Browse KAT5 in pancancer
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01853 MOZ/SAS family PF11717 RNA binding activity-knot of a chromodomain |
||||||||||
| Function |
Catalytic subunit of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome-DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Directly acetylates and activates ATM. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AFZ from the nucleosome. In case of HIV-1 infection, interaction with the viral Tat protein leads to KAT5 polyubiquitination and targets it to degradation. Relieves NR1D2-mediated inhibition of APOC3 expression by acetylating NR1D2. Promotes FOXP3 acetylation and positively regulates its transcriptional repressor activity (PubMed:17360565). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000726 non-recombinational repair GO:0000729 DNA double-strand break processing GO:0000731 DNA synthesis involved in DNA repair GO:0000732 strand displacement GO:0001818 negative regulation of cytokine production GO:0006260 DNA replication GO:0006302 double-strand break repair GO:0006303 double-strand break repair via nonhomologous end joining GO:0006310 DNA recombination GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006978 DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator GO:0009314 response to radiation GO:0009755 hormone-mediated signaling pathway GO:0010212 response to ionizing radiation GO:0010498 proteasomal protein catabolic process GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0032355 response to estradiol GO:0032623 interleukin-2 production GO:0032663 regulation of interleukin-2 production GO:0032703 negative regulation of interleukin-2 production GO:0042770 signal transduction in response to DNA damage GO:0042772 DNA damage response, signal transduction resulting in transcription GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043401 steroid hormone mediated signaling pathway GO:0043543 protein acylation GO:0048545 response to steroid hormone GO:0071383 cellular response to steroid hormone stimulus GO:0071392 cellular response to estradiol stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071897 DNA biosynthetic process GO:0072331 signal transduction by p53 class mediator GO:1901796 regulation of signal transduction by p53 class mediator GO:1901983 regulation of protein acetylation GO:1901985 positive regulation of protein acetylation GO:1904837 beta-catenin-TCF complex assembly |
| Molecular Function |
GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0034212 peptide N-acetyltransferase activity GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0050681 androgen receptor binding GO:0051427 hormone receptor binding GO:0061733 peptide-lysine-N-acetyltransferase activity GO:0070491 repressing transcription factor binding |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000812 Swr1 complex GO:0005667 transcription factor complex GO:0031248 protein acetyltransferase complex GO:0032777 Piccolo NuA4 histone acetyltransferase complex GO:0035267 NuA4 histone acetyltransferase complex GO:0043189 H4/H2A histone acetyltransferase complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0097346 INO80-type complex GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-69620: Cell Cycle Checkpoints R-HSA-2559583: Cellular Senescence R-HSA-2262752: Cellular responses to stress R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2559586: DNA Damage/Telomere Stress Induced Senescence R-HSA-5693606: DNA Double Strand Break Response R-HSA-5693532: DNA Double-Strand Break Repair R-HSA-73894: DNA Repair R-HSA-201722: Formation of the beta-catenin R-HSA-69481: G2/M Checkpoints R-HSA-69473: G2/M DNA damage checkpoint R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-5693567: HDR through Homologous Recombination (HR) or Single Strand Annealing (SSA) R-HSA-5685942: HDR through Homologous Recombination (HRR) R-HSA-5685938: HDR through Single Strand Annealing (SSA) R-HSA-5693579: Homologous DNA Pairing and Strand Exchange R-HSA-5693538: Homology Directed Repair R-HSA-5693571: Nonhomologous End-Joining (NHEJ) R-HSA-5693616: Presynaptic phase of homologous DNA pairing and strand exchange R-HSA-5693607: Processing of DNA double-strand break ends R-HSA-5693565: Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804756: Regulation of TP53 Activity through Phosphorylation R-HSA-5693537: Resolution of D-Loop Structures R-HSA-5693568: Resolution of D-loop Structures through Holliday Junction Intermediates R-HSA-5693554: Resolution of D-loop Structures through Synthesis-Dependent Strand Annealing (SDSA) R-HSA-5693548: Sensing of DNA Double Strand Breaks R-HSA-162582: Signal Transduction R-HSA-195721: Signaling by Wnt R-HSA-201681: TCF dependent signaling in response to WNT R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for KAT5. |
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
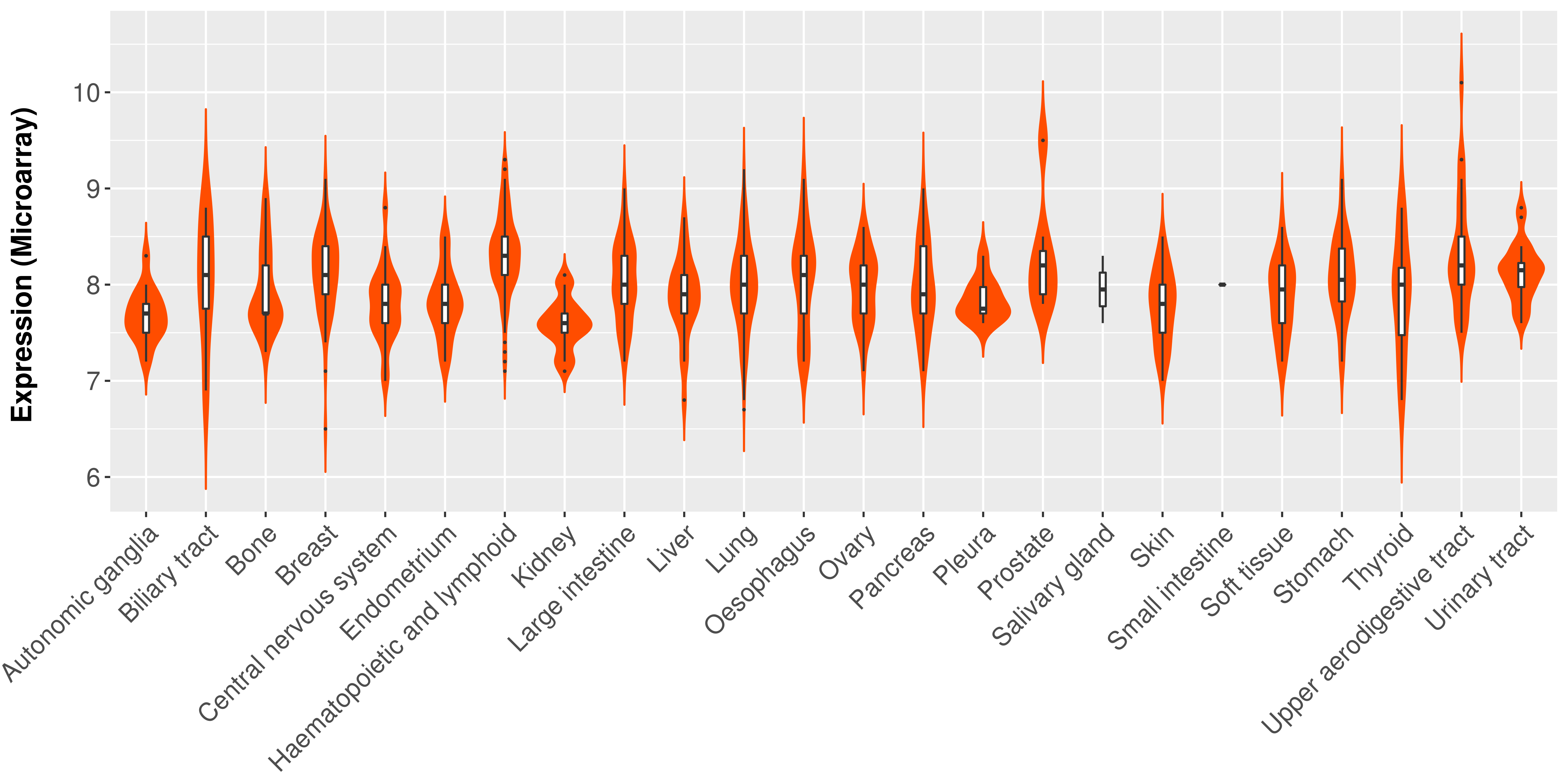
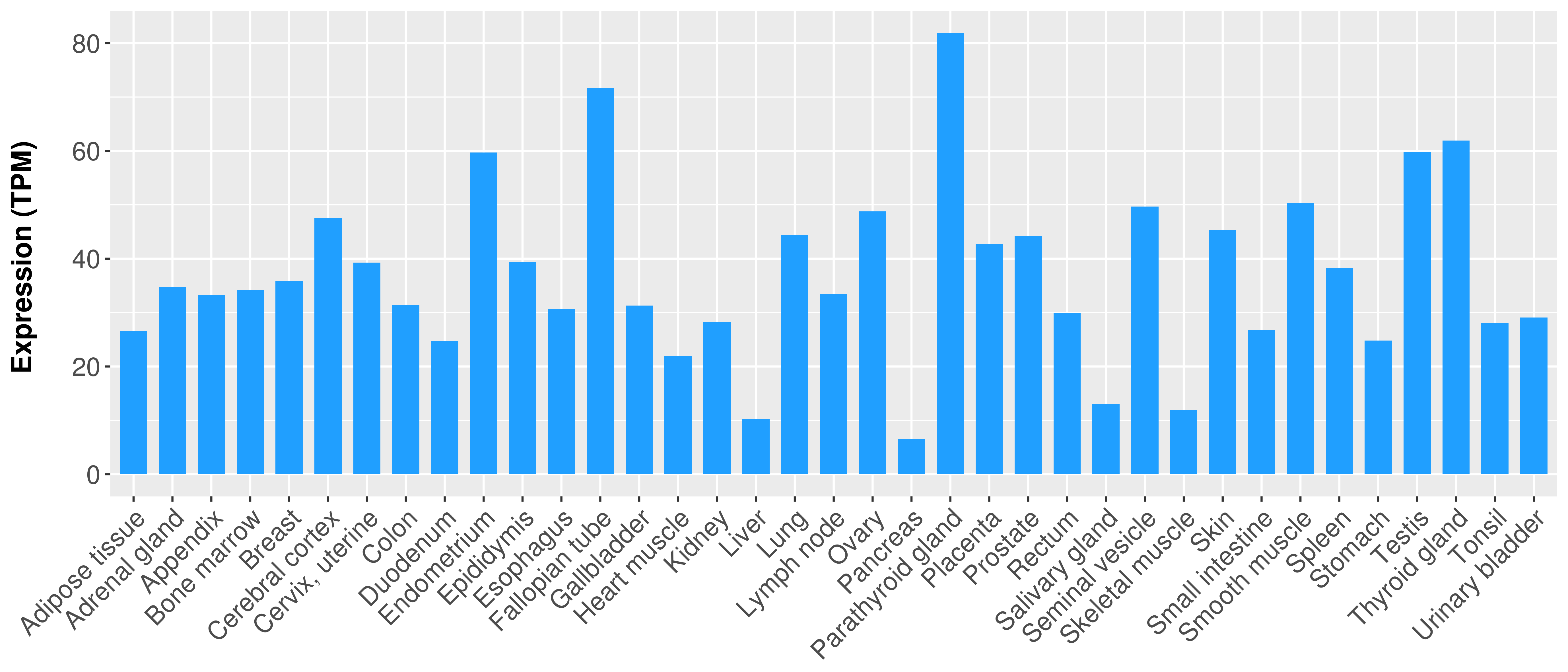
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
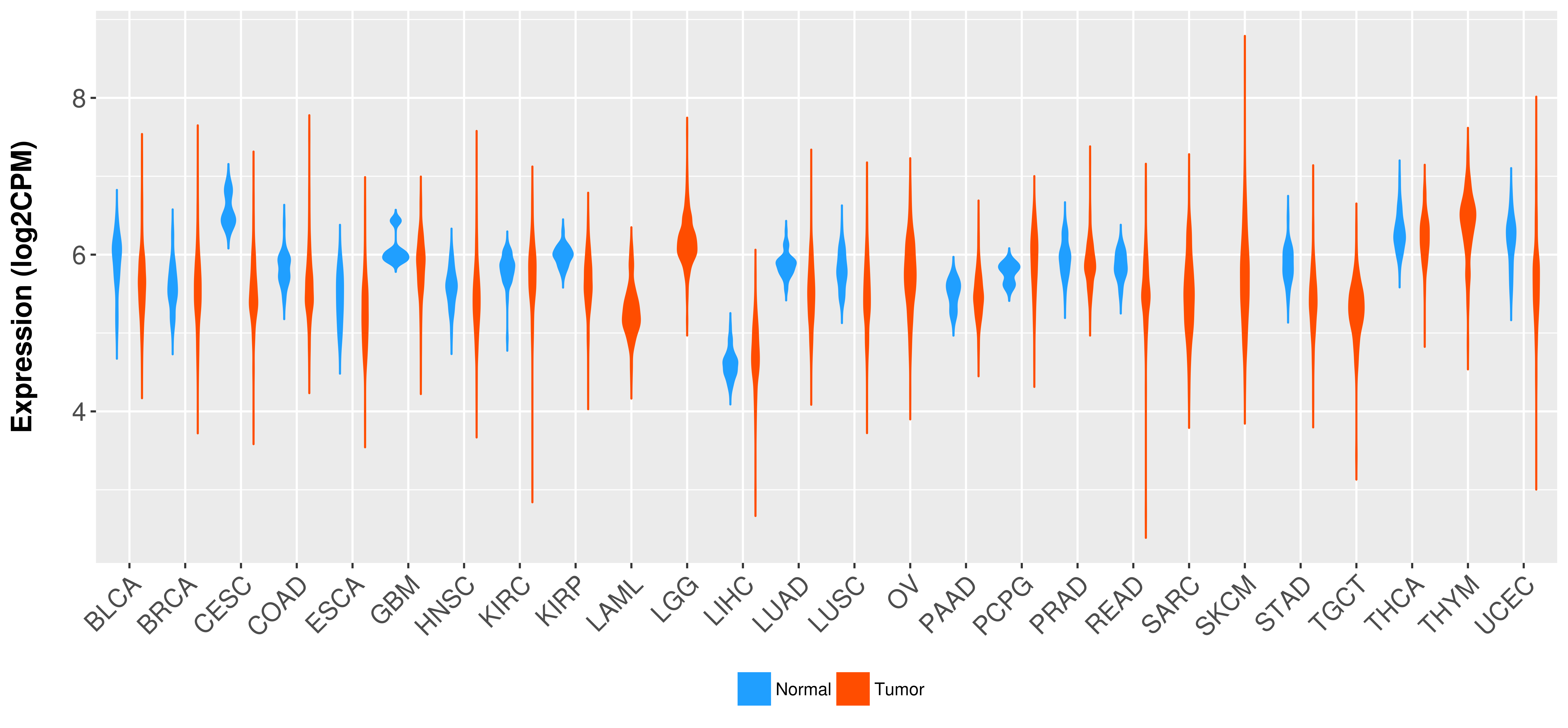
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
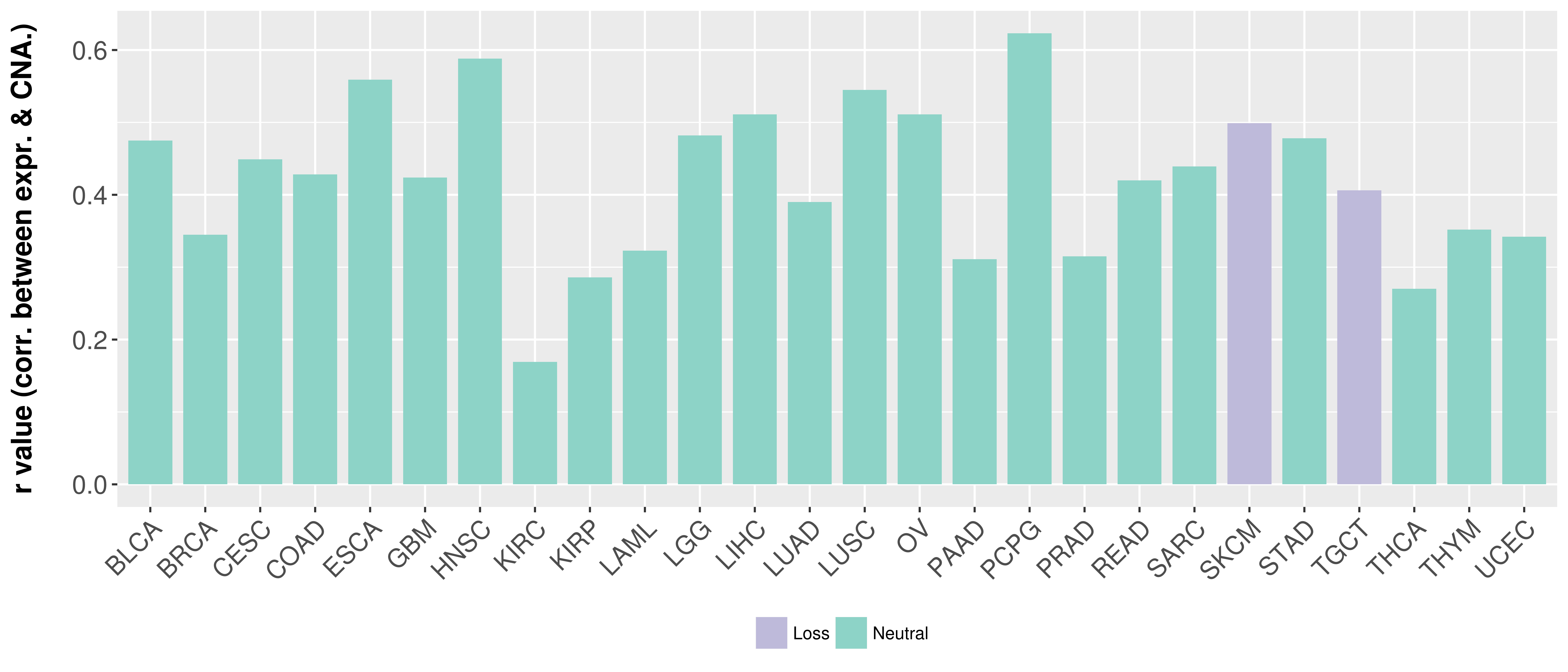
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
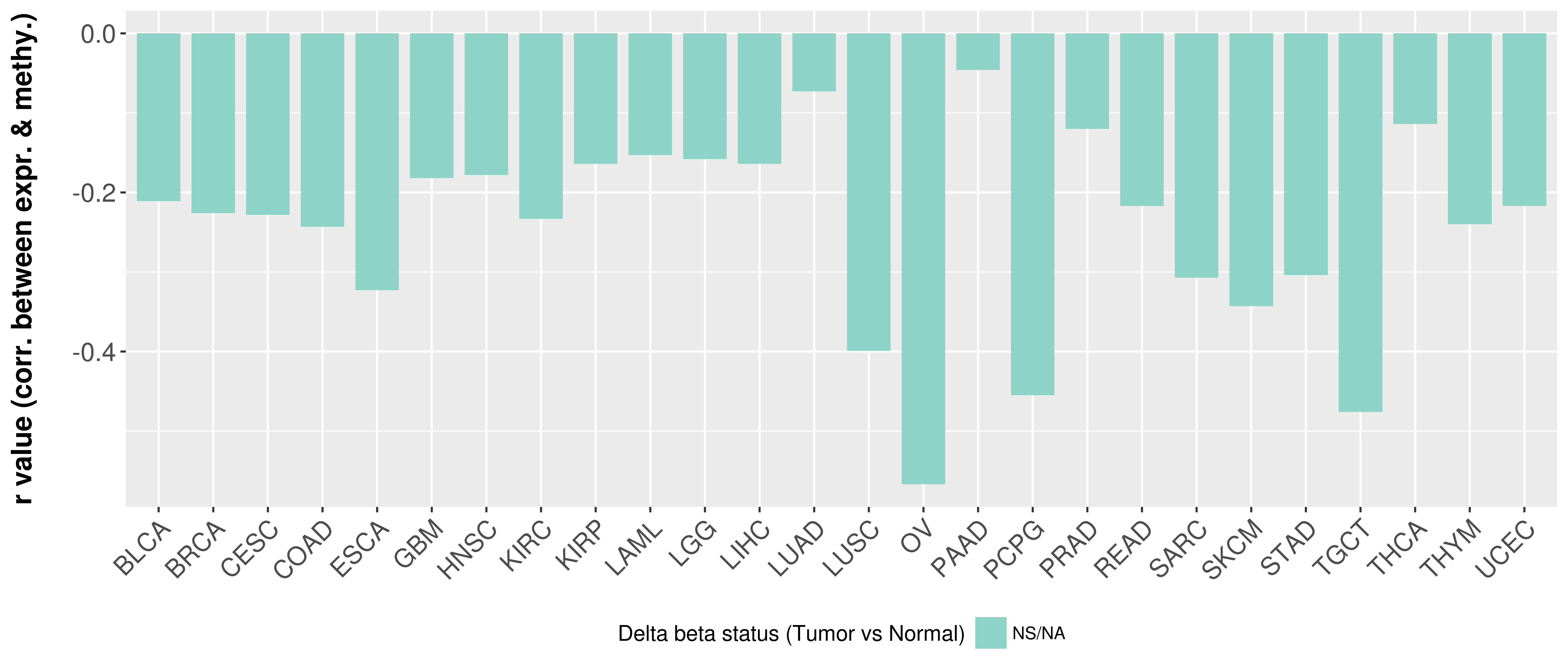
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
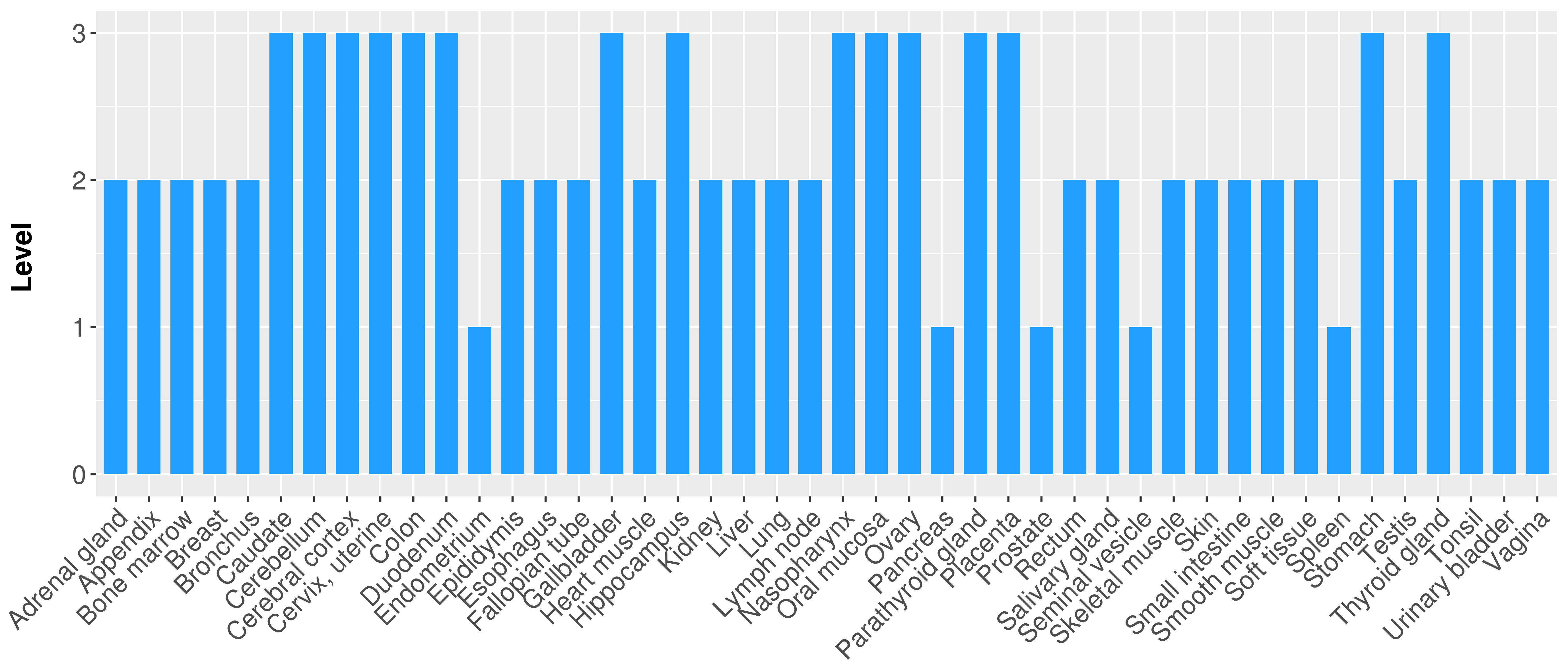
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |

Association between expresson and subtype.
|
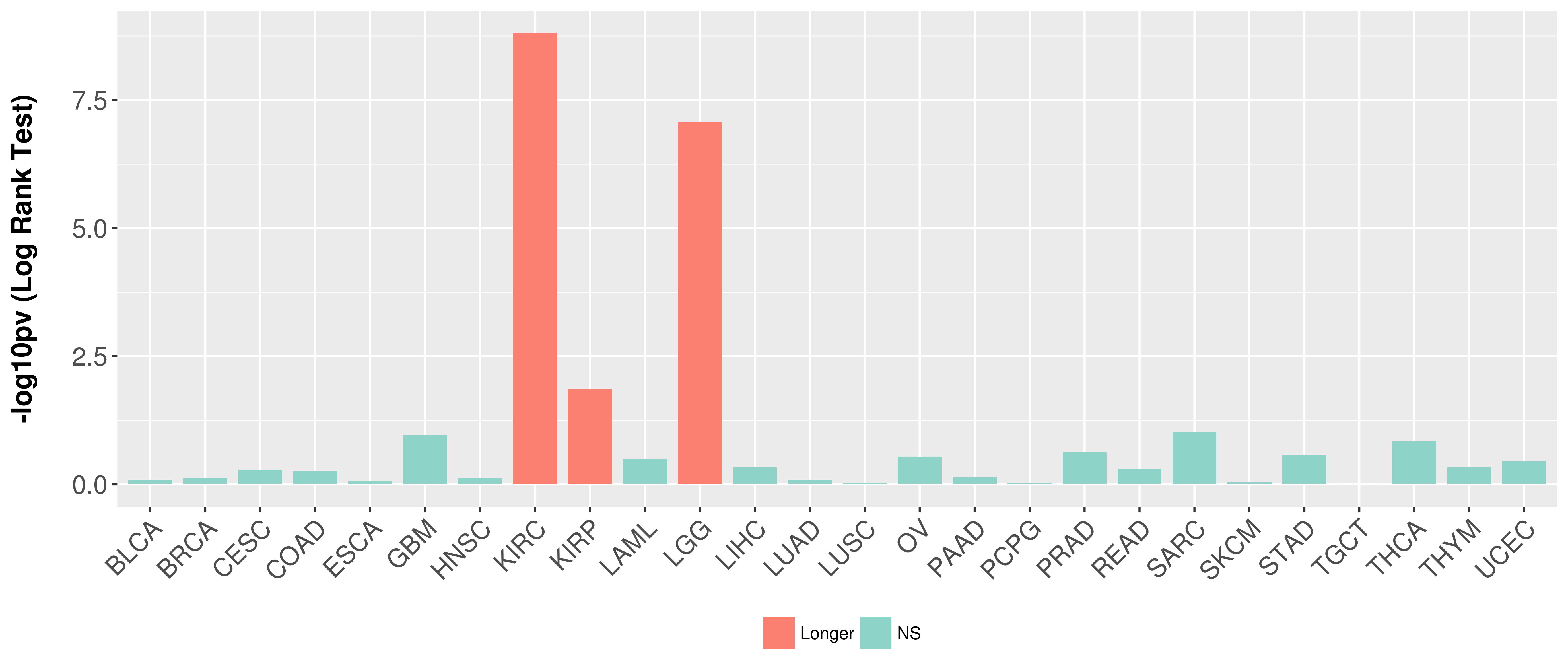
Overall survival analysis based on expression.
|
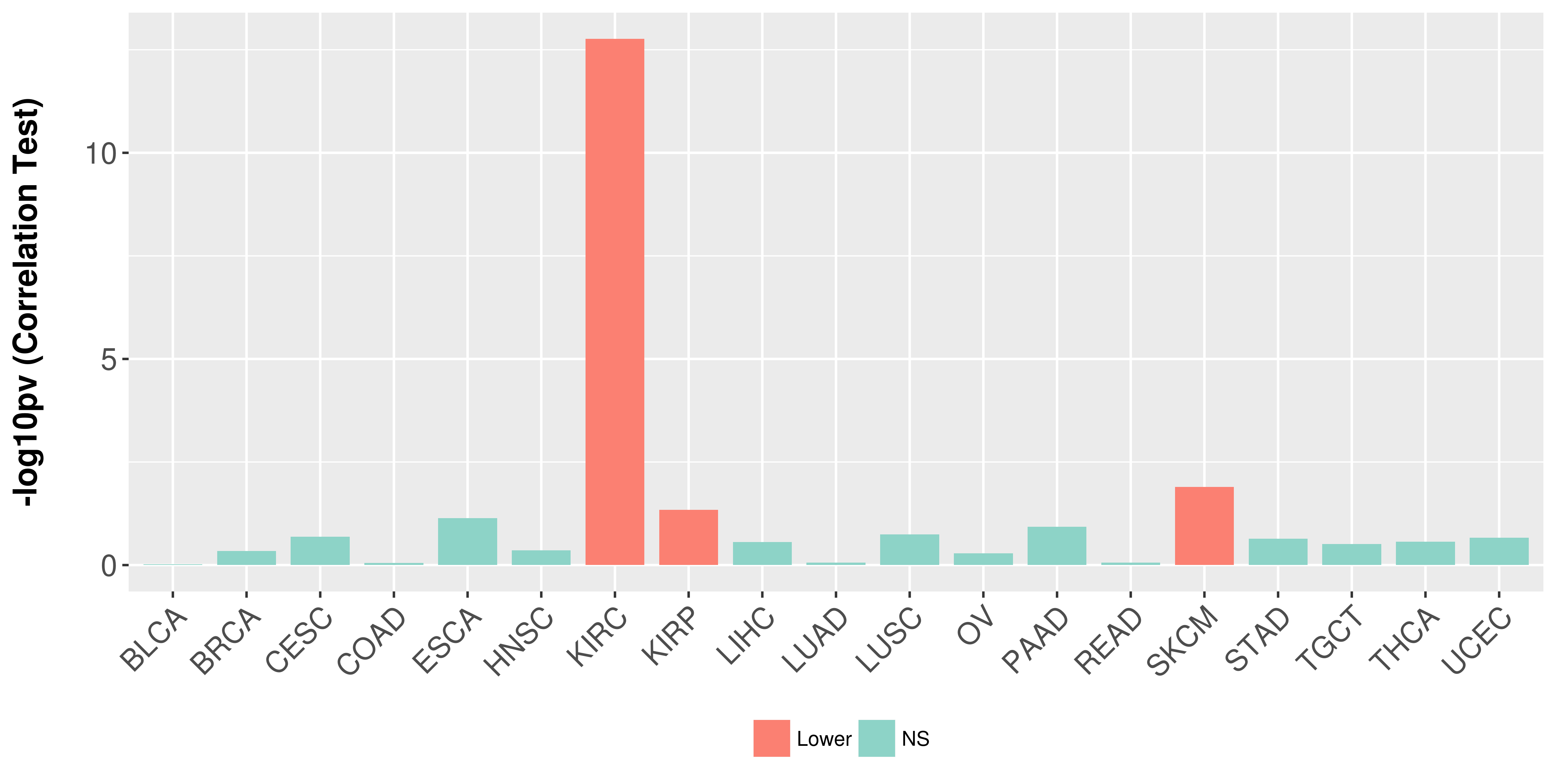
Association between expresson and stage.
|
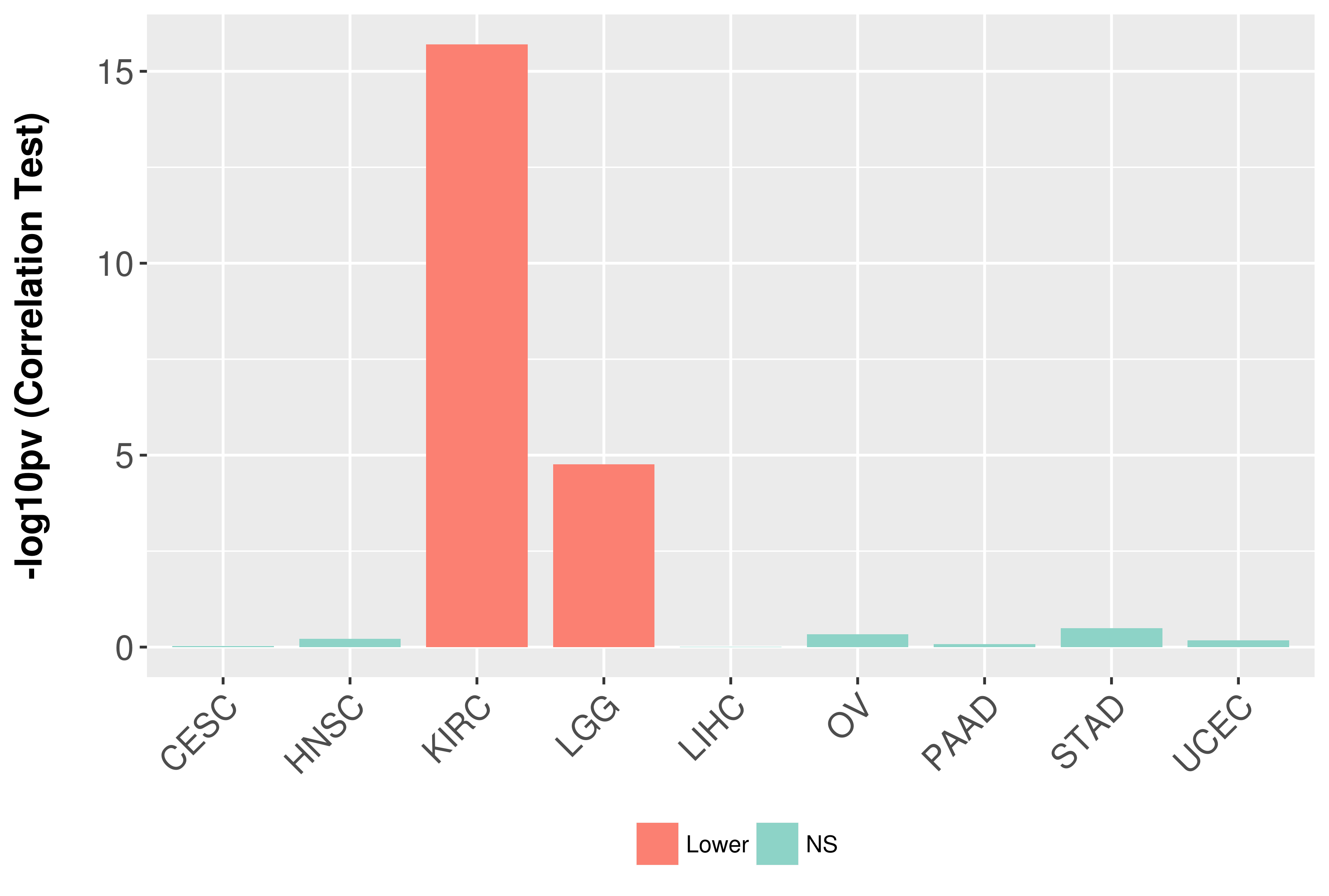
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
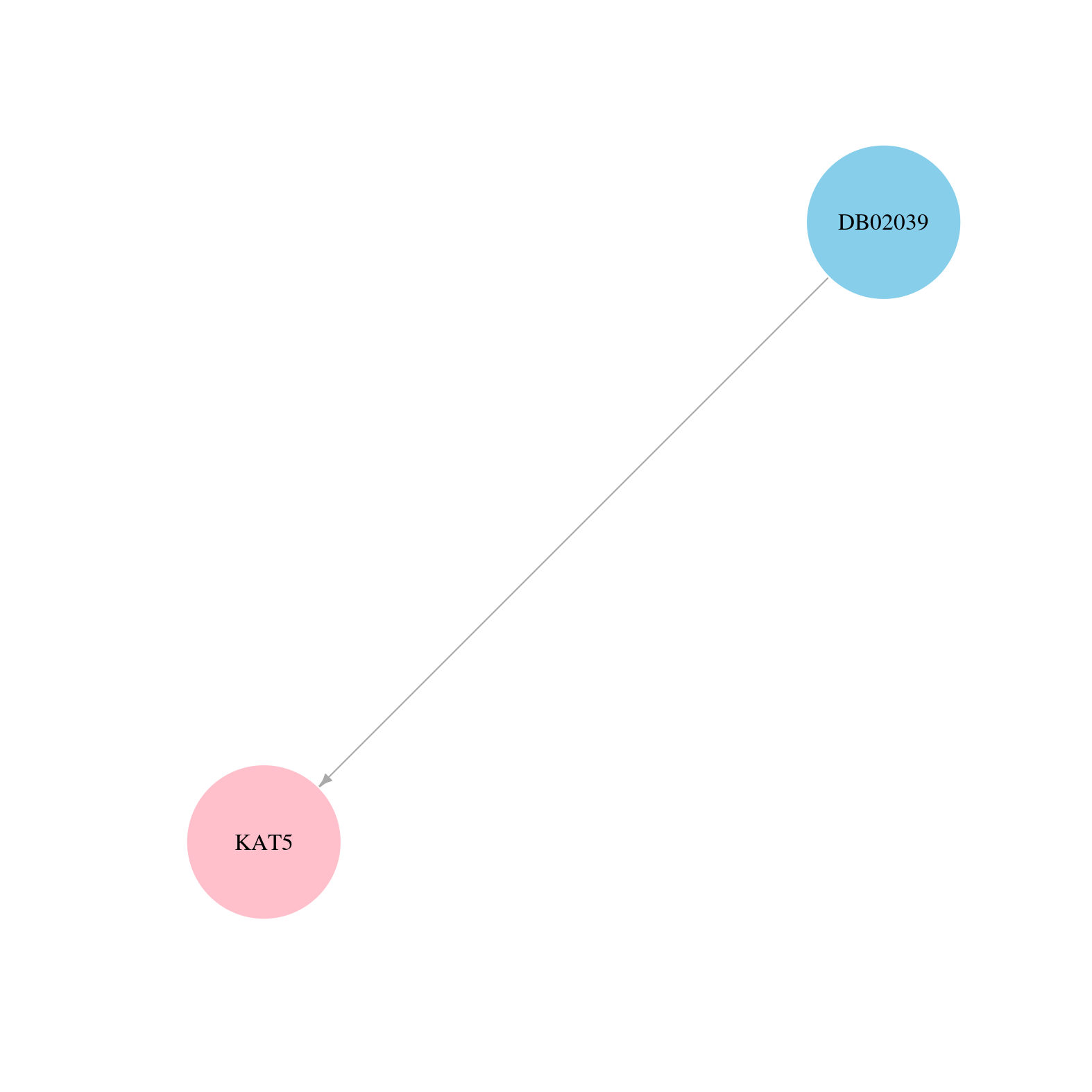
| Content |
> Drugs from DrugBank database |

|
| Summary | |
|---|---|
| Symbol | KAT5 |
| Name | lysine acetyltransferase 5 |
| Aliases | TIP60; HTATIP1; Tat interacting protein, 60kDa; K-acetyltransferase 5; HTATIP; HIV-1 Tat interactive protein ...... |
| Location | 11q13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for KAT5. |