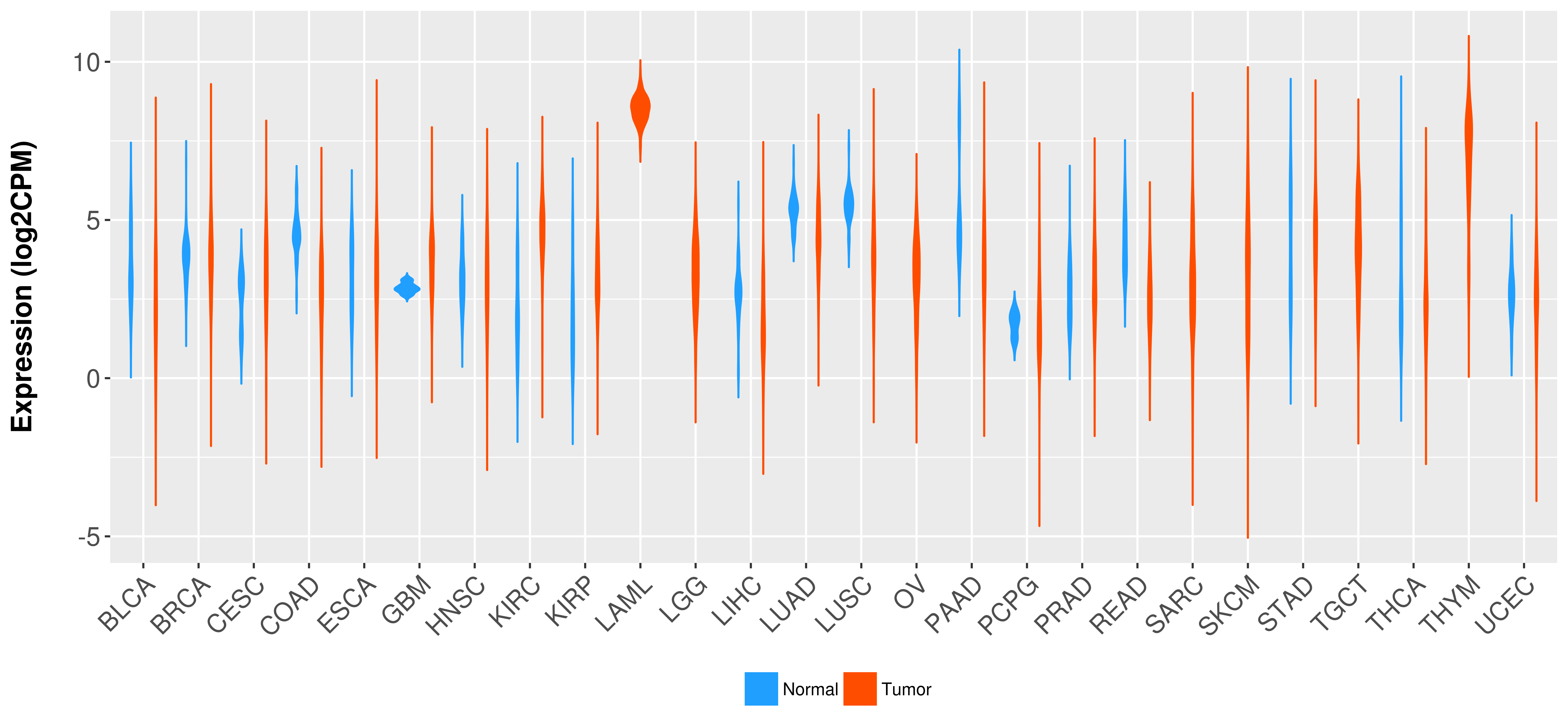
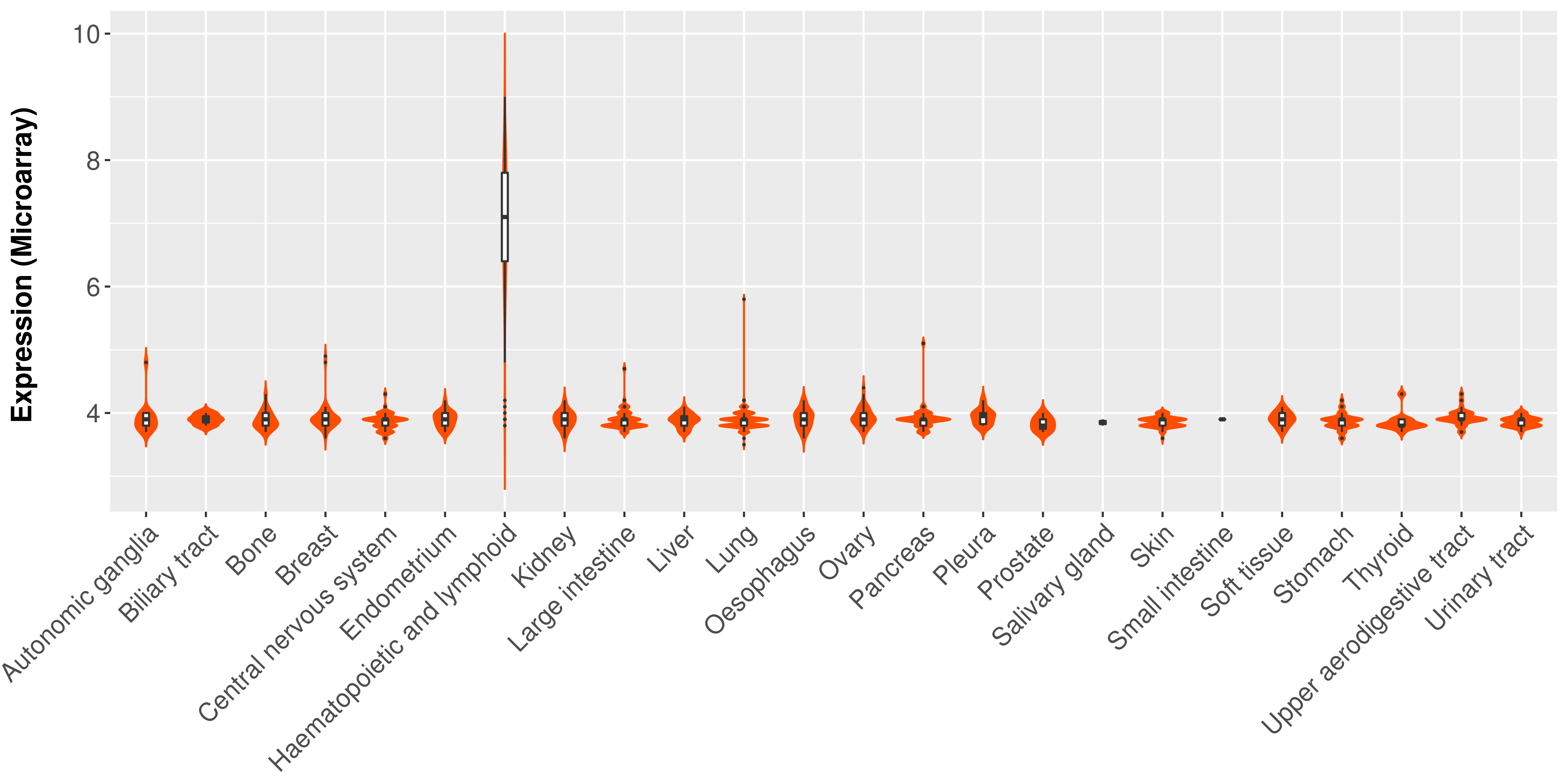
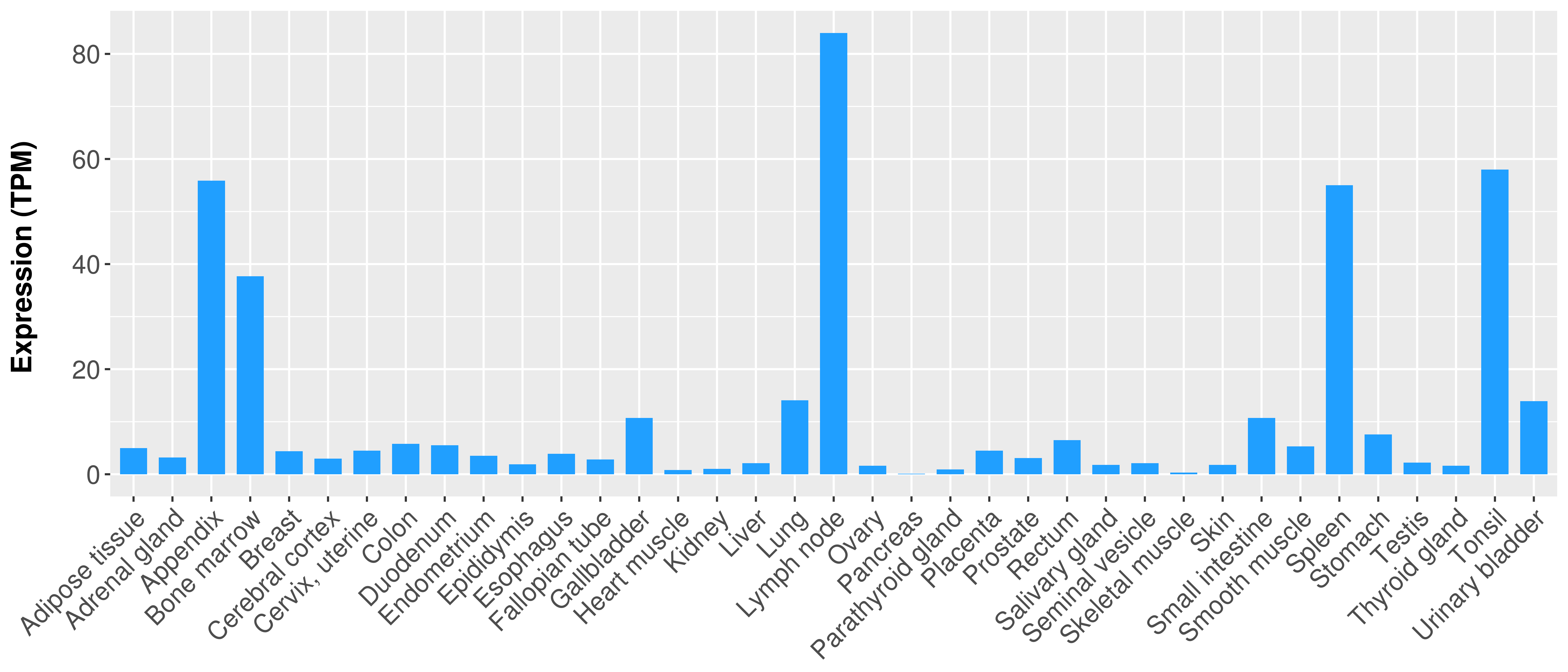
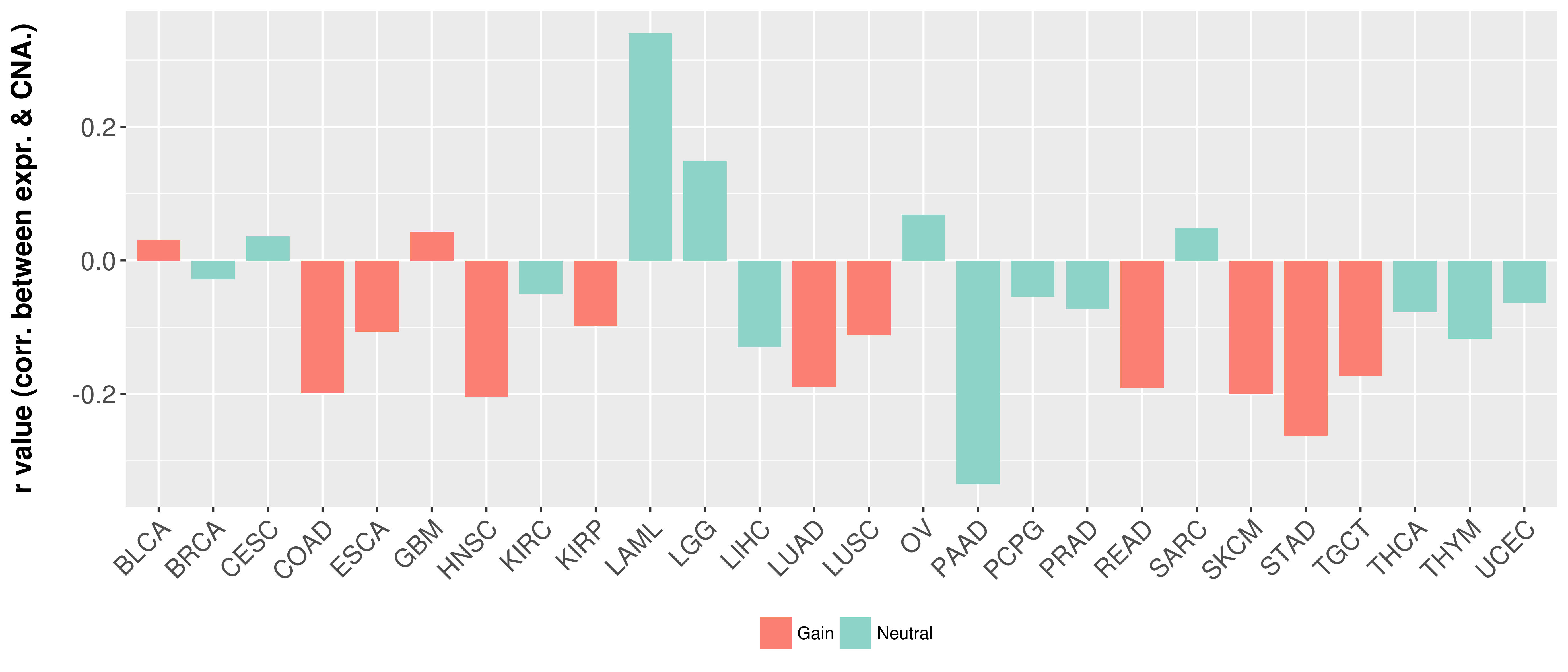
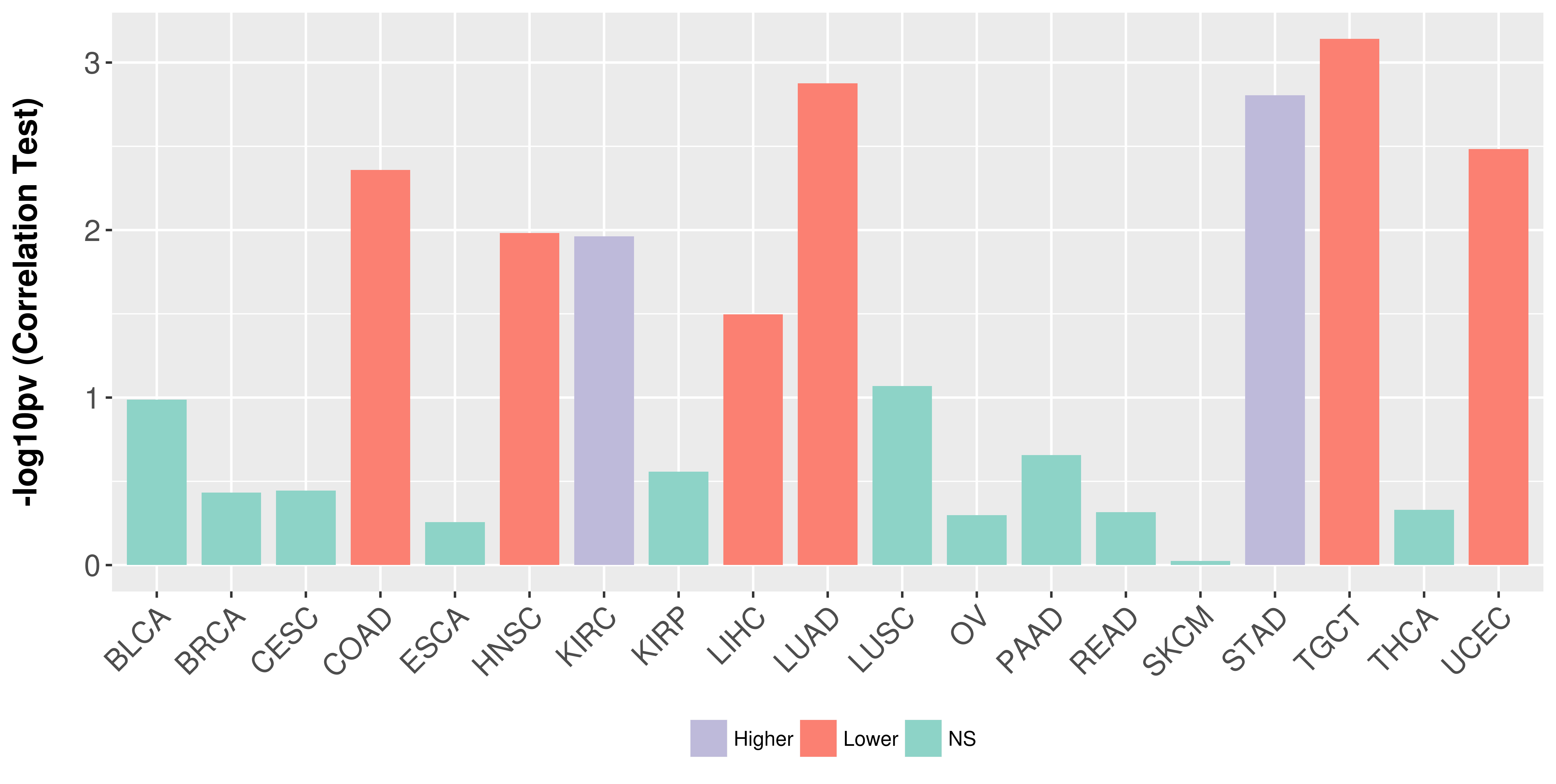
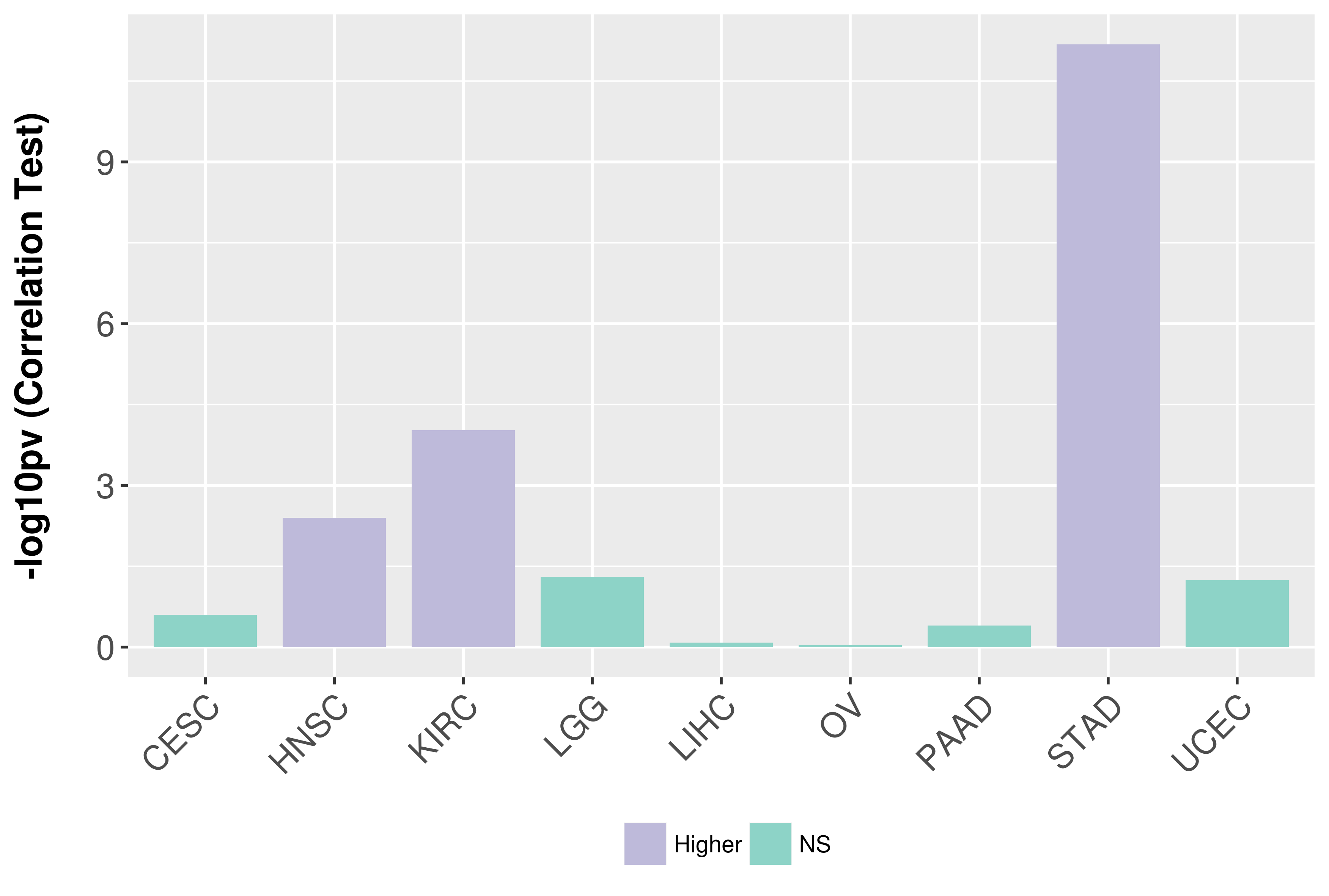
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4172535 | c.? | p.? | Unknown | Skin
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328011 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4989854 | c.? | p.M476>PR | Complex - insertion inframe | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86975 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1238061 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87254 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86975 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87297 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM4989857 | c.? | p.V341fs | Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1238061 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328008 | c.851-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87253 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87263 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86971 | c.1-?_715+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM303909 | c.?_?del? | p.?fs | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM303909 | c.?_?del? | p.?fs | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM86972 | c.1-?_421+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86975 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328503 | c.161-31360_850+4104>TCC | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86968 | c.? | p.L117fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86978 | c.41-?_715+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86975 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85910 | c.1004_1005insC | p.G337fs*152 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87263 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86967 | c.? | p.H224fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328502 | c.161-31337_850+4078>GGGG | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM133012 | c.41-?_160+?del120 | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM133032 | c.161-?_715+?del555 | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86974 | c.1-?_589+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86971 | c.1-?_715+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86969 | c.? | p.S402fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86975 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4989853 | c.? | p.F471>LRA | Complex - insertion inframe | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM303909 | c.?_?del? | p.?fs | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87260 | c.41-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87253 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86974 | c.1-?_589+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86974 | c.1-?_589+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87249 | c.1520C>A | p.S507* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87296 | c.1-?_421+?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM26582 | c.1297C>A | p.R433S | Substitution - Missense | Lung
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85908 | c.? | p.R111* | Substitution - Nonsense | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87254 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86942 | c.1-?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86972 | c.1-?_421+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328008 | c.851-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87302 | c.308_309ins23 | p.R111fs*15 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM249841 | c.472G>A | p.G158S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM249842 | c.87G>C | p.E29D | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM87296 | c.1-?_421+?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4606322 | c.551G>C | p.R184P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86968 | c.? | p.L117fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4989855 | c.? | p.Q62fs | Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4989852 | c.? | p.C495Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM87254 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86970 | c.? | p.E504fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86966 | c.? | p.G158S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87297 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328501 | c.161-31337_850+4069del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM303909 | c.?_?del? | p.?fs | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM86978 | c.41-?_715+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87302 | c.308_309ins23 | p.R111fs*15 | Insertion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86942 | c.1-?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1238061 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87263 | c.1-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86942 | c.1-?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4989856 | c.? | p.R143Q | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM1238061 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86979 | c.41-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM328011 | c.? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM4606322 | c.551G>C | p.R184P | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM85913 | c.1-?_160+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86969 | c.? | p.S402fs*? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5985127 | c.? | p.E251K | Substitution - Missense | Salivary_gland
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM86976 | c.1-?_40+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM133032 | c.161-?_715+?del555 | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM1716600 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM86973 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM87295 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85916 | c.?_?del? | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85914 | c.1_1560del1560 | p.0? | Whole gene deletion | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85911 | c.161-?_1560+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM5022462 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85912 | c.1-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|
| COSM85915 | c.161-?_850+?del | p.? | Unknown | Haematopoietic_and_lymphoid_tissue
|