Browse LMNA in pancancer
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00038 Intermediate filament protein PF00932 Lamin Tail Domain |
||||||||||
| Function |
Lamins are components of the nuclear lamina, a fibrous layer on the nucleoplasmic side of the inner nuclear membrane, which is thought to provide a framework for the nuclear envelope and may also interact with chromatin. Lamin A and C are present in equal amounts in the lamina of mammals. Plays an important role in nuclear assembly, chromatin organization, nuclear membrane and telomere dynamics. Required for normal development of peripheral nervous system and skeletal muscle and for muscle satellite cell proliferation. Required for osteoblastogenesis and bone formation. Also prevents fat infiltration of muscle and bone marrow, helping to maintain the volume and strength of skeletal muscle and bone.; FUNCTION: Prelamin-A/C can accelerate smooth muscle cell senescence. It acts to disrupt mitosis and induce DNA damage in vascular smooth muscle cells (VSMCs), leading to mitotic failure, genomic instability, and premature senescence. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000226 microtubule cytoskeleton organization GO:0001666 response to hypoxia GO:0001836 release of cytochrome c from mitochondria GO:0006606 protein import into nucleus GO:0006913 nucleocytoplasmic transport GO:0006984 ER-nucleus signaling pathway GO:0006986 response to unfolded protein GO:0006991 response to sterol depletion GO:0006997 nucleus organization GO:0006998 nuclear envelope organization GO:0007077 mitotic nuclear envelope disassembly GO:0007084 mitotic nuclear envelope reassembly GO:0007163 establishment or maintenance of cell polarity GO:0007507 heart development GO:0007568 aging GO:0007569 cell aging GO:0008637 apoptotic mitochondrial changes GO:0010463 mesenchymal cell proliferation GO:0010464 regulation of mesenchymal cell proliferation GO:0010639 negative regulation of organelle organization GO:0010821 regulation of mitochondrion organization GO:0010823 negative regulation of mitochondrion organization GO:0014706 striated muscle tissue development GO:0017038 protein import GO:0030397 membrane disassembly GO:0030951 establishment or maintenance of microtubule cytoskeleton polarity GO:0030952 establishment or maintenance of cytoskeleton polarity GO:0030968 endoplasmic reticulum unfolded protein response GO:0031468 nuclear envelope reassembly GO:0032933 SREBP signaling pathway GO:0034504 protein localization to nucleus GO:0034620 cellular response to unfolded protein GO:0034976 response to endoplasmic reticulum stress GO:0035051 cardiocyte differentiation GO:0035105 sterol regulatory element binding protein import into nucleus GO:0035966 response to topologically incorrect protein GO:0035967 cellular response to topologically incorrect protein GO:0036293 response to decreased oxygen levels GO:0036294 cellular response to decreased oxygen levels GO:0036498 IRE1-mediated unfolded protein response GO:0042692 muscle cell differentiation GO:0044744 protein targeting to nucleus GO:0048738 cardiac muscle tissue development GO:0051081 nuclear envelope disassembly GO:0051146 striated muscle cell differentiation GO:0051169 nuclear transport GO:0051170 nuclear import GO:0055001 muscle cell development GO:0055002 striated muscle cell development GO:0055006 cardiac cell development GO:0055007 cardiac muscle cell differentiation GO:0055012 ventricular cardiac muscle cell differentiation GO:0055013 cardiac muscle cell development GO:0055015 ventricular cardiac muscle cell development GO:0060537 muscle tissue development GO:0070482 response to oxygen levels GO:0071453 cellular response to oxygen levels GO:0071456 cellular response to hypoxia GO:0071501 cellular response to sterol depletion GO:0072201 negative regulation of mesenchymal cell proliferation GO:0090199 regulation of release of cytochrome c from mitochondria GO:0090201 negative regulation of release of cytochrome c from mitochondria GO:0090342 regulation of cell aging GO:0090343 positive regulation of cell aging GO:0097191 extrinsic apoptotic signaling pathway GO:1900180 regulation of protein localization to nucleus GO:1902593 single-organism nuclear import GO:2001233 regulation of apoptotic signaling pathway GO:2001234 negative regulation of apoptotic signaling pathway GO:2001236 regulation of extrinsic apoptotic signaling pathway GO:2001237 negative regulation of extrinsic apoptotic signaling pathway |
| Molecular Function | - |
| Cellular Component |
GO:0005635 nuclear envelope GO:0005638 lamin filament GO:0005652 nuclear lamina GO:0005882 intermediate filament GO:0016604 nuclear body GO:0016607 nuclear speck GO:0031965 nuclear membrane GO:0034399 nuclear periphery GO:0045111 intermediate filament cytoskeleton |
| KEGG |
hsa04210 Apoptosis |
| Reactome |
R-HSA-109581: Apoptosis R-HSA-111465: Apoptotic cleavage of cellular proteins R-HSA-75153: Apoptotic execution phase R-HSA-352238: Breakdown of the nuclear lamina R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-4419969: Depolymerisation of the Nuclear Lamina R-HSA-8862803: Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-381070: IRE1alpha activates chaperones R-HSA-2995383: Initiation of Nuclear Envelope Reformation R-HSA-68886: M Phase R-HSA-1500620: Meiosis R-HSA-1221632: Meiotic synapsis R-HSA-392499: Metabolism of proteins R-HSA-68882: Mitotic Anaphase R-HSA-2555396: Mitotic Metaphase and Anaphase R-HSA-68875: Mitotic Prophase R-HSA-8863678: Neurodegenerative Diseases R-HSA-2980766: Nuclear Envelope Breakdown R-HSA-2995410: Nuclear Envelope Reassembly R-HSA-6802957: Oncogenic MAPK signaling R-HSA-5357801: Programmed Cell Death R-HSA-6802952: Signaling by BRAF and RAF fusions R-HSA-381119: Unfolded Protein Response (UPR) R-HSA-381038: XBP1(S) activates chaperone genes |
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|

|
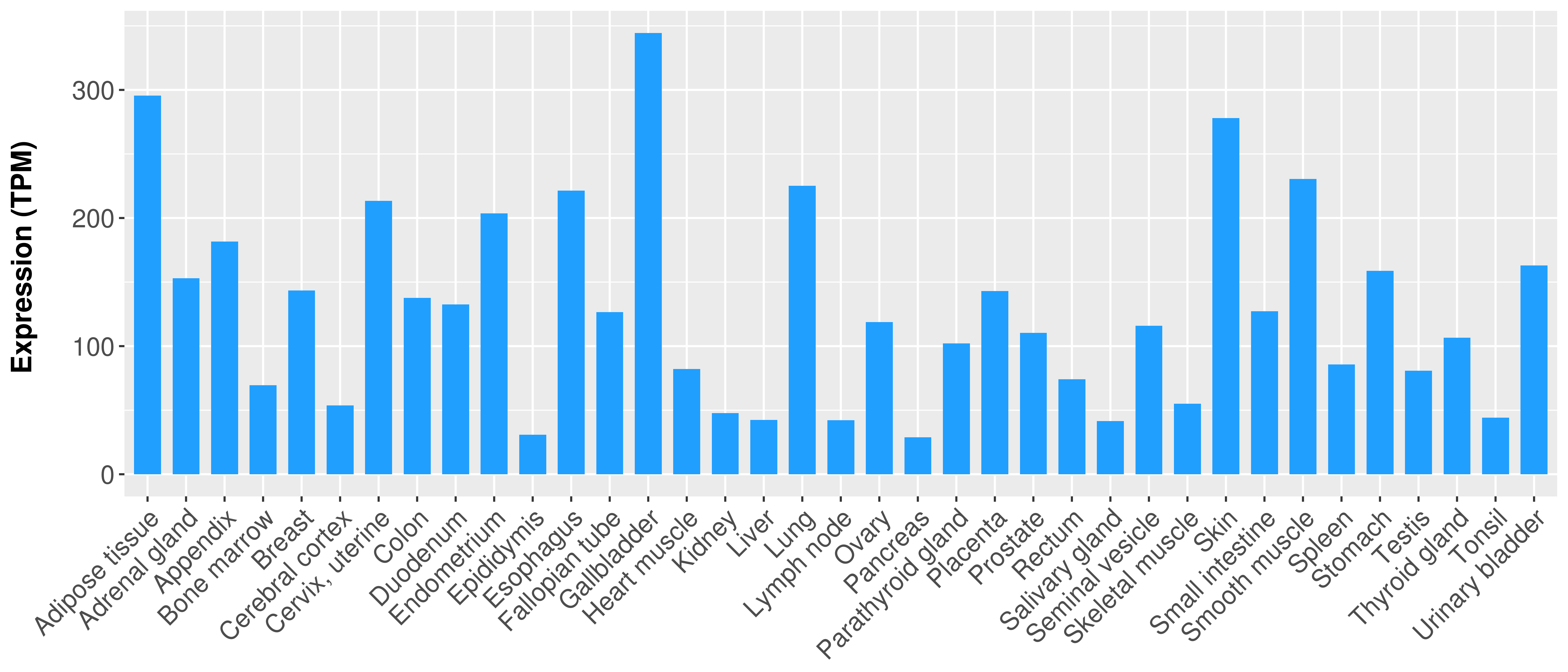
|
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
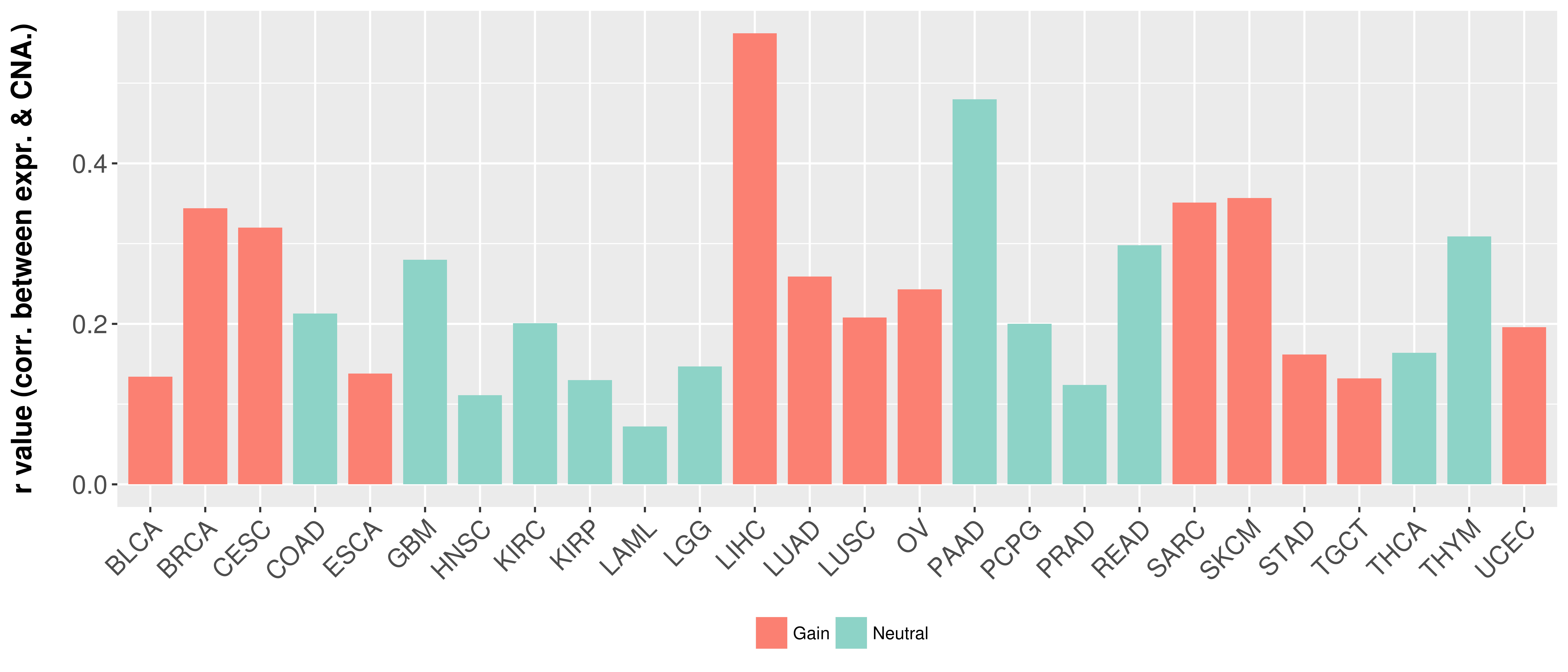
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |

Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
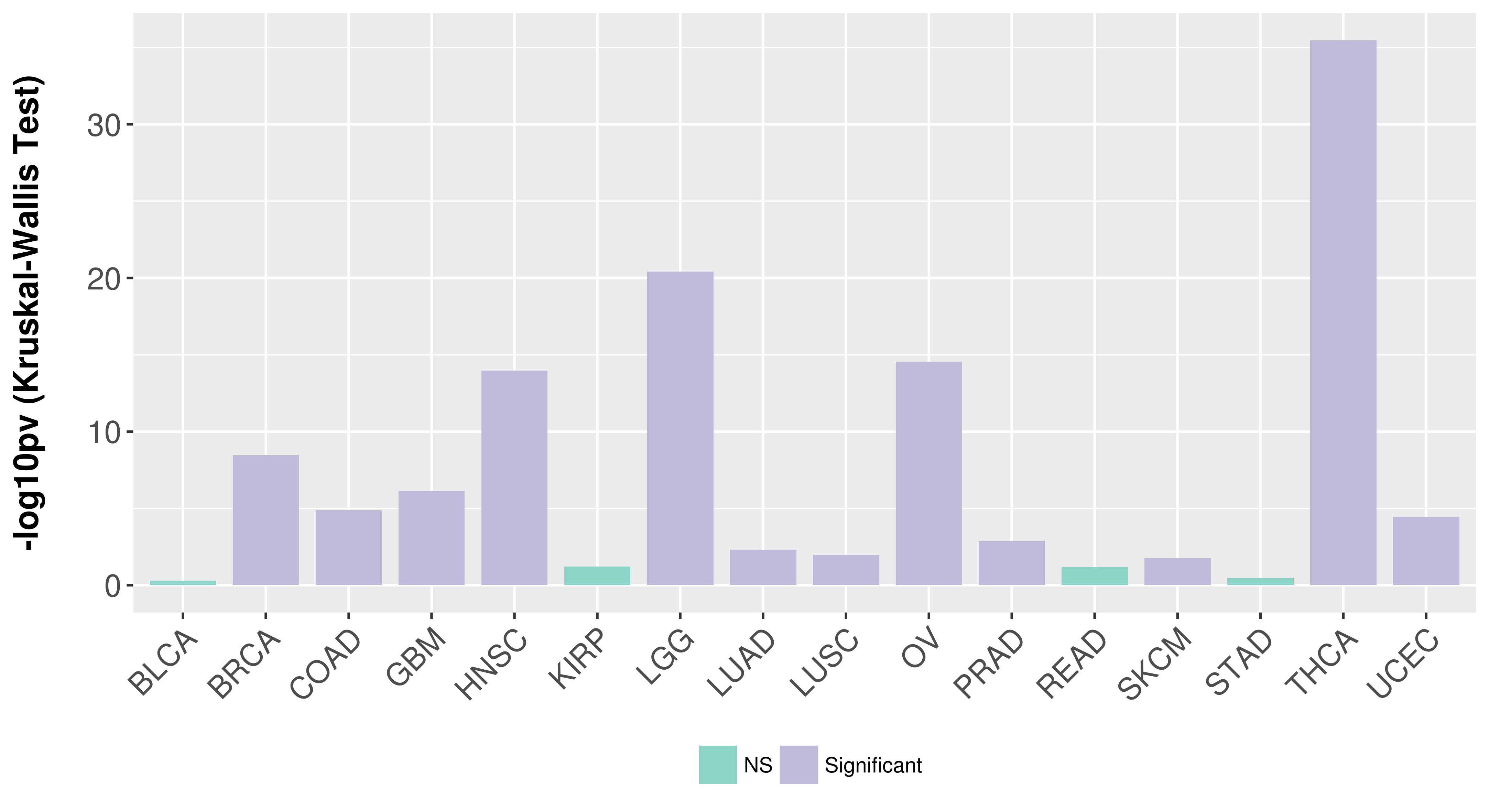
Association between expresson and subtype.
|
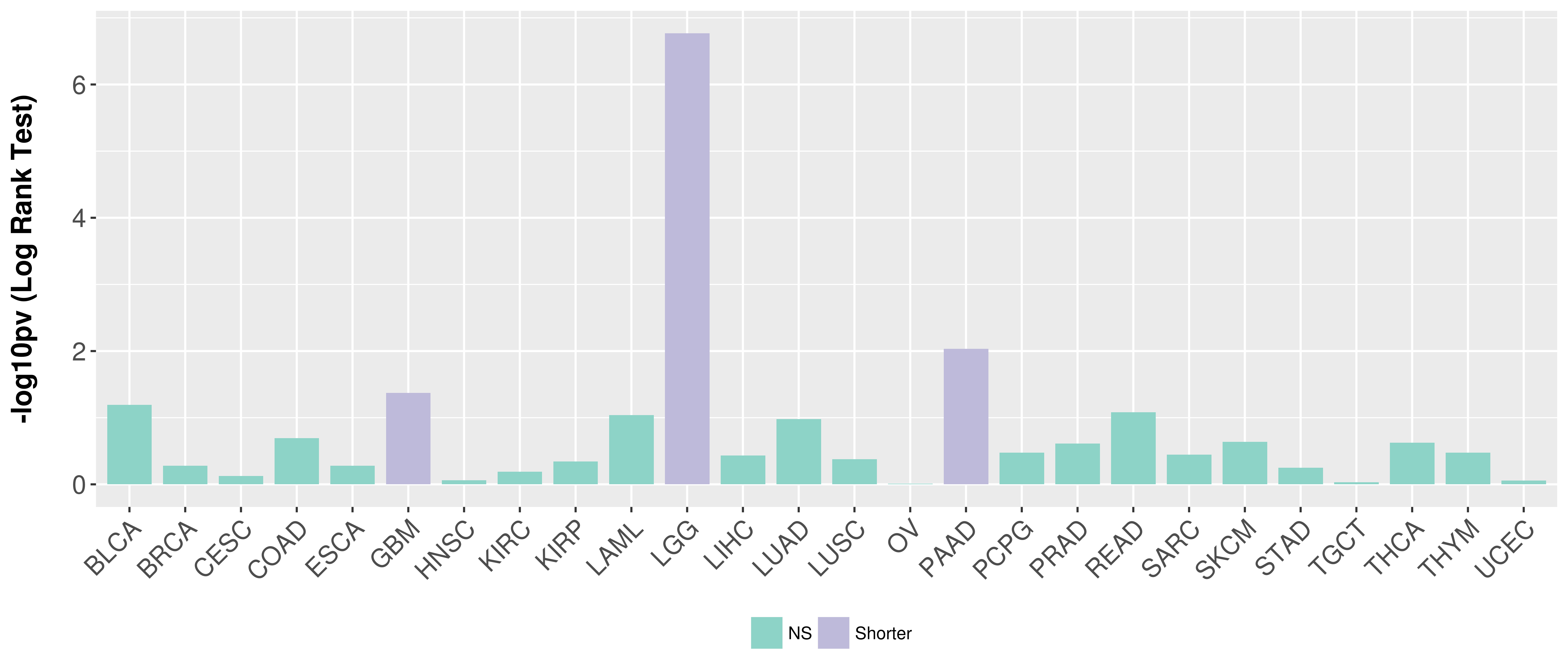
Overall survival analysis based on expression.
|
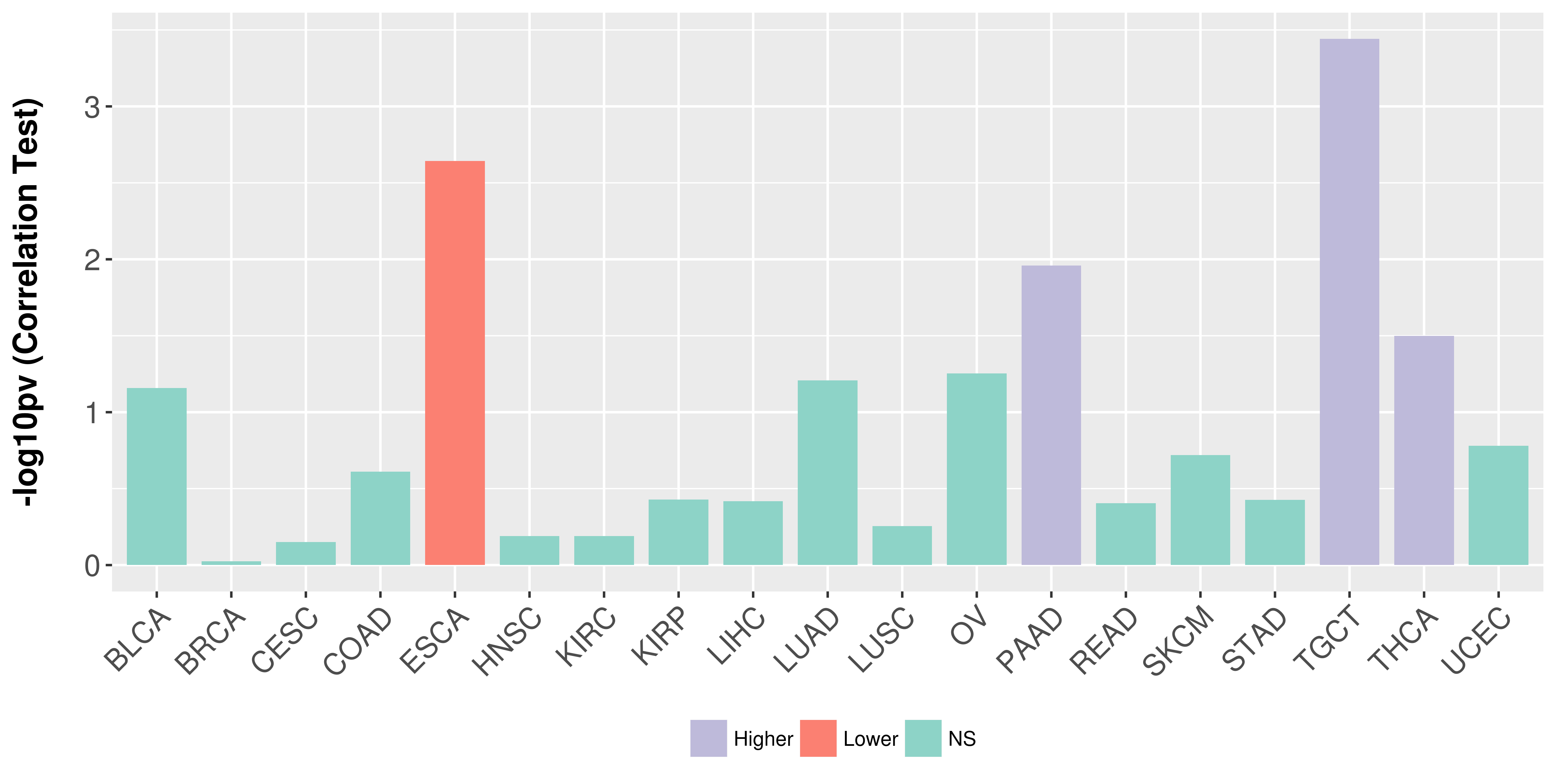
Association between expresson and stage.
|
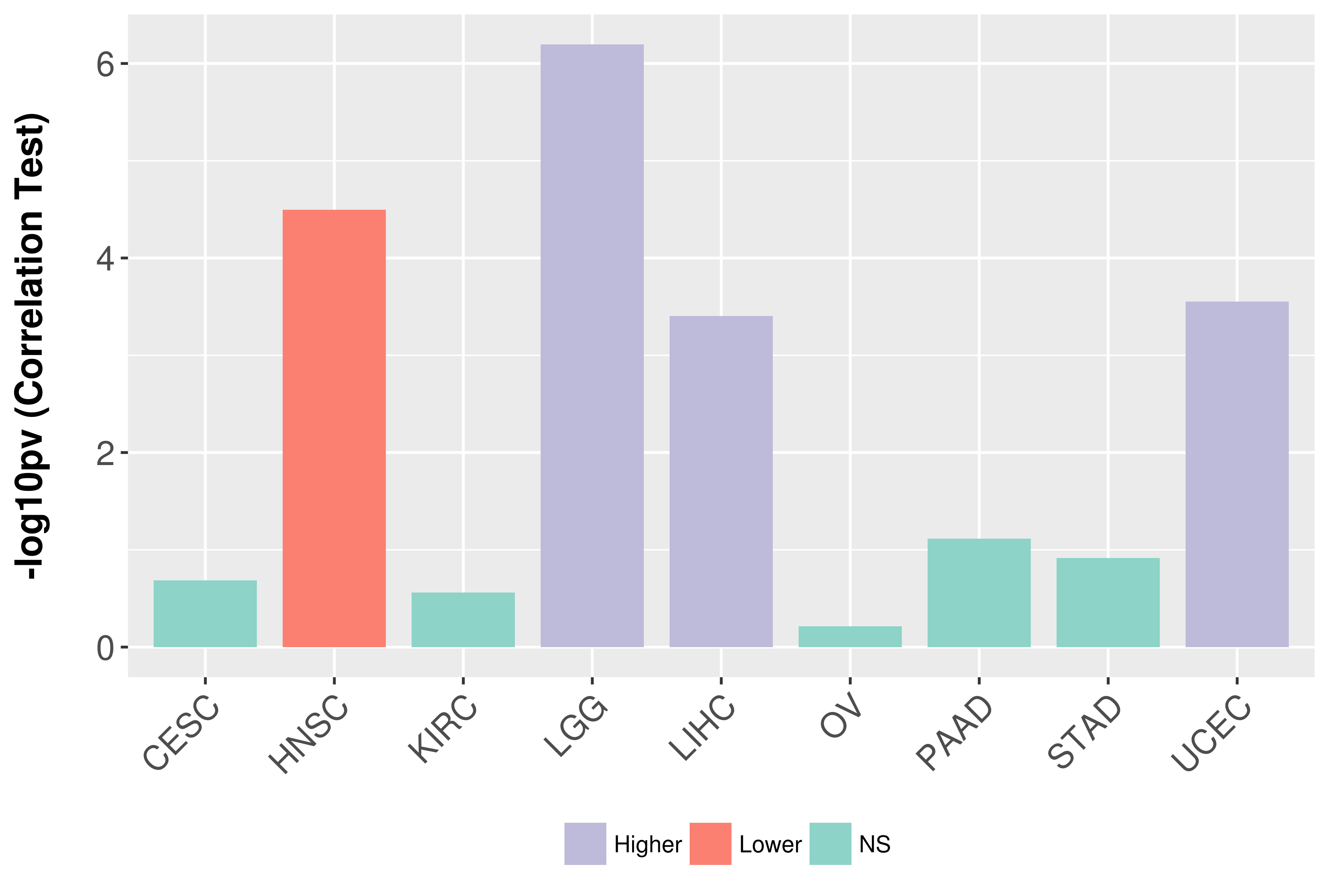
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for LMNA. |
| Summary | |
|---|---|
| Symbol | LMNA |
| Name | lamin A/C |
| Aliases | LMN1; CMD1A; LGMD1B; LMNL1; cardiomyopathy, dilated 1A (autosomal dominant); limb girdle muscular dystrophy ...... |
| Location | 1q22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|