Browse KAT8 in pancancer
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01853 MOZ/SAS family PF11717 RNA binding activity-knot of a chromodomain |
||||||||||
| Function |
Histone acetyltransferase which may be involved in transcriptional activation. May influence the function of ATM. As part of the MSL complex it is involved in acetylation of nucleosomal histone H4 producing specifically H4K16ac. As part of the NSL complex it may be involved in acetylation of nucleosomal histone H4 on several lysine residues. That activity is less specific than the one of the MSL complex. Can also acetylate TP53/p53 at 'Lys-120'. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006914 autophagy GO:0010506 regulation of autophagy GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0030099 myeloid cell differentiation GO:0043543 protein acylation GO:0043967 histone H4 acetylation GO:0043981 histone H4-K5 acetylation GO:0043982 histone H4-K8 acetylation GO:0043984 histone H4-K16 acetylation |
| Molecular Function |
GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0010485 H4 histone acetyltransferase activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0034212 peptide N-acetyltransferase activity GO:0035064 methylated histone binding GO:0042393 histone binding GO:0043995 histone acetyltransferase activity (H4-K5 specific) GO:0043996 histone acetyltransferase activity (H4-K8 specific) GO:0046972 histone acetyltransferase activity (H4-K16 specific) GO:0061733 peptide-lysine-N-acetyltransferase activity |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000775 chromosome, centromeric region GO:0000776 kinetochore GO:0031248 protein acetyltransferase complex GO:0034708 methyltransferase complex GO:0035097 histone methyltransferase complex GO:0044665 MLL1/2 complex GO:0071339 MLL1 complex GO:0072487 MSL complex GO:0098687 chromosomal region GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214847: HATs acetylate histones |
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for KAT8. |
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
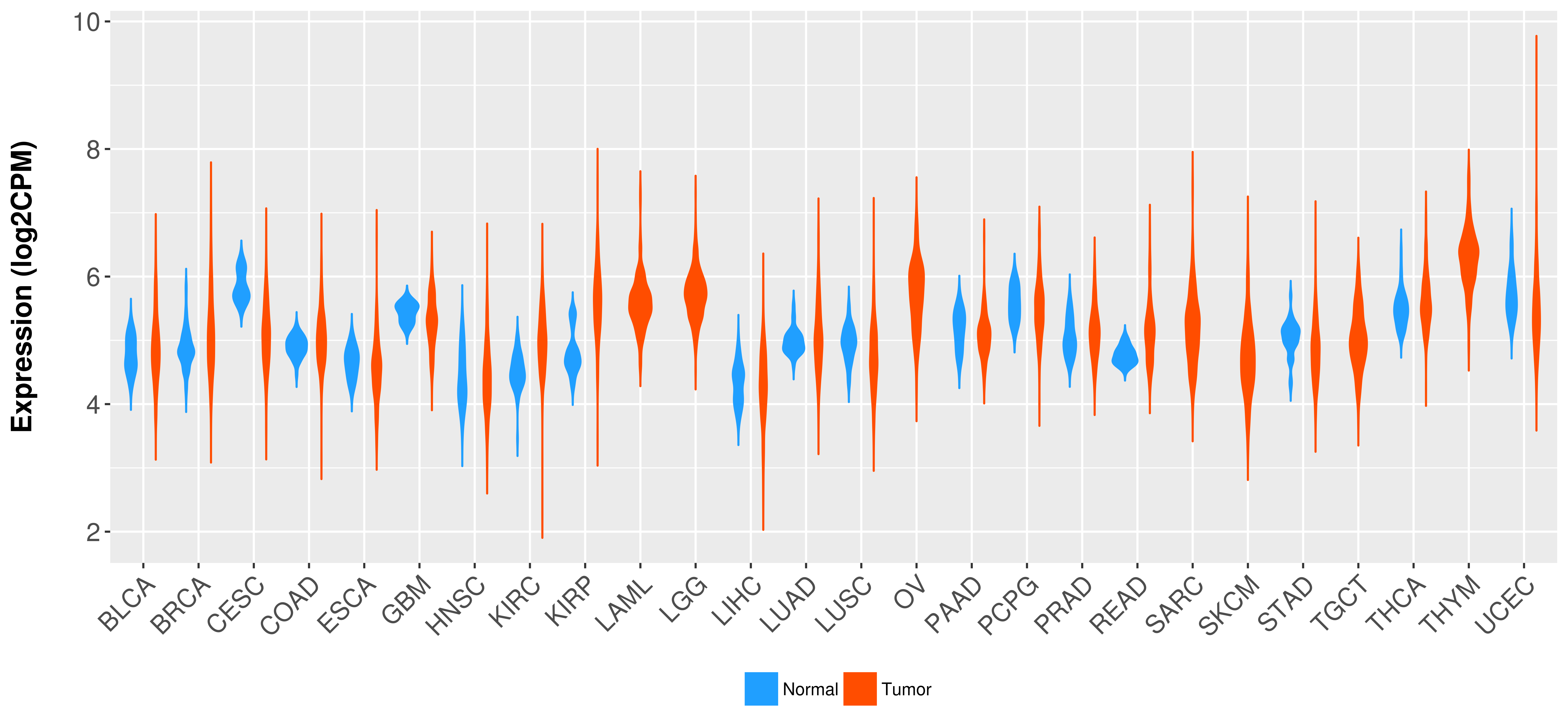
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
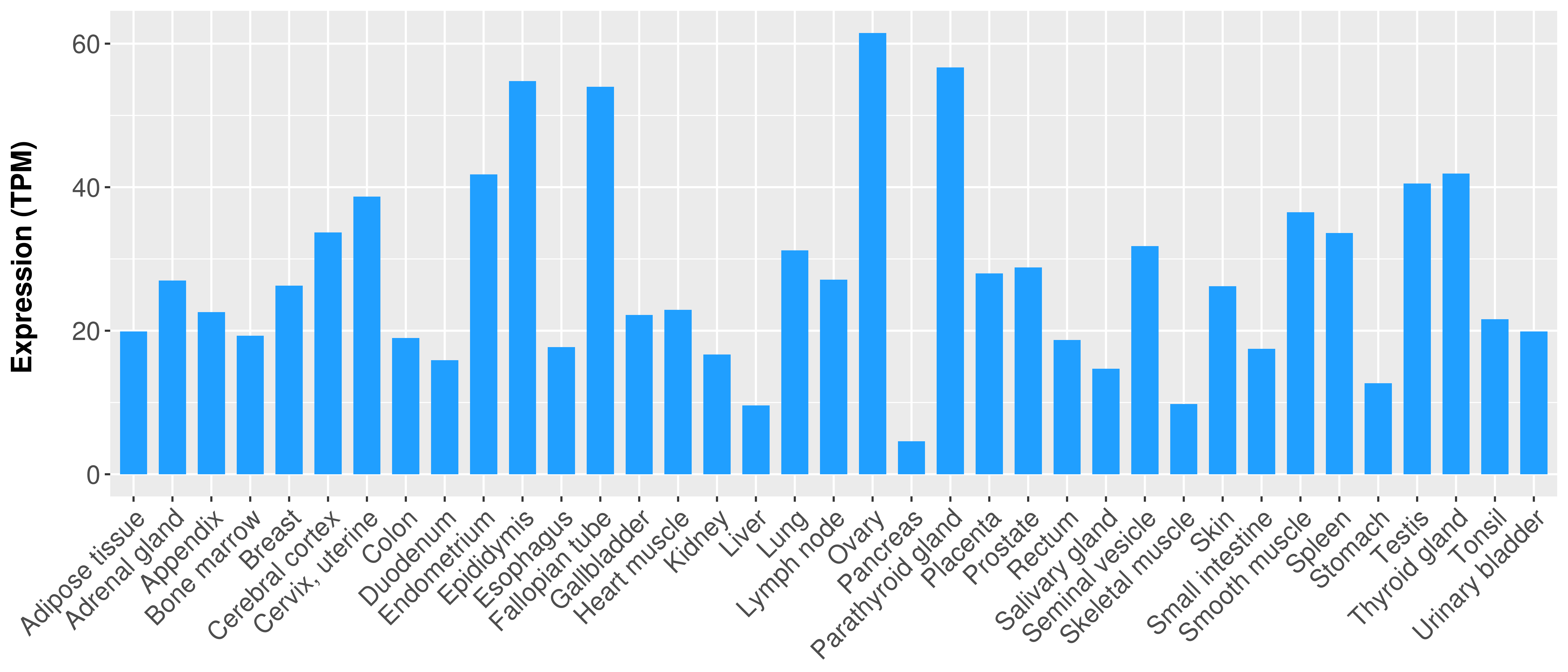
|
|
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
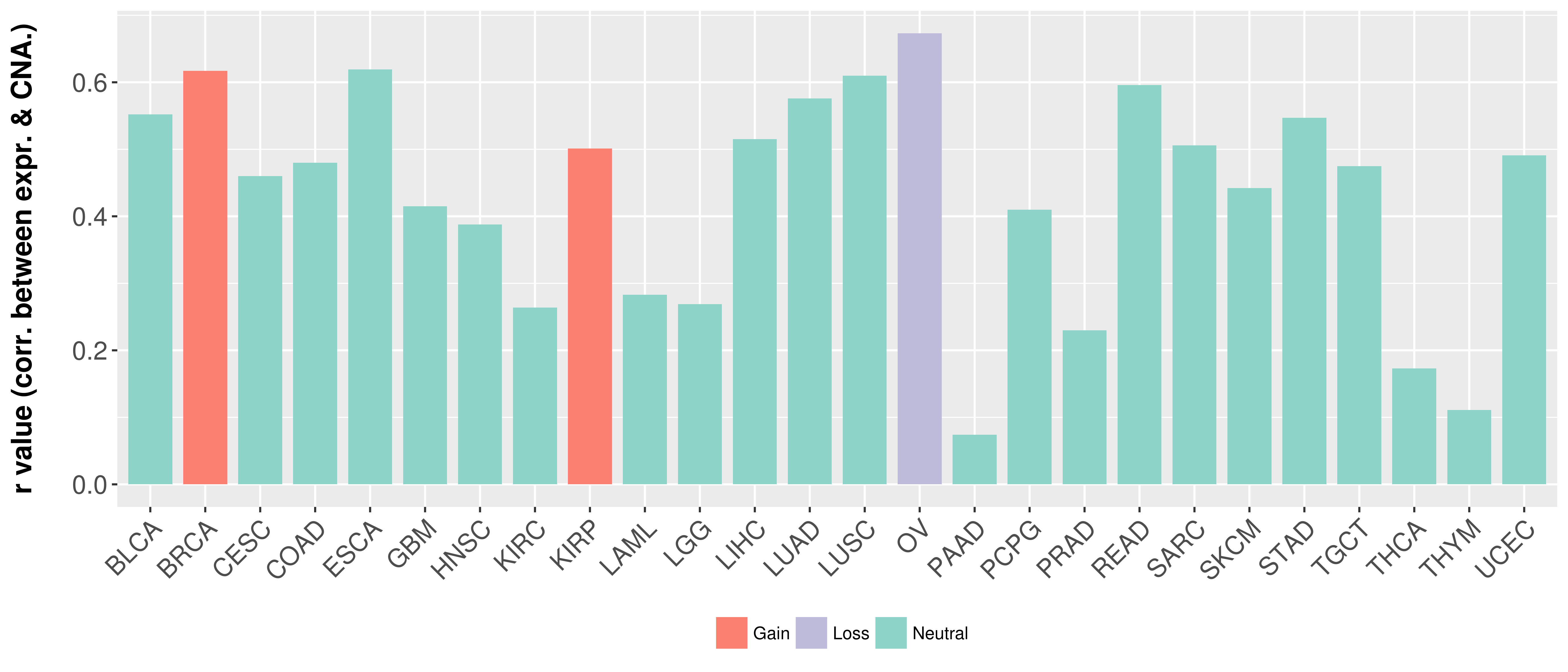
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
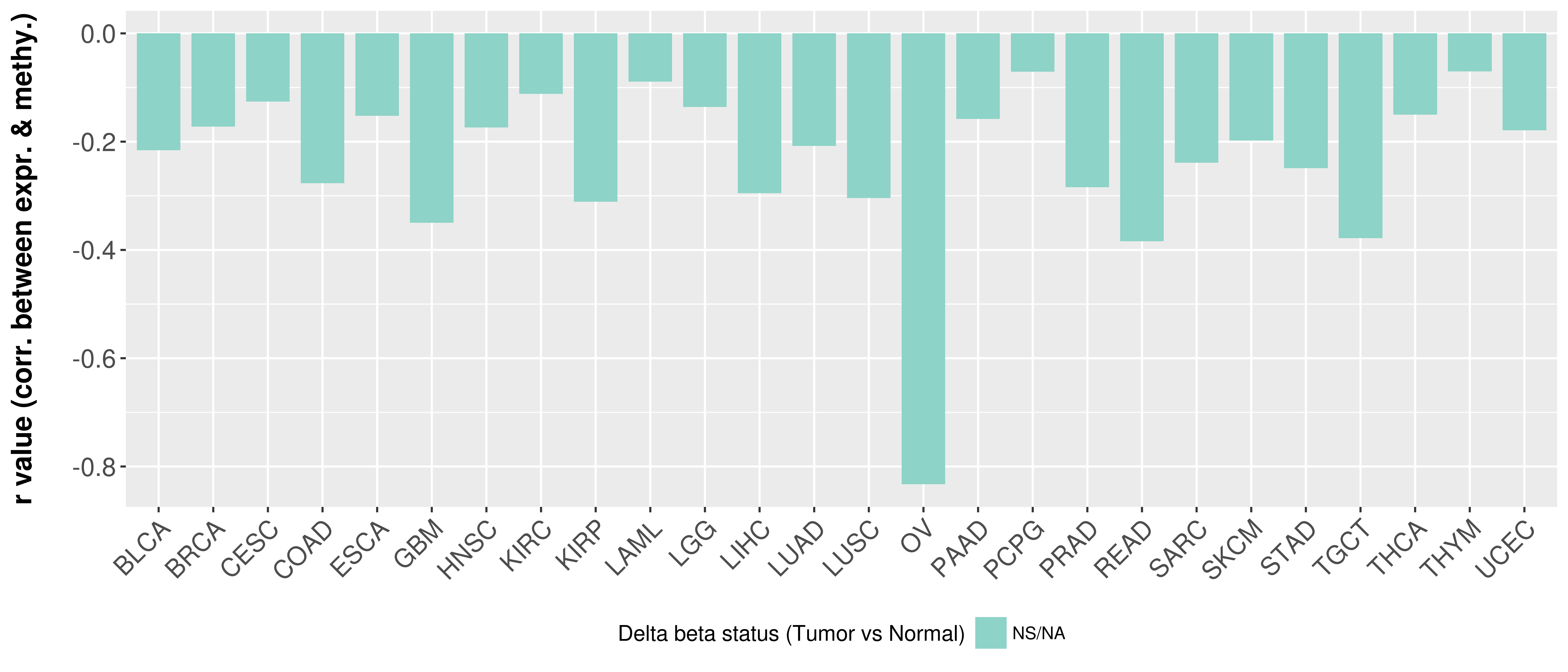
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
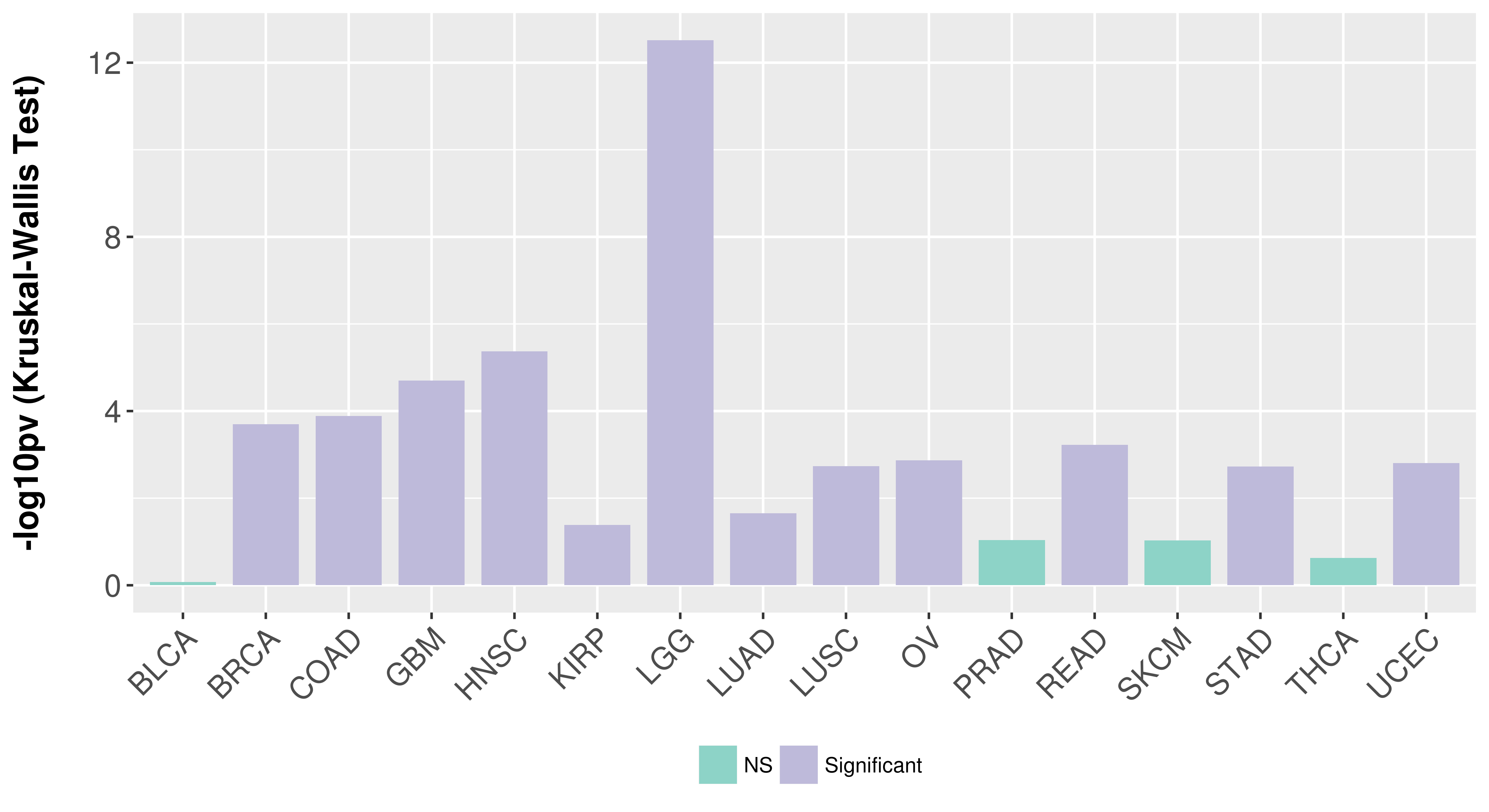
Association between expresson and subtype.
|
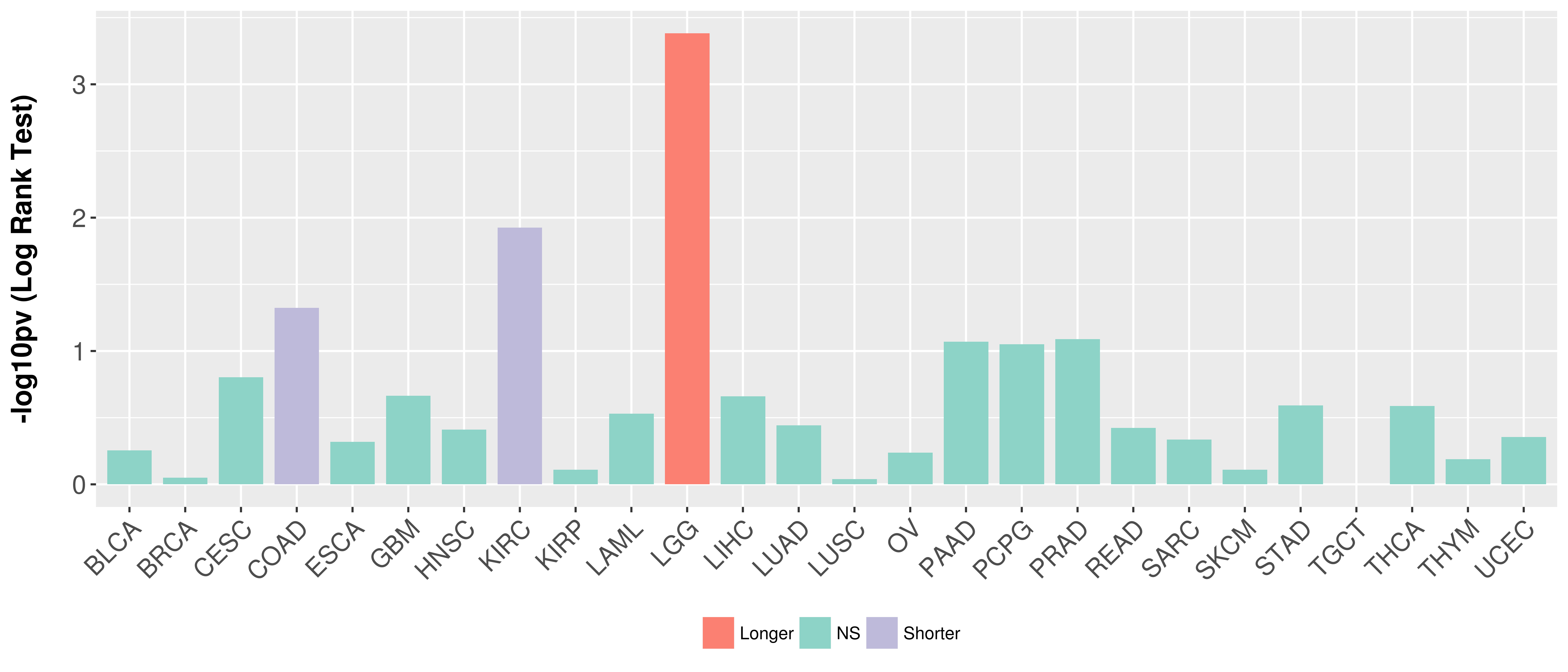
Overall survival analysis based on expression.
|
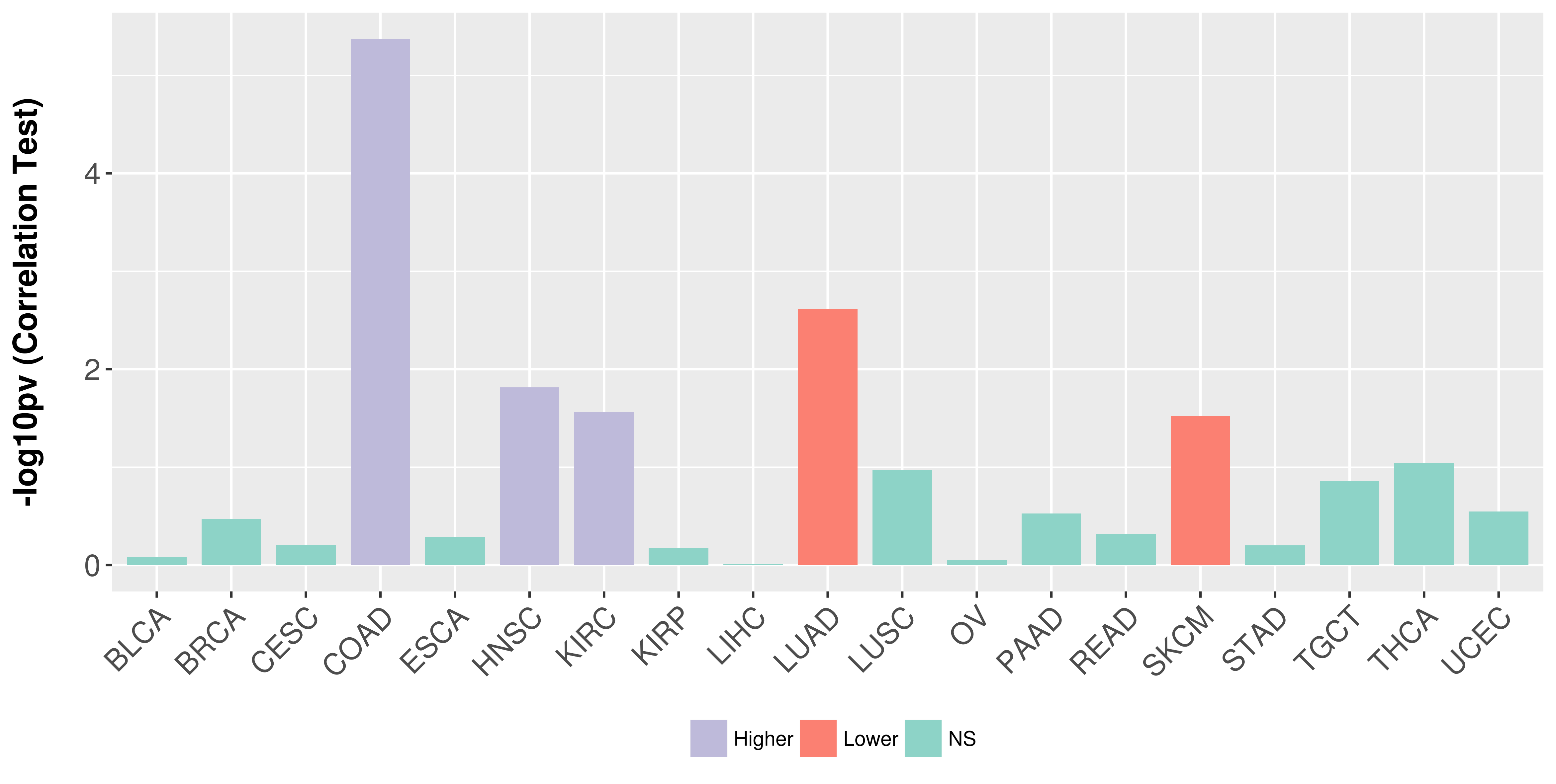
Association between expresson and stage.
|
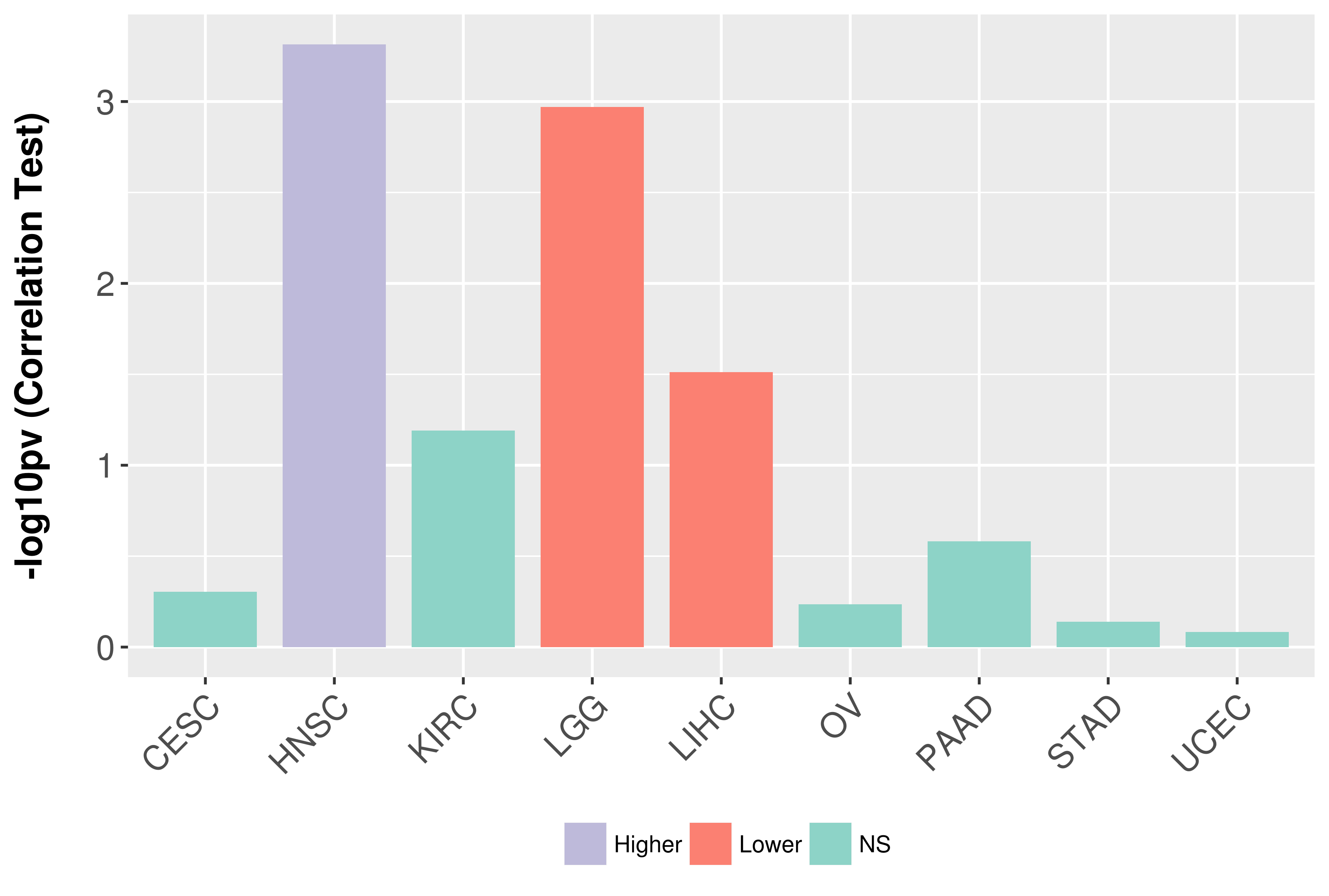
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for KAT8. |
| Summary | |
|---|---|
| Symbol | KAT8 |
| Name | lysine acetyltransferase 8 |
| Aliases | MOF; FLJ14040; hMOF; ZC2HC8; MYST1; MYST histone acetyltransferase 1; MOZ, YBF2/SAS3, SAS2 and TIP60 protein ...... |
| Location | 16p11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for KAT8. |
|