Browse KAT2B in pancancer
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00583 Acetyltransferase (GNAT) family PF00439 Bromodomain PF06466 PCAF (P300/CBP-associated factor) N-terminal domain |
||||||||||
| Function |
Functions as a histone acetyltransferase (HAT) to promote transcriptional activation. Has significant histone acetyltransferase activity with core histones (H3 and H4), and also with nucleosome core particles. Also acetylates non-histone proteins, such as ACLY. Inhibits cell-cycle progression and counteracts the mitogenic activity of the adenoviral oncoprotein E1A. In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat's transactivating activity and may help inducing chromatin remodeling of proviral genes. Acts as a circadian transcriptional coactivator which enhances the activity of the circadian transcriptional activators: NPAS2-ARNTL/BMAL1 and CLOCK-ARNTL/BMAL1 heterodimers. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000079 regulation of cyclin-dependent protein serine/threonine kinase activity GO:0001933 negative regulation of protein phosphorylation GO:0005996 monosaccharide metabolic process GO:0006006 glucose metabolic process GO:0006094 gluconeogenesis GO:0006109 regulation of carbohydrate metabolic process GO:0006111 regulation of gluconeogenesis GO:0006338 chromatin remodeling GO:0006352 DNA-templated transcription, initiation GO:0006367 transcription initiation from RNA polymerase II promoter GO:0006469 negative regulation of protein kinase activity GO:0006471 protein ADP-ribosylation GO:0006473 protein acetylation GO:0006474 N-terminal protein amino acid acetylation GO:0006475 internal protein amino acid acetylation GO:0006486 protein glycosylation GO:0007050 cell cycle arrest GO:0007219 Notch signaling pathway GO:0009100 glycoprotein metabolic process GO:0009101 glycoprotein biosynthetic process GO:0010559 regulation of glycoprotein biosynthetic process GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010676 positive regulation of cellular carbohydrate metabolic process GO:0010835 regulation of protein ADP-ribosylation GO:0010906 regulation of glucose metabolic process GO:0010907 positive regulation of glucose metabolic process GO:0016051 carbohydrate biosynthetic process GO:0016570 histone modification GO:0016573 histone acetylation GO:0018076 N-terminal peptidyl-lysine acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019318 hexose metabolic process GO:0019319 hexose biosynthetic process GO:0031365 N-terminal protein amino acid modification GO:0032868 response to insulin GO:0032869 cellular response to insulin stimulus GO:0033673 negative regulation of kinase activity GO:0035947 regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter GO:0035948 positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter GO:0040029 regulation of gene expression, epigenetic GO:0042326 negative regulation of phosphorylation GO:0043255 regulation of carbohydrate biosynthetic process GO:0043413 macromolecule glycosylation GO:0043434 response to peptide hormone GO:0043543 protein acylation GO:0043966 histone H3 acetylation GO:0043970 histone H3-K9 acetylation GO:0044262 cellular carbohydrate metabolic process GO:0044283 small molecule biosynthetic process GO:0044723 single-organism carbohydrate metabolic process GO:0045722 positive regulation of gluconeogenesis GO:0045736 negative regulation of cyclin-dependent protein serine/threonine kinase activity GO:0045786 negative regulation of cell cycle GO:0045815 positive regulation of gene expression, epigenetic GO:0045913 positive regulation of carbohydrate metabolic process GO:0046364 monosaccharide biosynthetic process GO:0048511 rhythmic process GO:0051348 negative regulation of transferase activity GO:0060049 regulation of protein glycosylation GO:0061647 histone H3-K9 modification GO:0070085 glycosylation GO:0071375 cellular response to peptide hormone stimulus GO:0071417 cellular response to organonitrogen compound GO:0071900 regulation of protein serine/threonine kinase activity GO:0071901 negative regulation of protein serine/threonine kinase activity GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1903018 regulation of glycoprotein metabolic process GO:1904029 regulation of cyclin-dependent protein kinase activity GO:1904030 negative regulation of cyclin-dependent protein kinase activity |
| Molecular Function |
GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0004468 lysine N-acetyltransferase activity, acting on acetyl phosphate as donor GO:0004857 enzyme inhibitor activity GO:0004860 protein kinase inhibitor activity GO:0004861 cyclin-dependent protein serine/threonine kinase inhibitor activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016538 cyclin-dependent protein serine/threonine kinase regulator activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0019207 kinase regulator activity GO:0019210 kinase inhibitor activity GO:0019887 protein kinase regulator activity GO:0030291 protein serine/threonine kinase inhibitor activity GO:0034212 peptide N-acetyltransferase activity GO:0042826 histone deacetylase binding GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000125 PCAF complex GO:0000775 chromosome, centromeric region GO:0000776 kinetochore GO:0005671 Ada2/Gcn5/Ada3 transcription activator complex GO:0015629 actin cytoskeleton GO:0030016 myofibril GO:0030017 sarcomere GO:0031248 protein acetyltransferase complex GO:0031672 A band GO:0031674 I band GO:0042641 actomyosin GO:0043292 contractile fiber GO:0044449 contractile fiber part GO:0070461 SAGA-type complex GO:0098687 chromosomal region GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG |
hsa04330 Notch signaling pathway hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-5250924: B-WICH complex positively regulates rRNA expression R-HSA-5576891: Cardiac conduction R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-5688426: Deubiquitination R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-392499: Metabolism of proteins R-HSA-5689901: Metalloprotease DUBs R-HSA-397014: Muscle contraction R-HSA-157052: NICD traffics to nucleus R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-350054: Notch-HLH transcription pathway R-HSA-5578768: Physiological factors R-HSA-5250913: Positive epigenetic regulation of rRNA expression R-HSA-597592: Post-translational protein modification R-HSA-1912422: Pre-NOTCH Expression and Processing R-HSA-1912408: Pre-NOTCH Transcription and Translation R-HSA-73854: RNA Polymerase I Promoter Clearance R-HSA-73864: RNA Polymerase I Transcription R-HSA-73762: RNA Polymerase I Transcription Initiation R-HSA-504046: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer R-HSA-2032785: YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
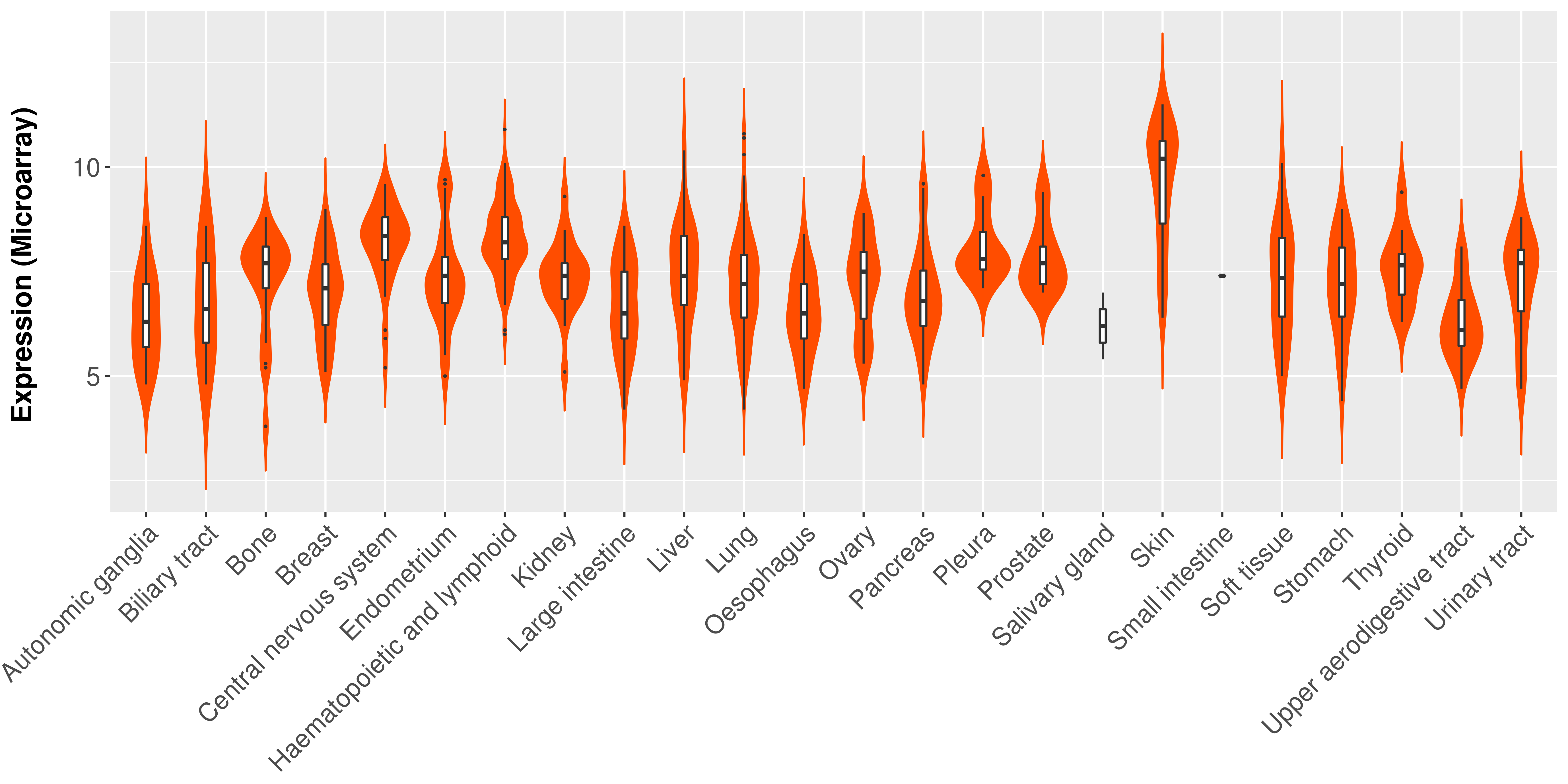
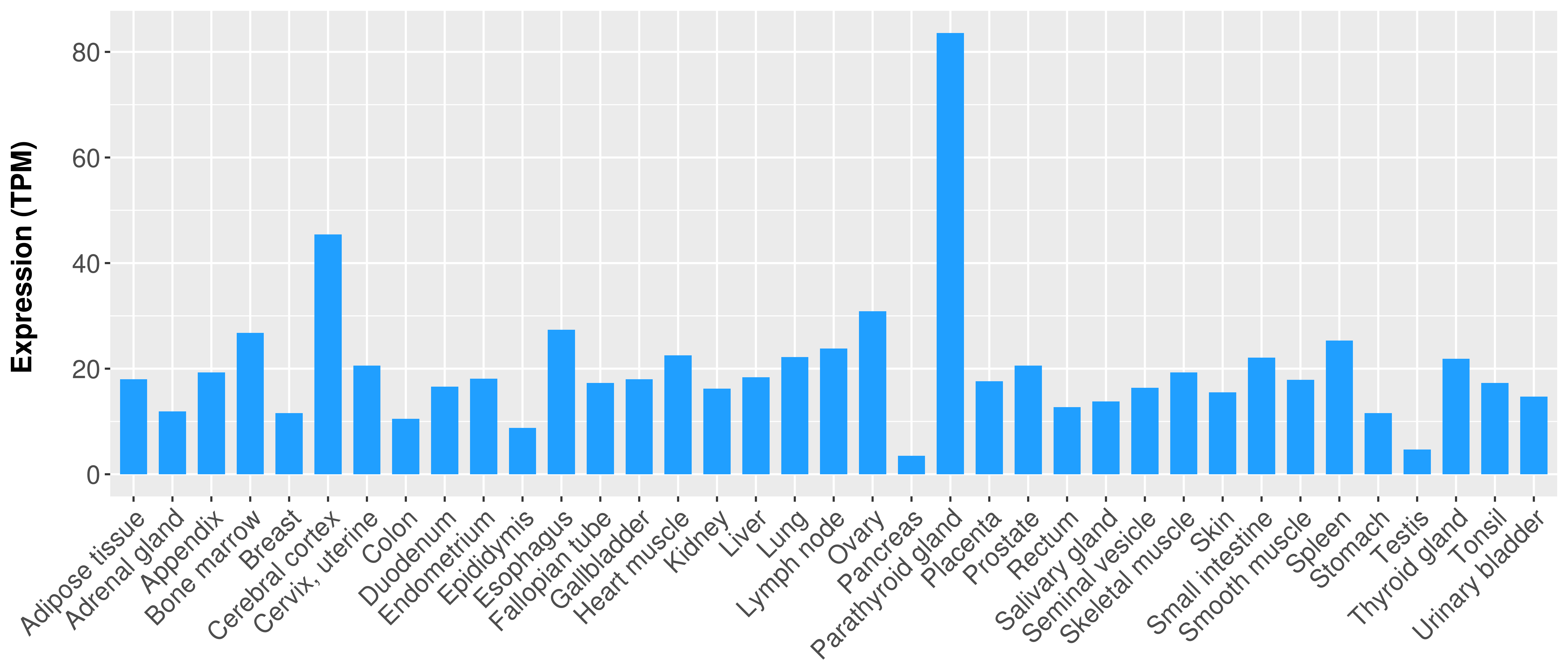
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
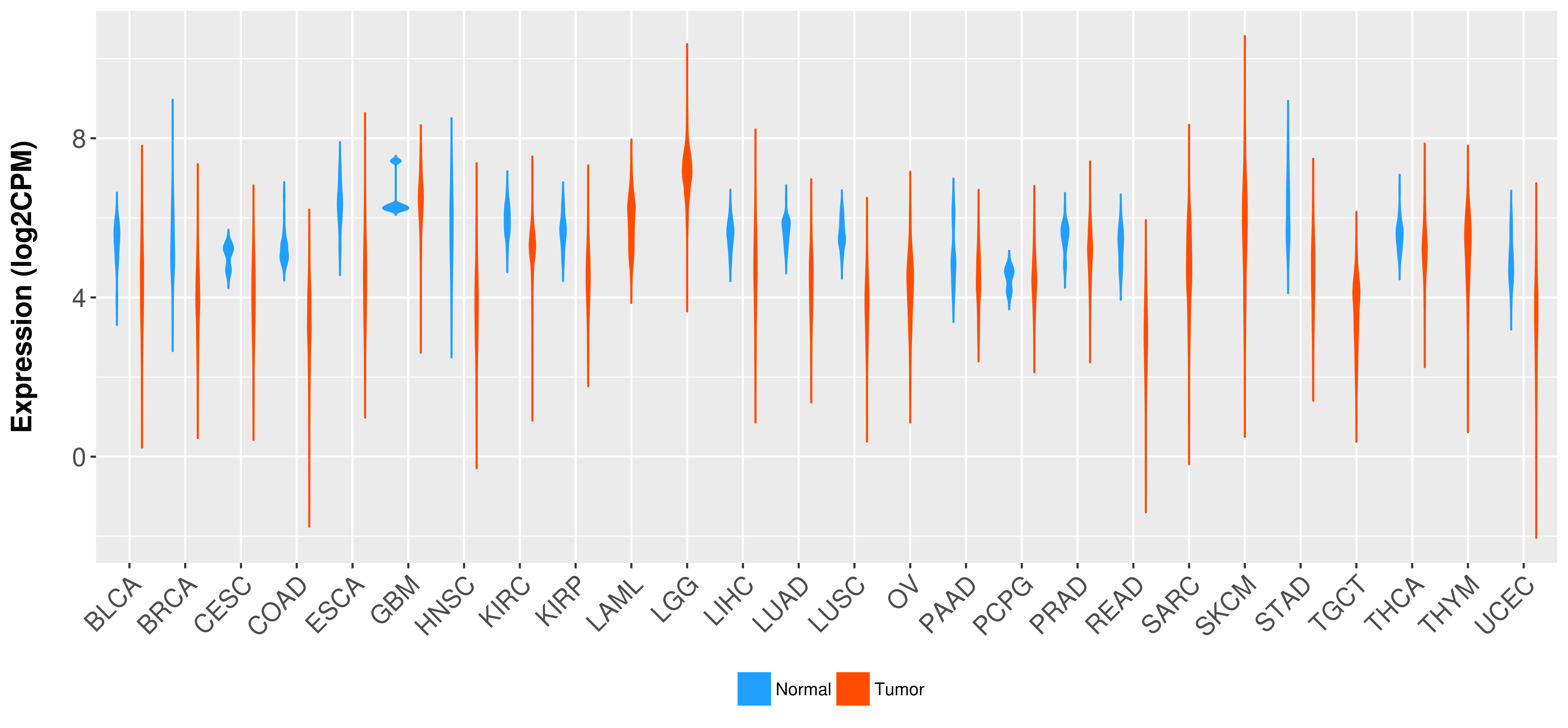
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
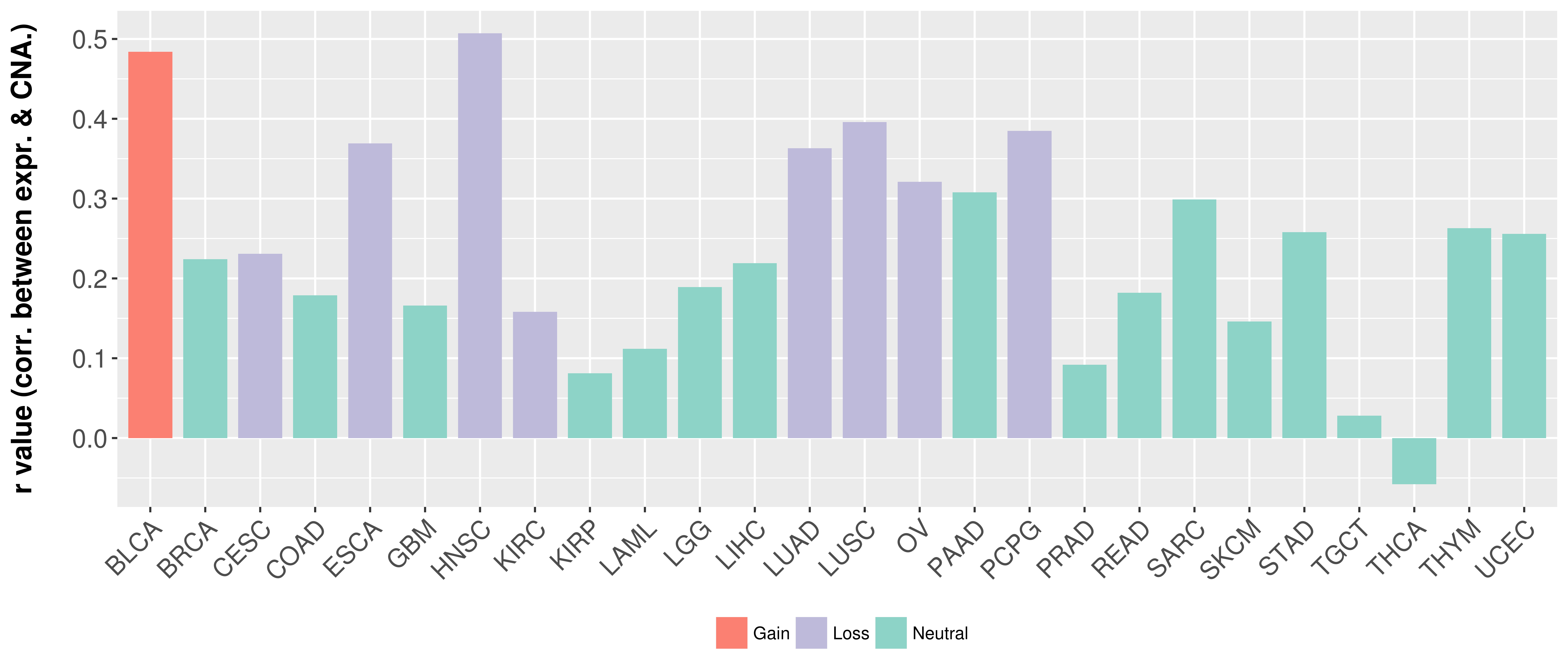
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
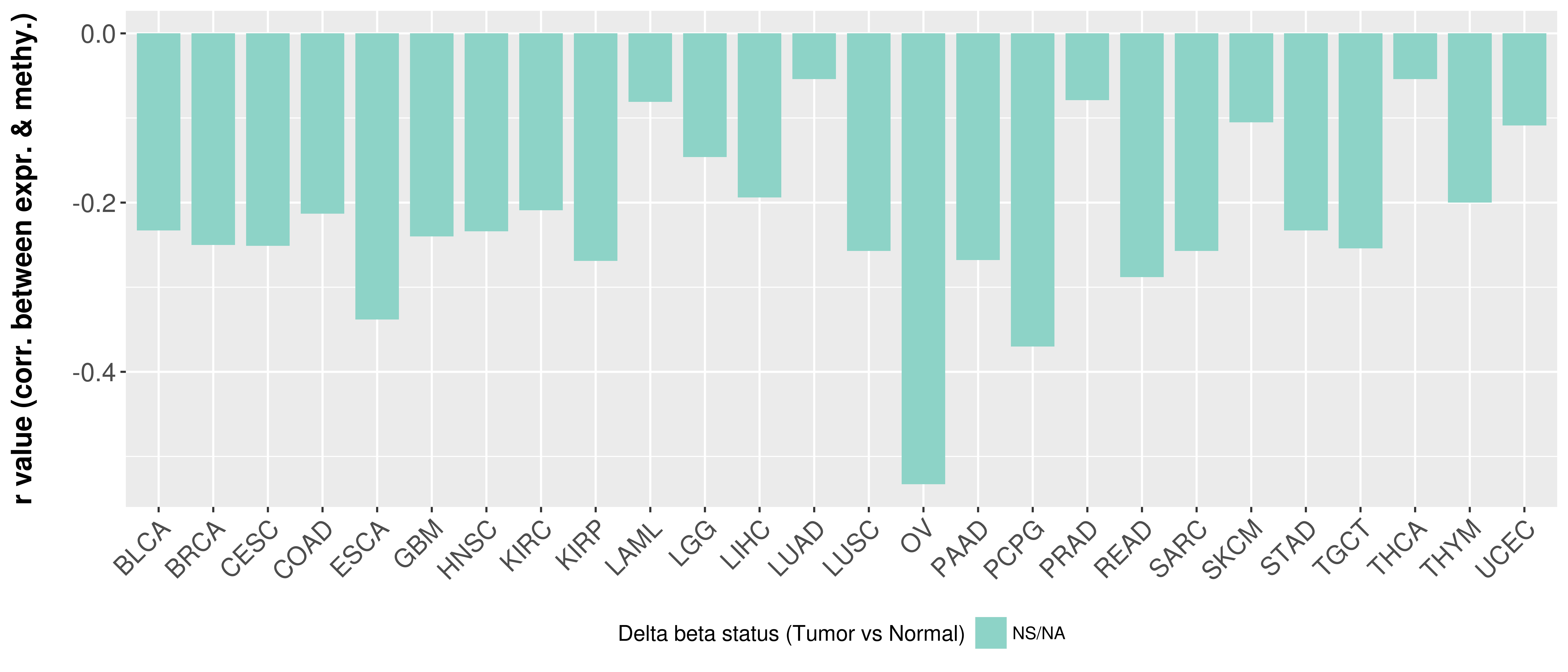
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
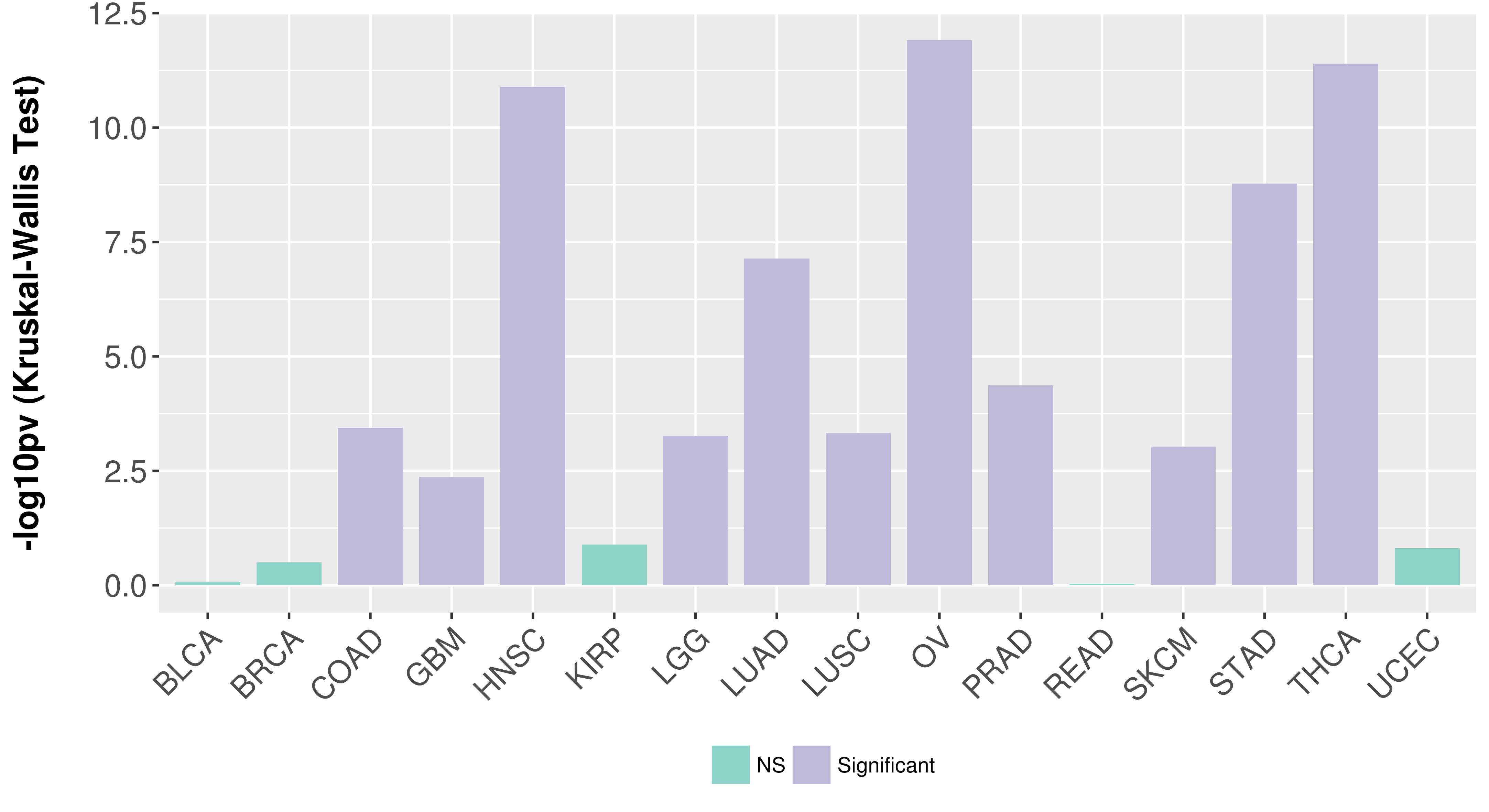
Association between expresson and subtype.
|
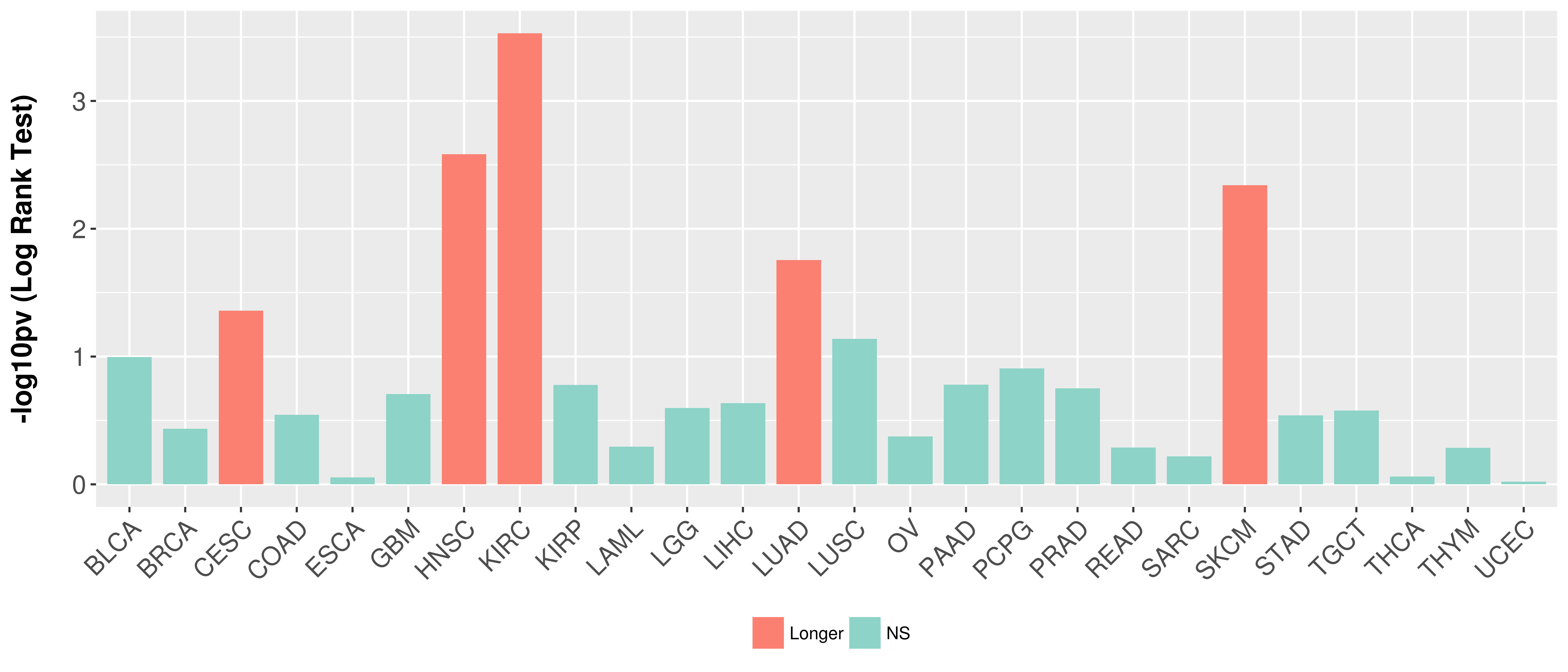
Overall survival analysis based on expression.
|
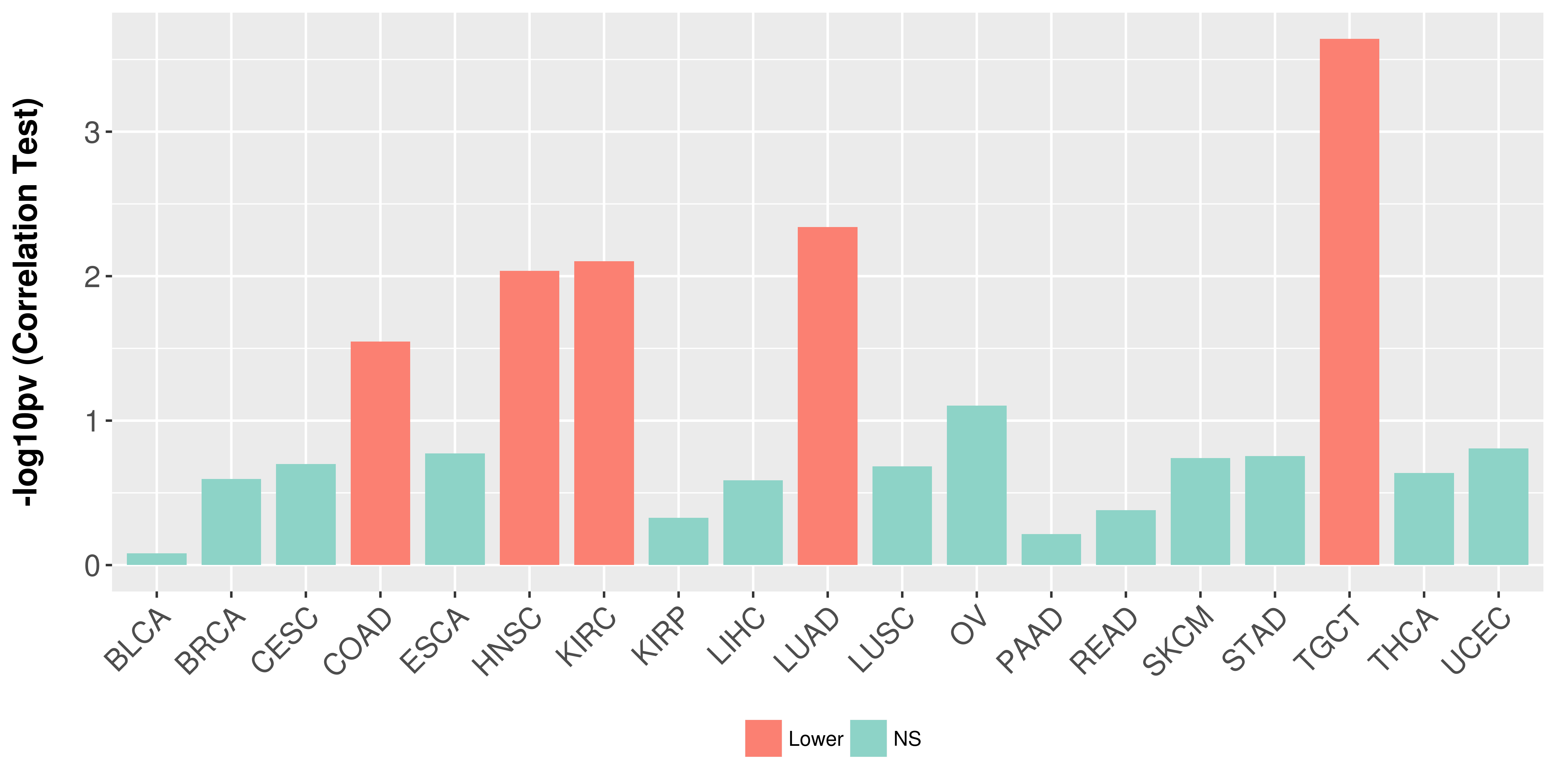
Association between expresson and stage.
|
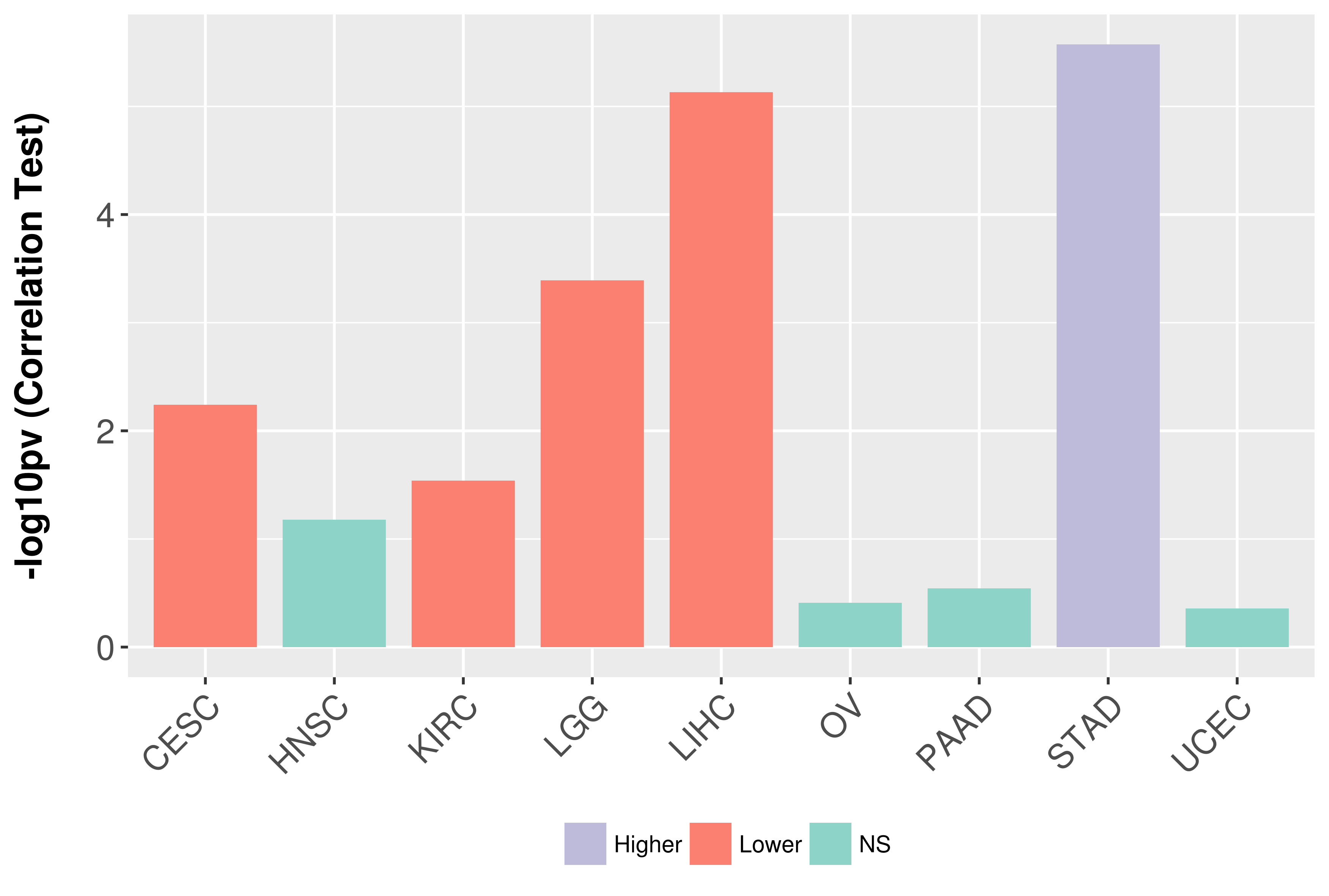
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
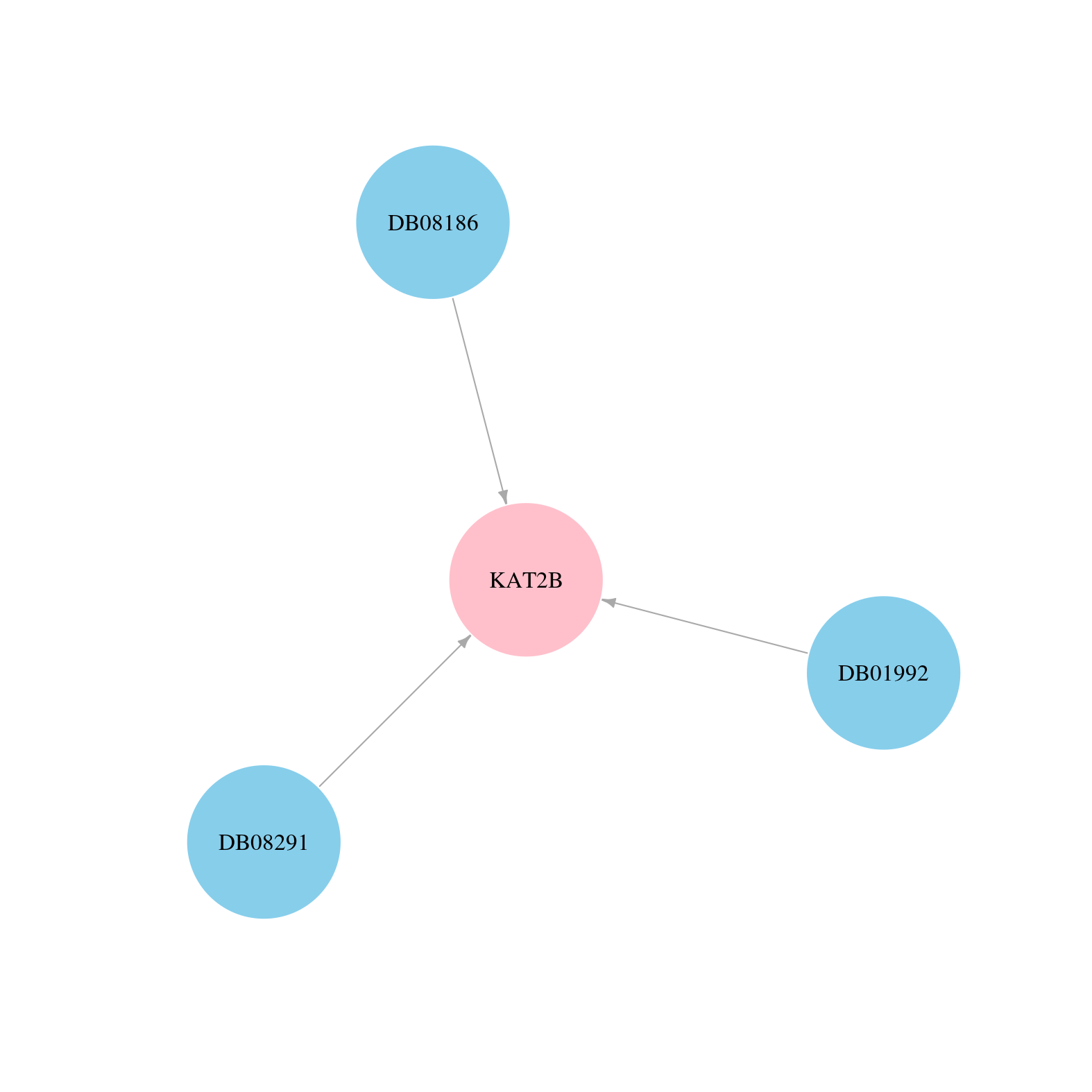
|
| Summary | |
|---|---|
| Symbol | KAT2B |
| Name | lysine acetyltransferase 2B |
| Aliases | P/CAF; GCN5L; PCAF; p300/CBP-associated factor; CAF; CREBBP-associated factor; histone acetylase PCAF; histo ...... |
| Location | 3p24.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|