Browse L3MBTL1 in pancancer
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02820 mbt repeat PF00536 SAM domain (Sterile alpha motif) PF01530 Zinc finger |
||||||||||
| Function |
Polycomb group (PcG) protein that specifically recognizes and binds mono- and dimethyllysine residues on target proteins, therey acting as a 'reader' of a network of post-translational modifications. PcG proteins maintain the transcriptionally repressive state of genes: acts as a chromatin compaction factor by recognizing and binding mono- and dimethylated histone H1b/HIST1H1E at 'Lys-26' (H1bK26me1 and H1bK26me2) and histone H4 at 'Lys-20' (H4K20me1 and H4K20me2), leading to condense chromatin and repress transcription. Recognizes and binds p53/TP53 monomethylated at 'Lys-382', leading to repress p53/TP53-target genes. Also recognizes and binds RB1/RB monomethylated at 'Lys-860'. Participates in the ETV6-mediated repression. Probably plays a role in cell proliferation. Overexpression induces multinucleated cells, suggesting that it is required to accomplish normal mitosis. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0007067 mitotic nuclear division GO:0007088 regulation of mitotic nuclear division GO:0007346 regulation of mitotic cell cycle GO:0030099 myeloid cell differentiation GO:0030219 megakaryocyte differentiation GO:0045637 regulation of myeloid cell differentiation GO:0045652 regulation of megakaryocyte differentiation GO:0051783 regulation of nuclear division GO:0072331 signal transduction by p53 class mediator GO:1901796 regulation of signal transduction by p53 class mediator GO:1903706 regulation of hemopoiesis |
| Molecular Function |
GO:0003682 chromatin binding GO:0031491 nucleosome binding GO:0031493 nucleosomal histone binding GO:0032093 SAM domain binding GO:0035064 methylated histone binding GO:0042393 histone binding |
| Cellular Component |
GO:0000785 chromatin GO:0000793 condensed chromosome |
| KEGG | - |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804760: Regulation of TP53 Activity through Methylation R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
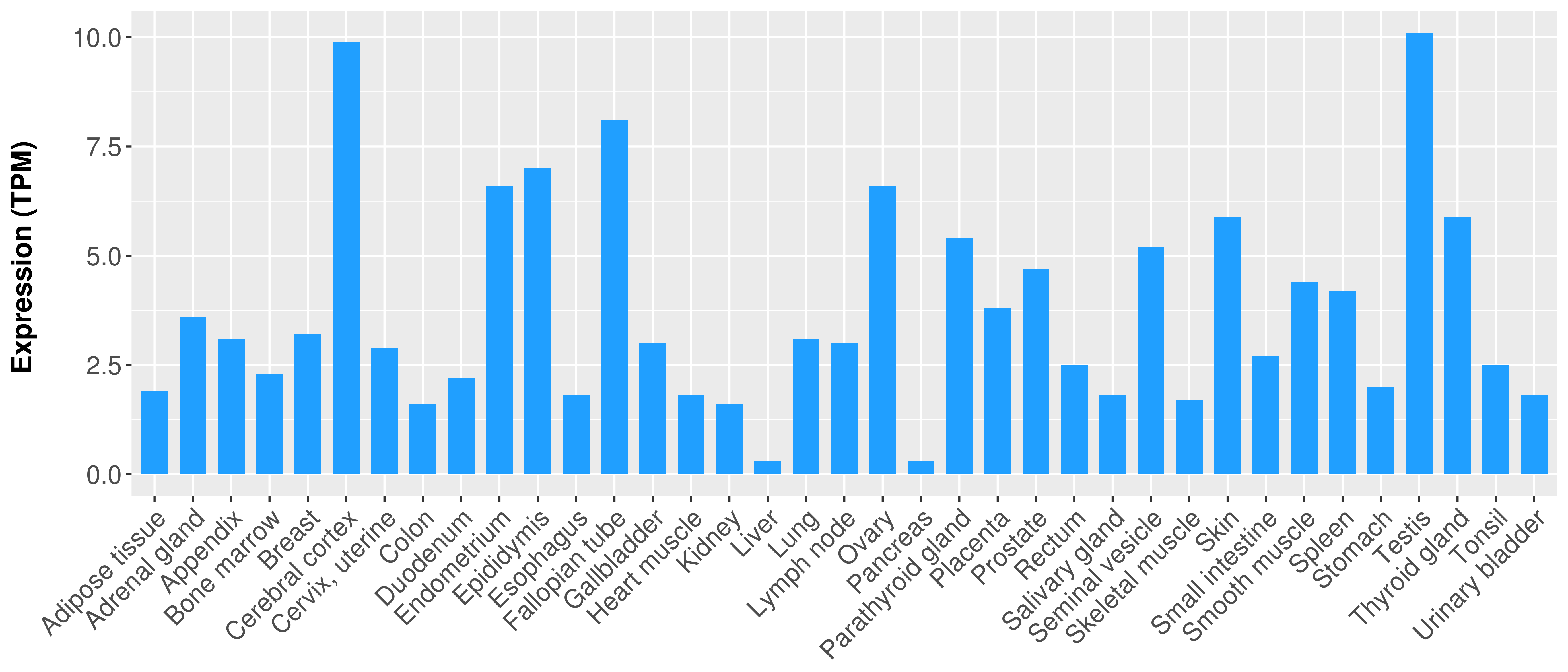
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
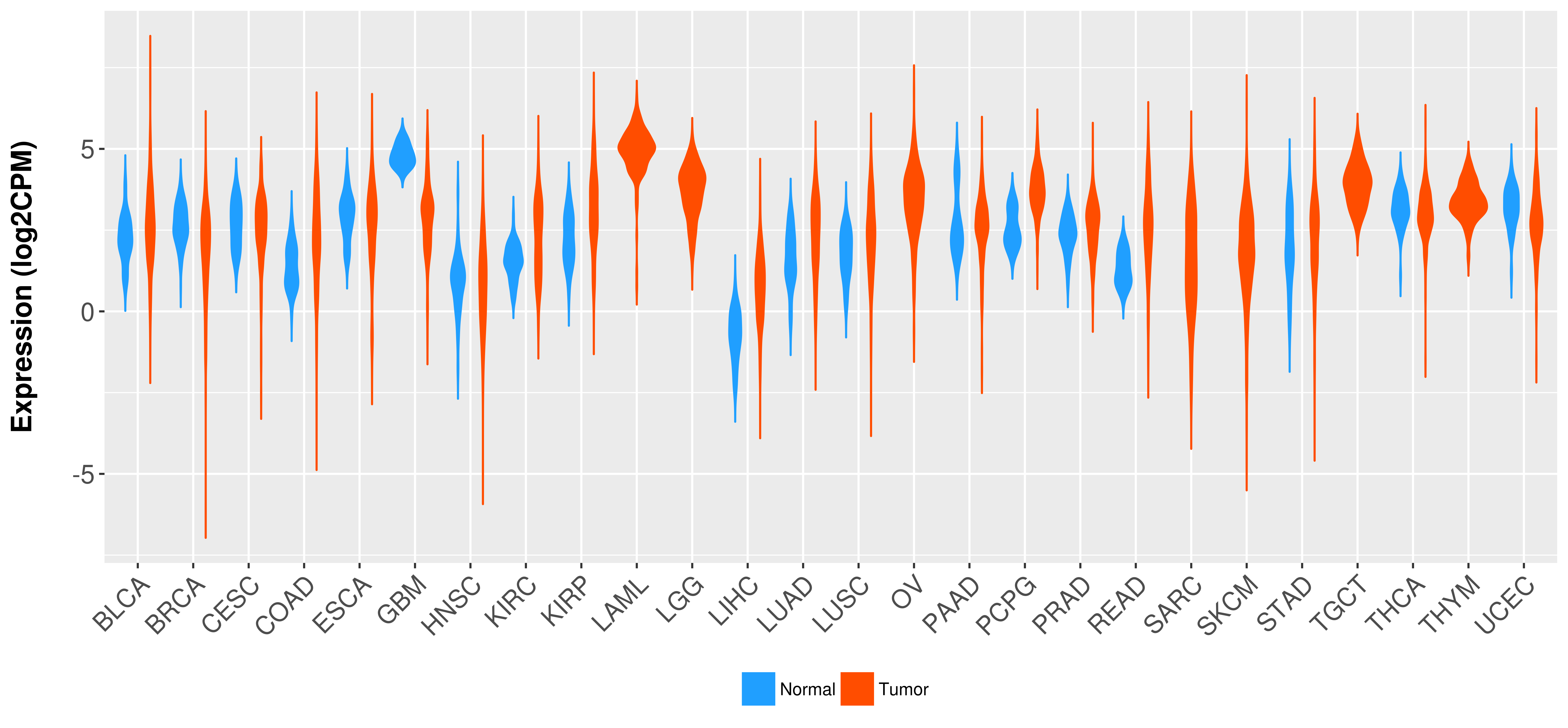
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
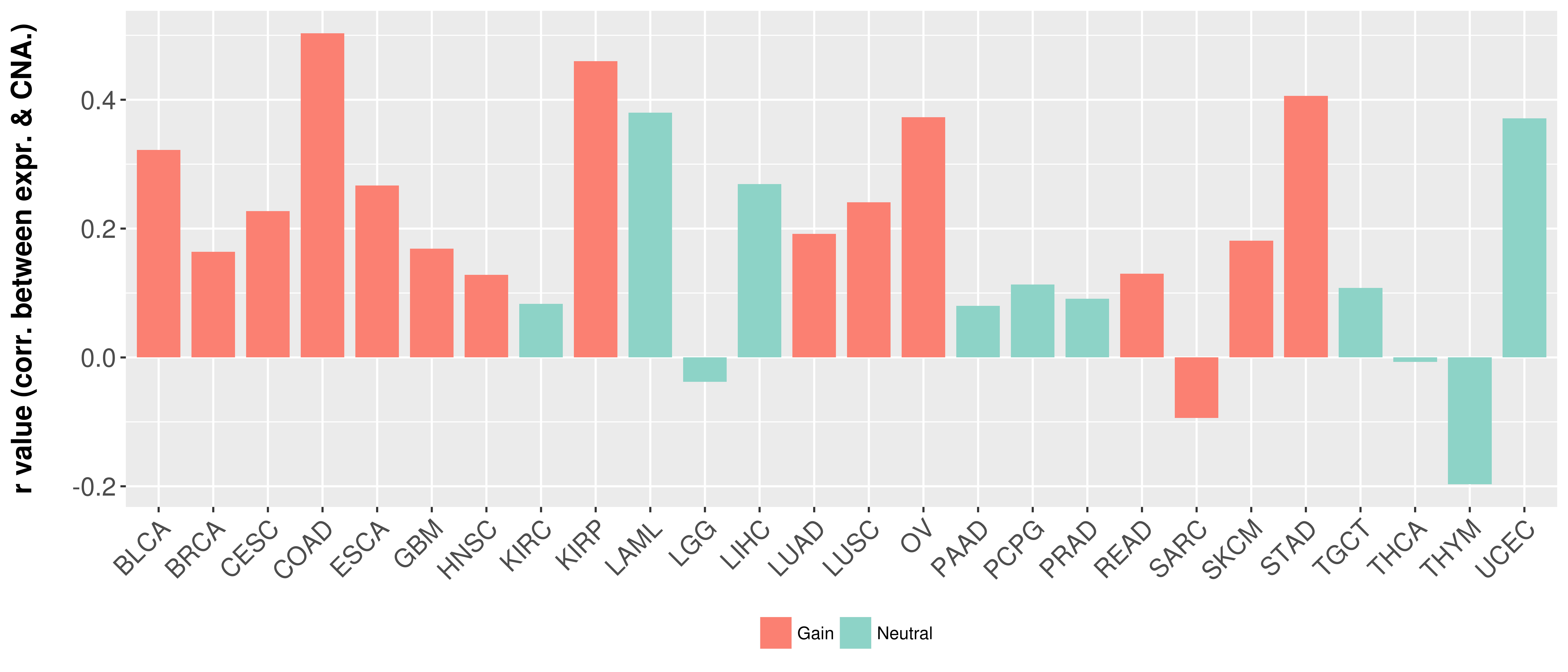
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
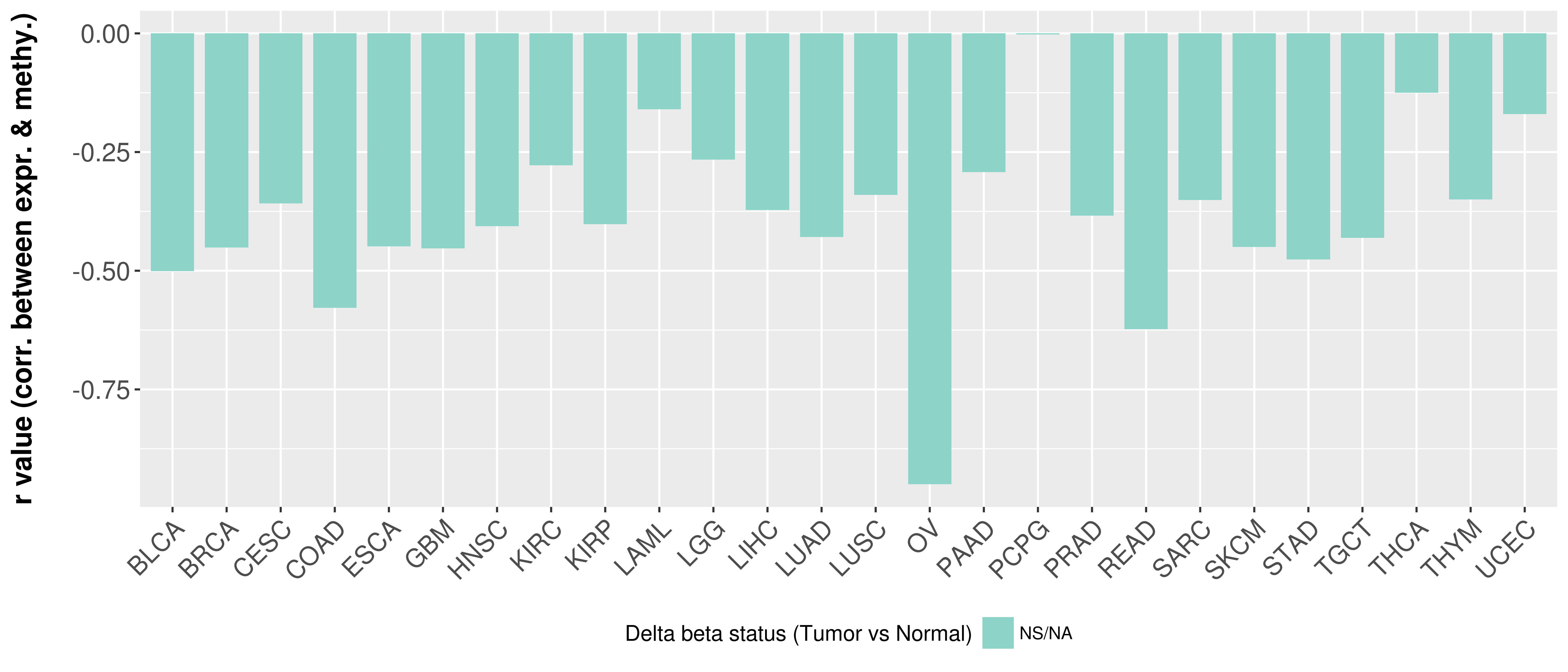
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
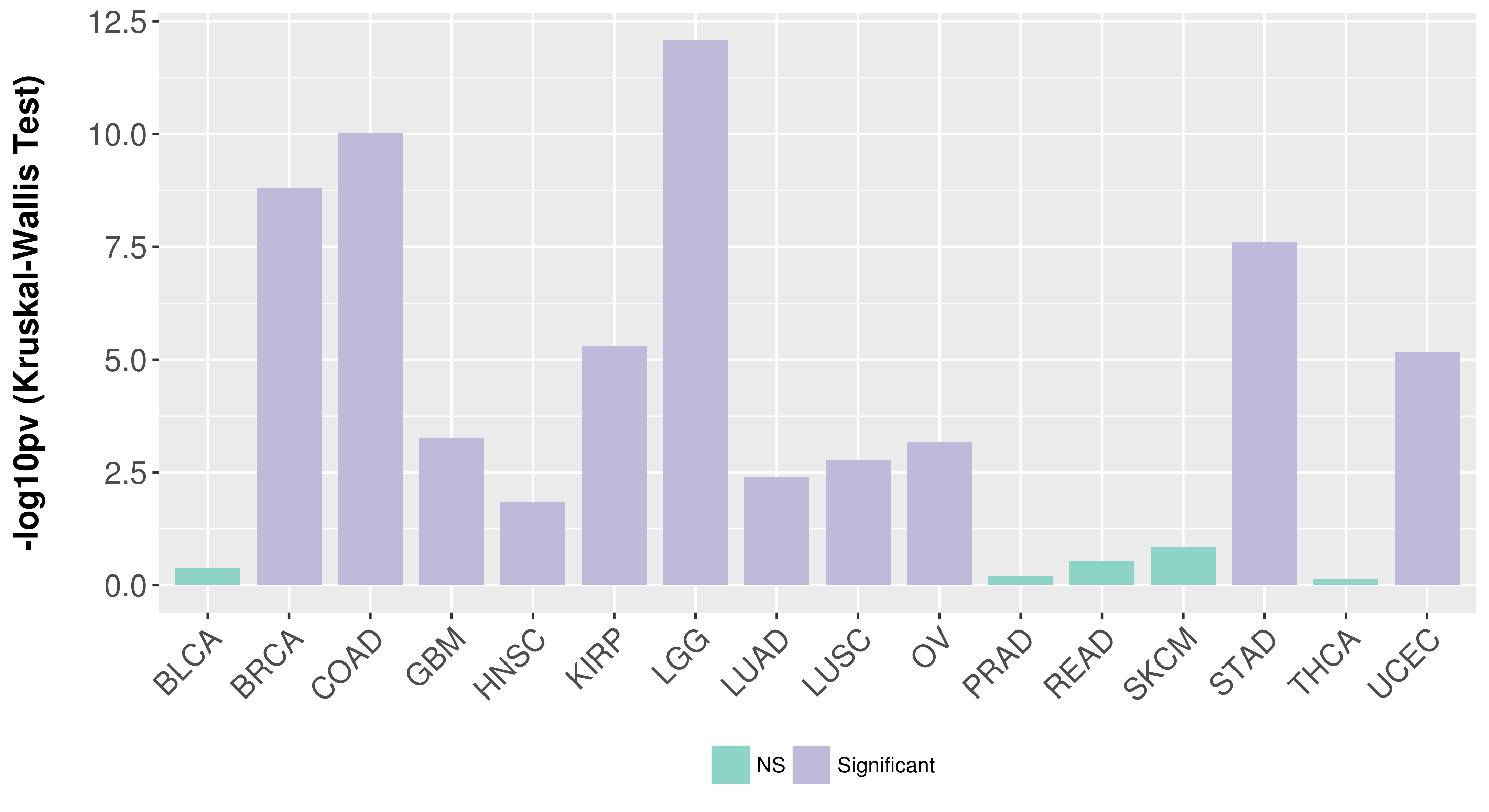
Association between expresson and subtype.
|
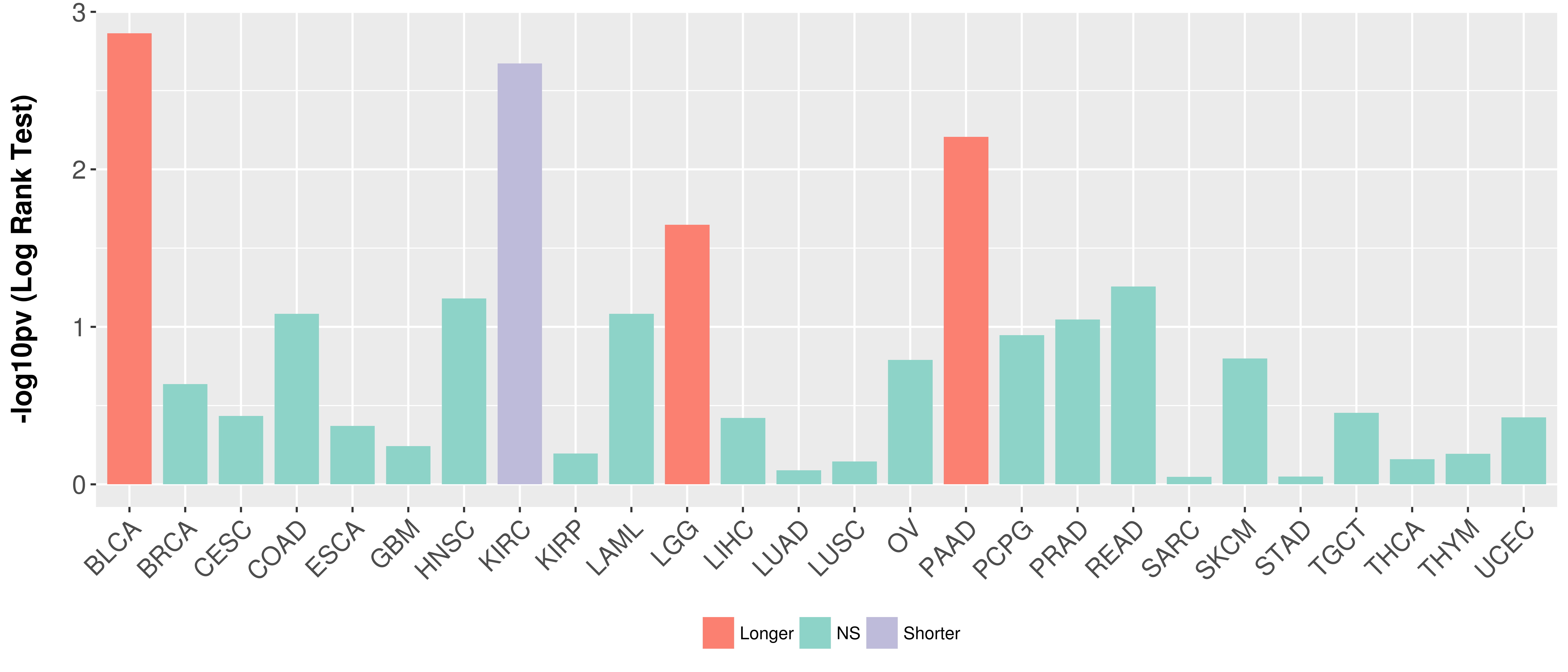
Overall survival analysis based on expression.
|
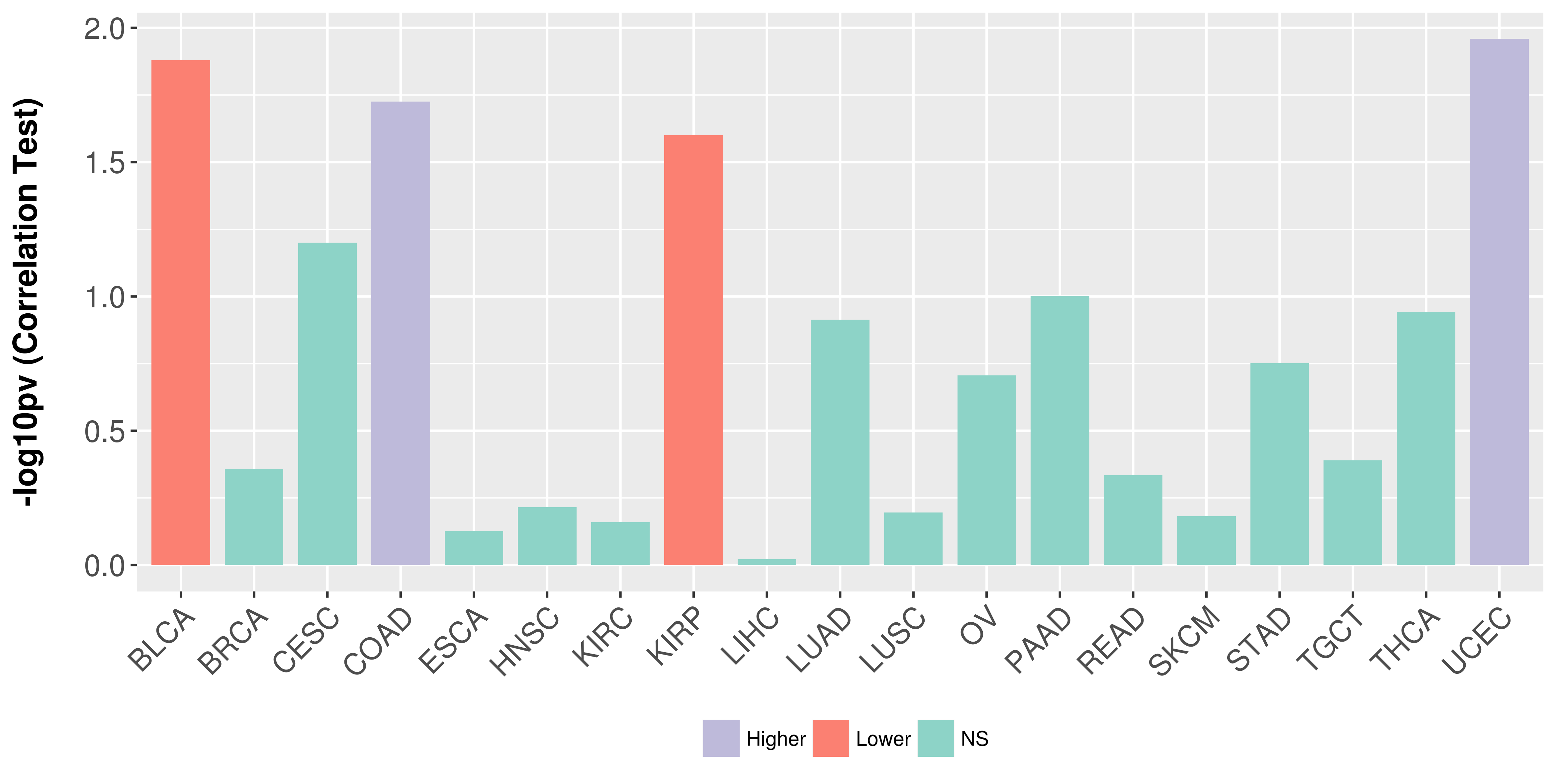
Association between expresson and stage.
|
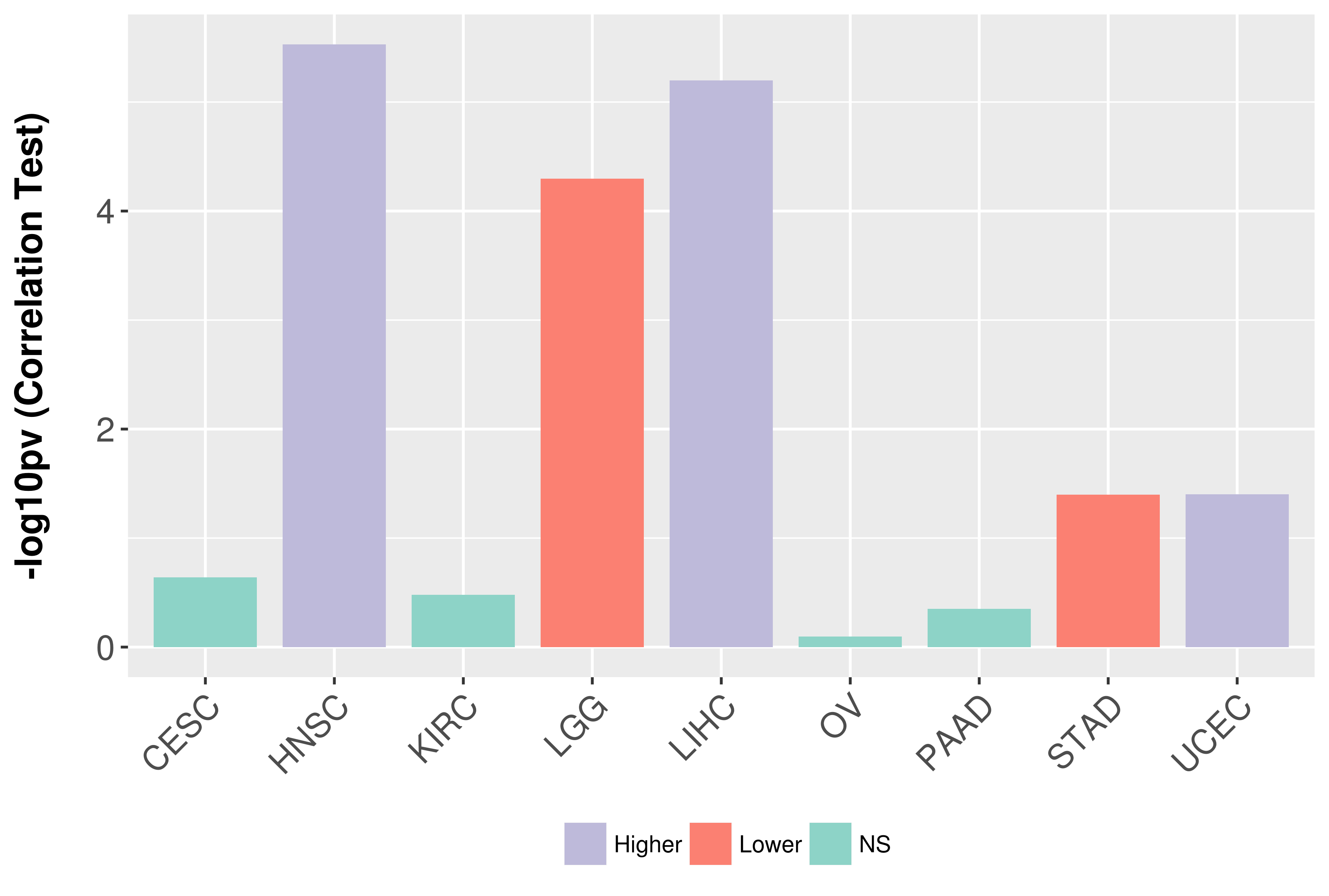
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
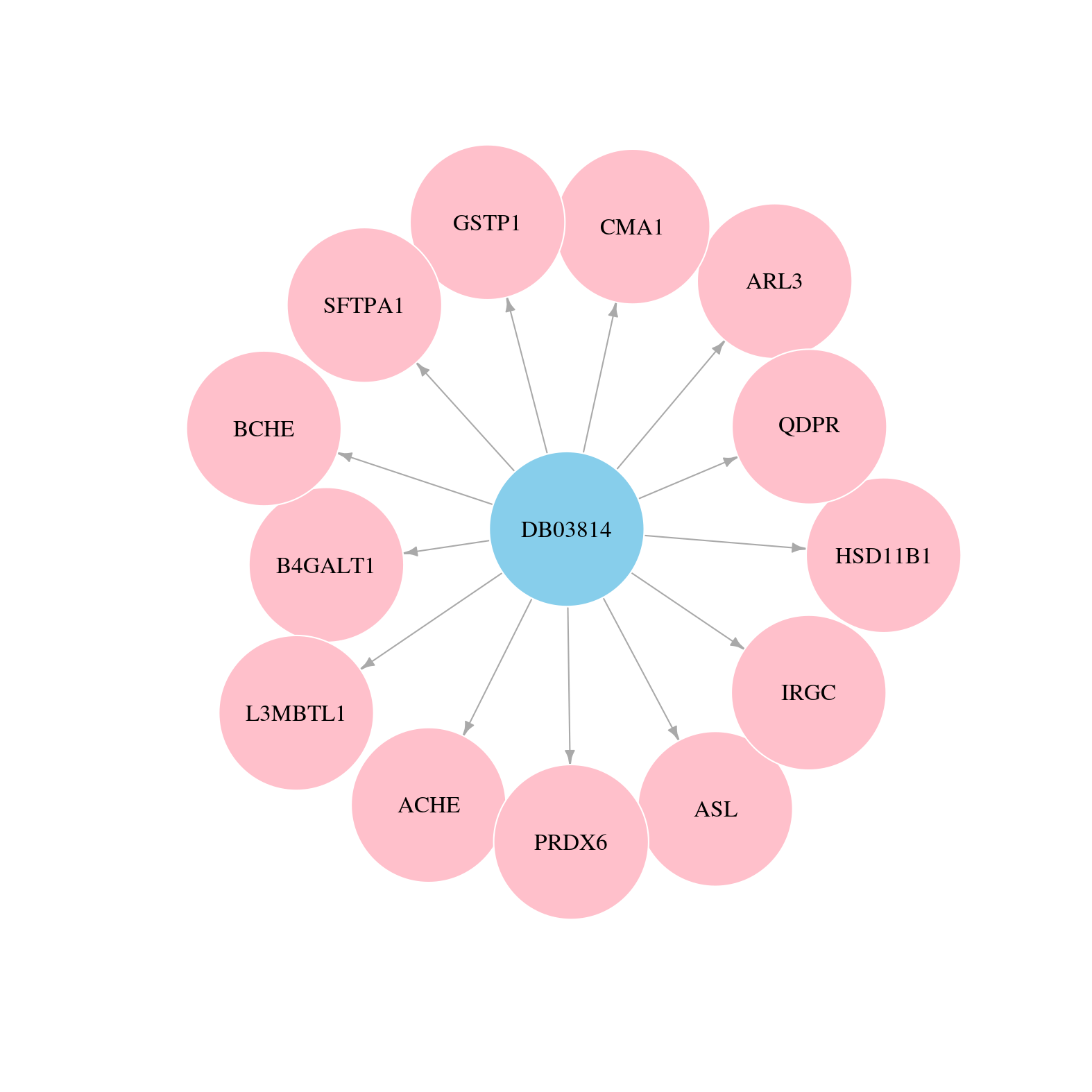
> Drugs from DrugBank database |
|
| Summary | |
|---|---|
| Symbol | L3MBTL1 |
| Name | l(3)mbt-like 1 (Drosophila) |
| Aliases | ZC2HC3; dJ138B7.3; DKFZp586P1522; KIAA0681; lethal (3) malignant brain tumor l(3); L3MBTL; l(3)mbt (Drosophi ...... |
| Location | 20q13.12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for L3MBTL1. |