Browse INO80B in pancancer
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF04795 PAPA-1-like conserved region PF04438 HIT zinc finger |
||||||||||
| Function |
Induces growth and cell cycle arrests at the G1 phase of the cell cycle. ; FUNCTION: Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006310 DNA recombination GO:0006338 chromatin remodeling |
| Molecular Function | - |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0031011 Ino80 complex GO:0033202 DNA helicase complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0097346 INO80-type complex |
| KEGG | - |
| Reactome |
R-HSA-5696394: DNA Damage Recognition in GG-NER R-HSA-73894: DNA Repair R-HSA-5688426: Deubiquitination R-HSA-5696399: Global Genome Nucleotide Excision Repair (GG-NER) R-HSA-392499: Metabolism of proteins R-HSA-5696398: Nucleotide Excision Repair R-HSA-597592: Post-translational protein modification R-HSA-5689603: UCH proteinases |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for INO80B. |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
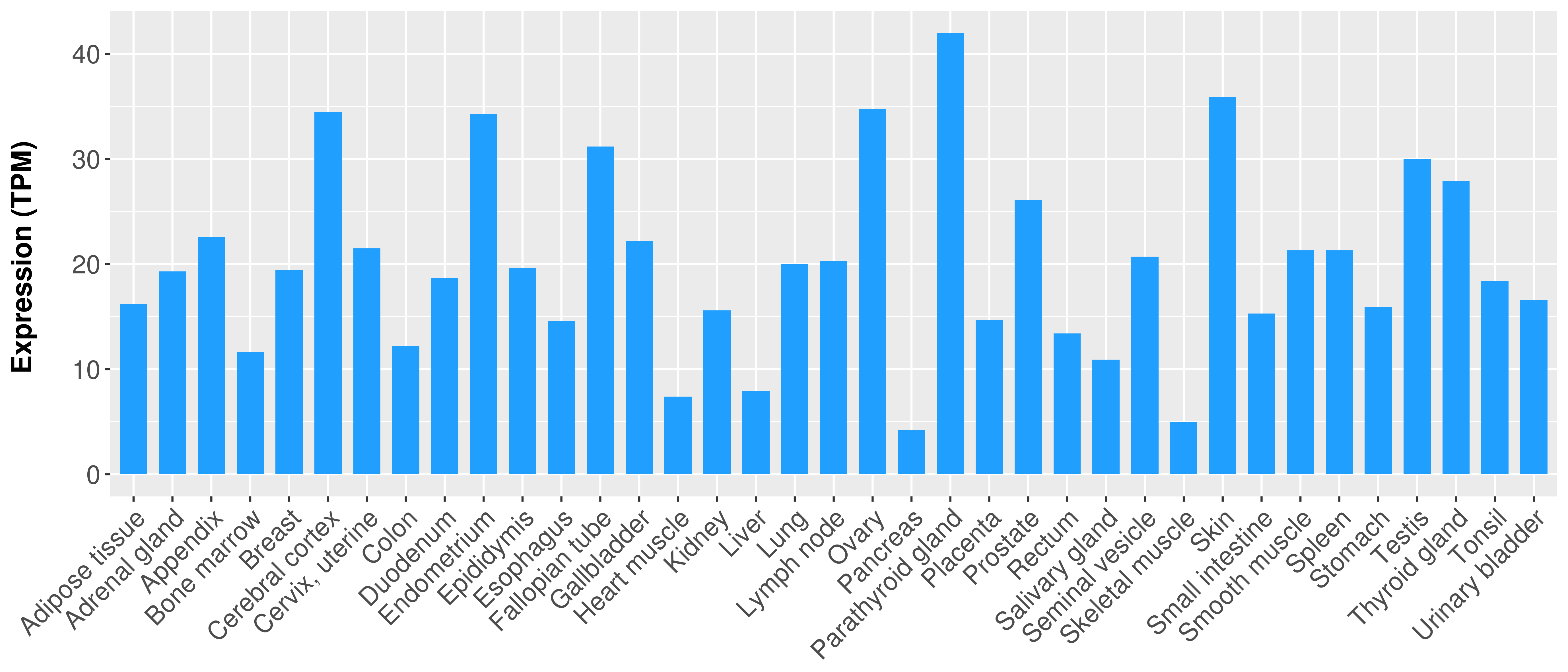
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
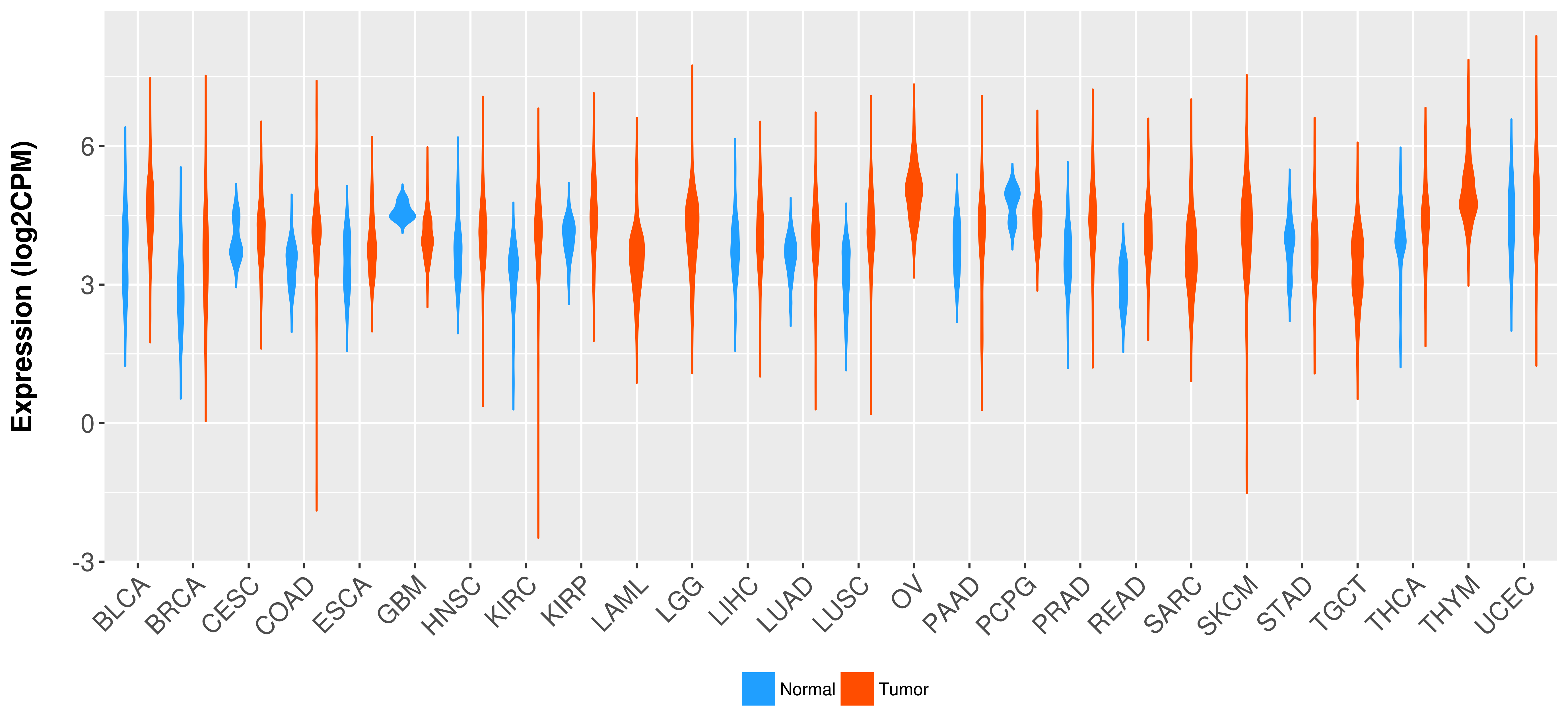
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
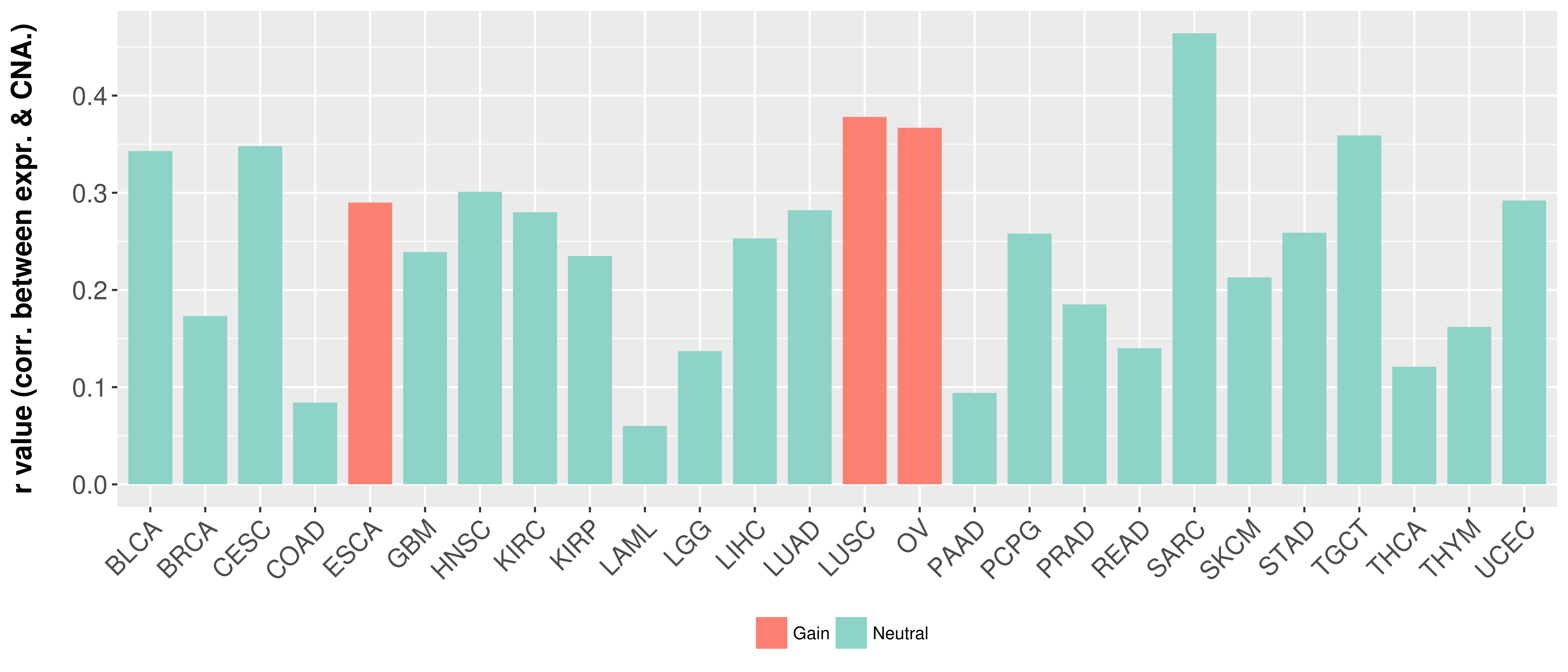
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
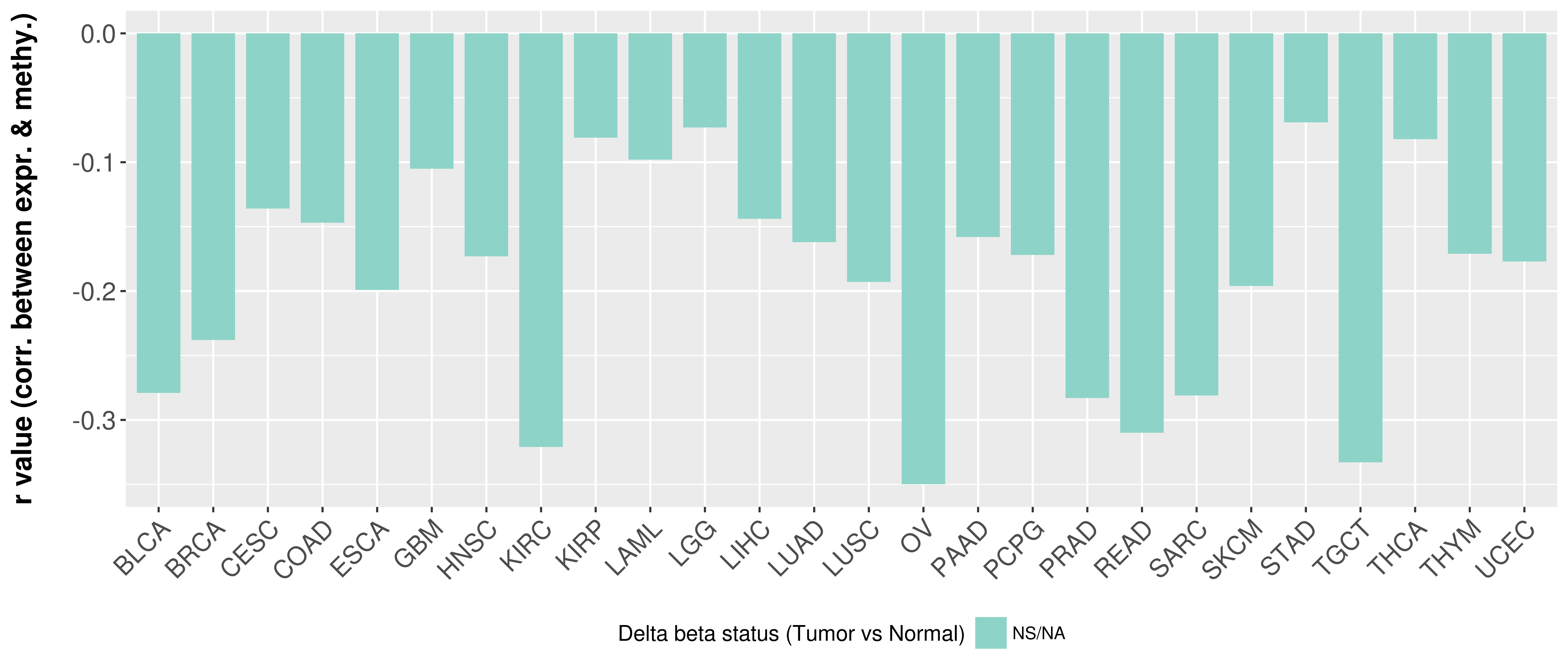
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
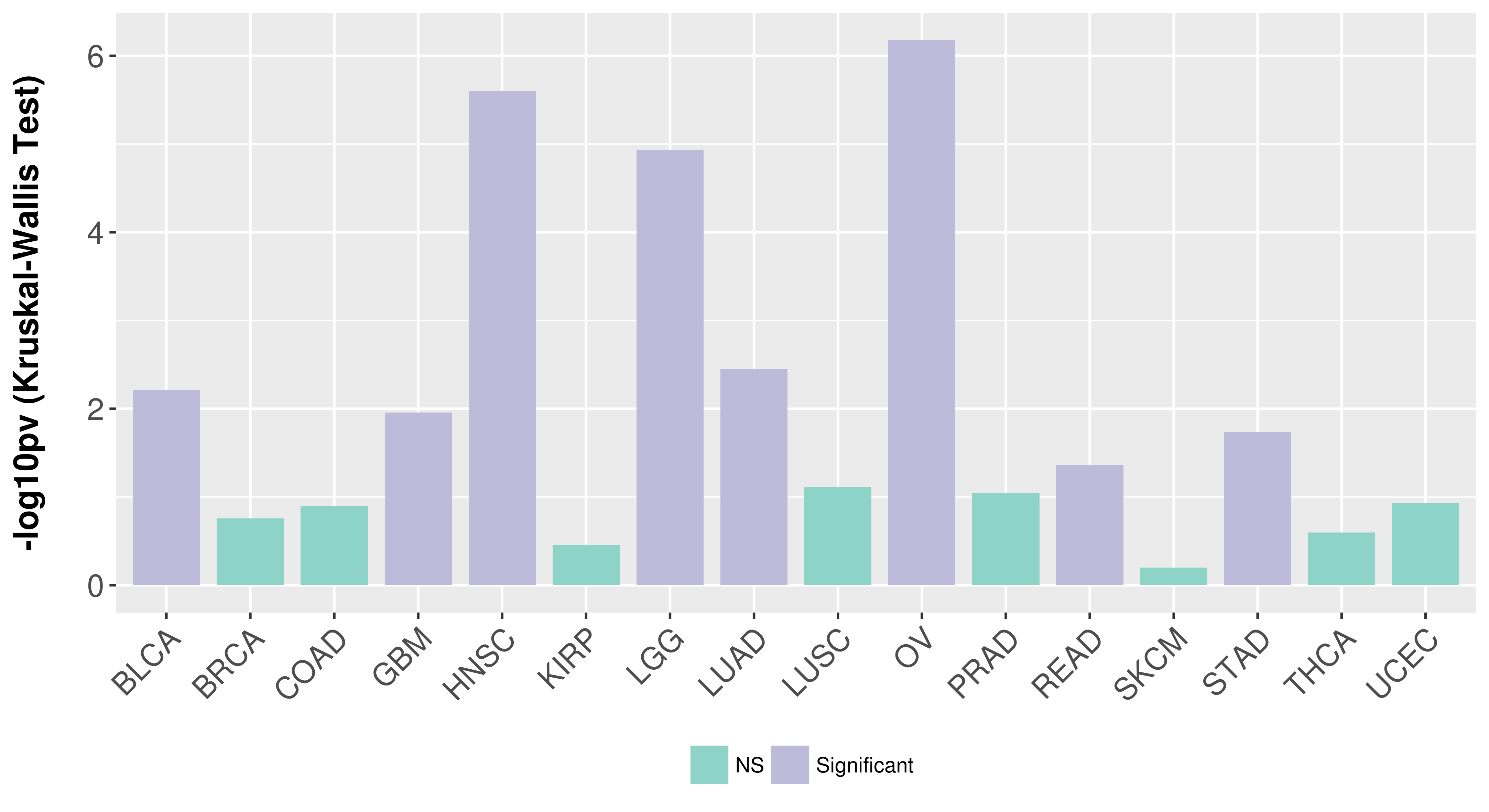
Association between expresson and subtype.
|
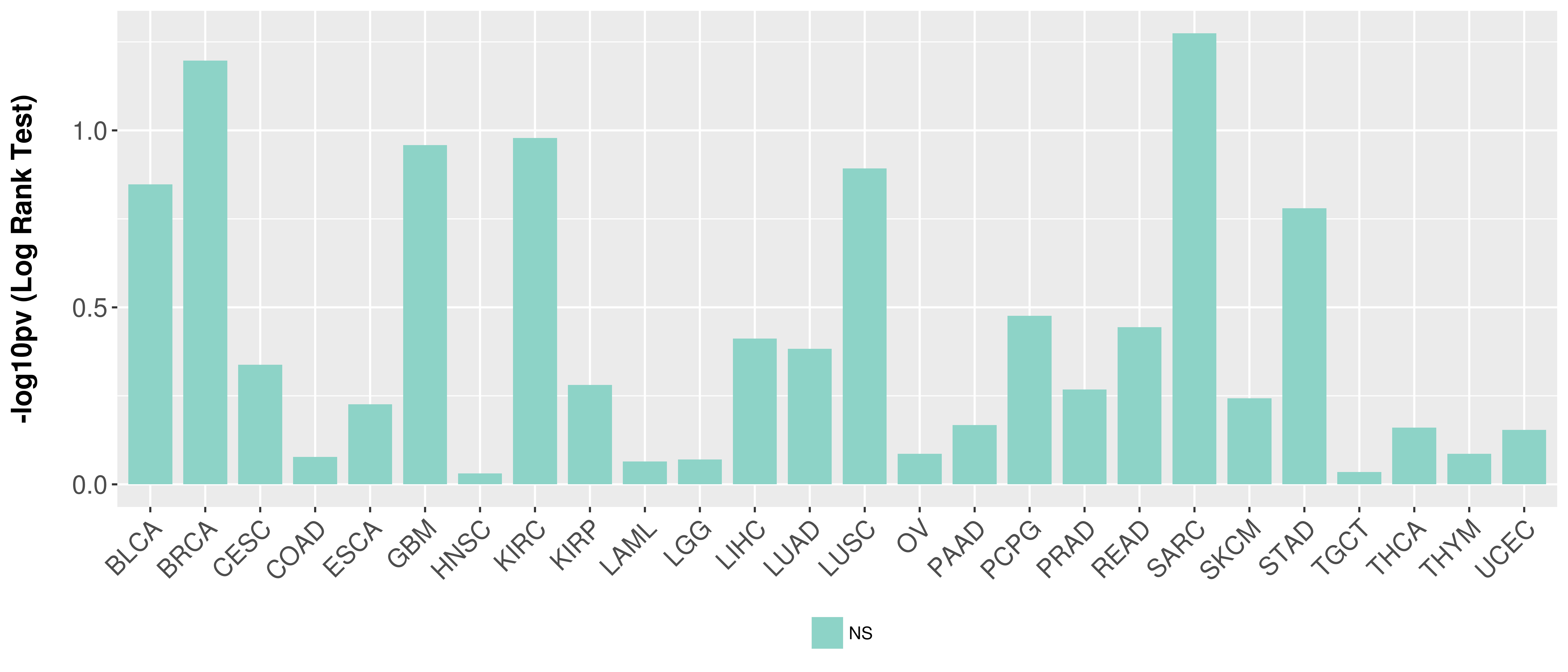
Overall survival analysis based on expression.
|
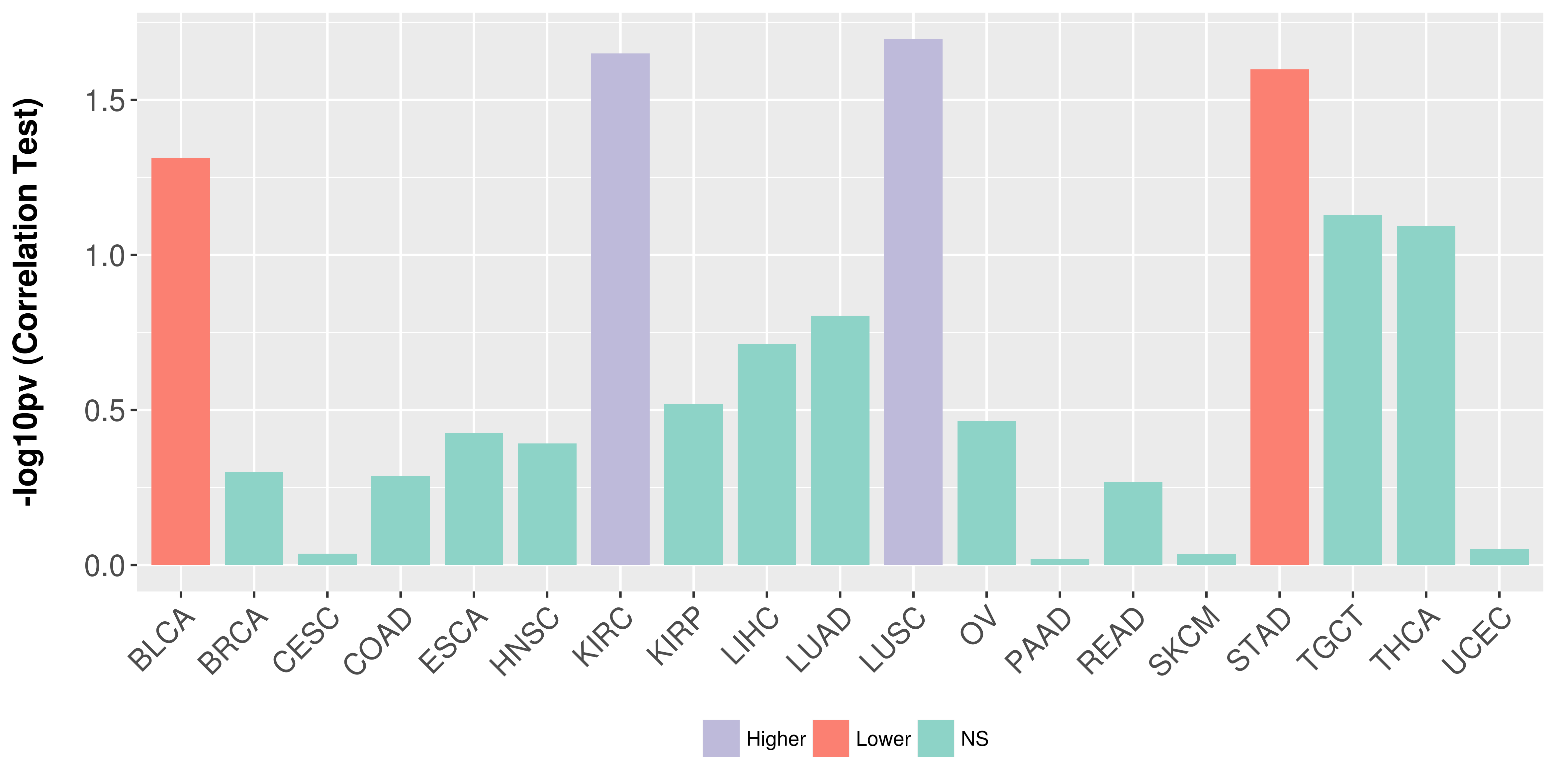
Association between expresson and stage.
|
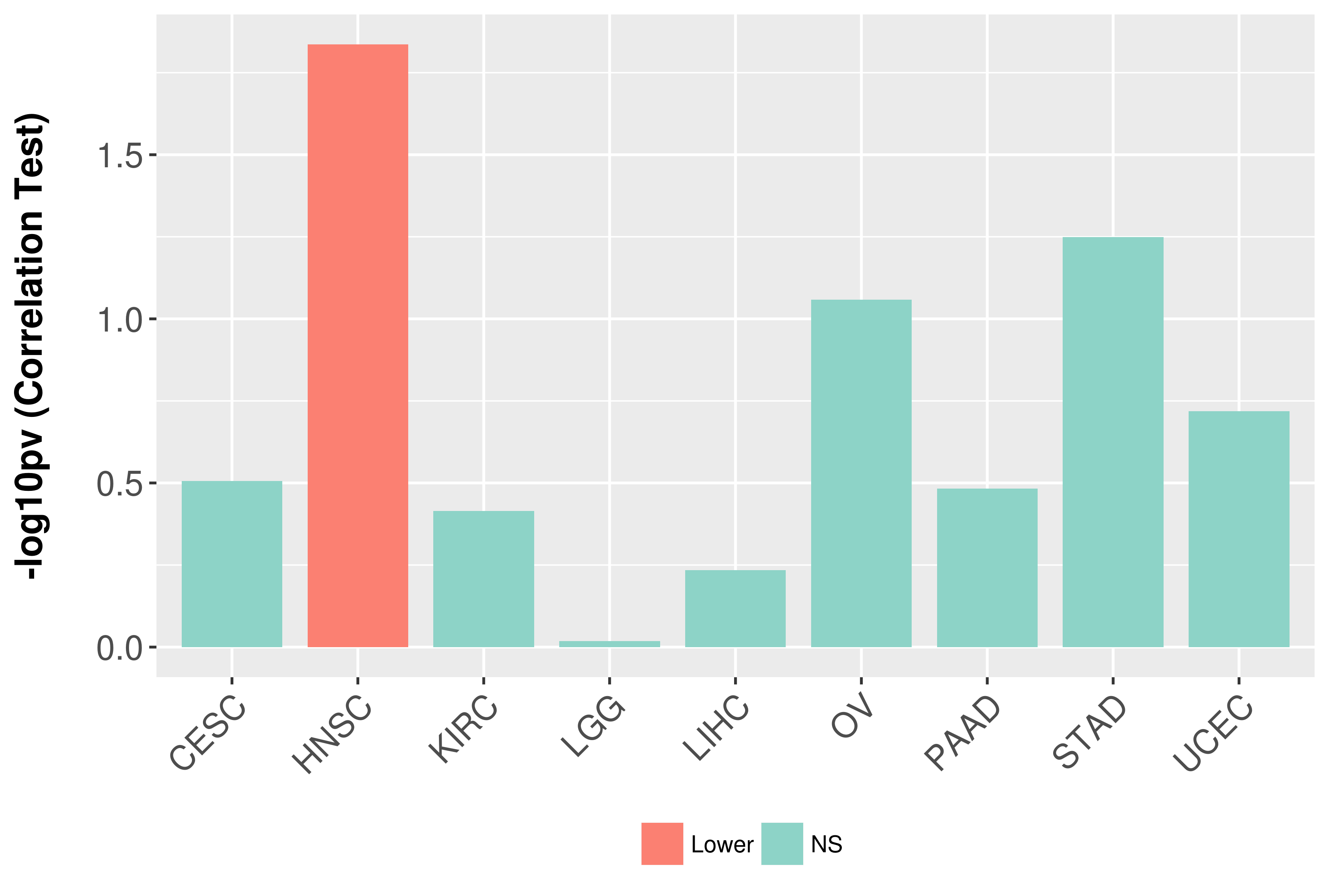
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for INO80B. |
| Summary | |
|---|---|
| Symbol | INO80B |
| Name | INO80 complex subunit B |
| Aliases | HMGIYL4; PAPA-1; hIes2; PAP-1BP; IES2; PAP-1 binding protein; IES2 homolog (S. cerevisiae); HMGA1L4; ZNHIT4; ...... |
| Location | 2p13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for INO80B. |