol | EYA1 |
| Name | EYA transcriptional coactivator and phosphatase 1 |
| Aliases | eyes absent (Drosophila) homolog 1; eyes absent homolog 1 (Drosophila); Eyes absent homolog 1 |
| Location | 8q13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for EYA1. |
|