Browse MECP2 in pancancer
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01429 Methyl-CpG binding domain |
||||||||||
| Function |
Chromosomal protein that binds to methylated DNA. It can bind specifically to a single methyl-CpG pair. It is not influenced by sequences flanking the methyl-CpGs. Mediates transcriptional repression through interaction with histone deacetylase and the corepressor SIN3A. Binds both 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC)-containing DNA, with a preference for 5-methylcytosine (5mC). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001662 behavioral fear response GO:0001666 response to hypoxia GO:0001964 startle response GO:0001976 neurological system process involved in regulation of systemic arterial blood pressure GO:0002087 regulation of respiratory gaseous exchange by neurological system process GO:0002209 behavioral defense response GO:0003013 circulatory system process GO:0003016 respiratory system process GO:0003073 regulation of systemic arterial blood pressure GO:0006020 inositol metabolic process GO:0006066 alcohol metabolic process GO:0006091 generation of precursor metabolites and energy GO:0006119 oxidative phosphorylation GO:0006122 mitochondrial electron transport, ubiquinol to cytochrome c GO:0006342 chromatin silencing GO:0006349 regulation of gene expression by genetic imprinting GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006479 protein methylation GO:0006520 cellular amino acid metabolic process GO:0006541 glutamine metabolic process GO:0006576 cellular biogenic amine metabolic process GO:0006644 phospholipid metabolic process GO:0006650 glycerophospholipid metabolic process GO:0007416 synapse assembly GO:0007585 respiratory gaseous exchange GO:0007611 learning or memory GO:0007612 learning GO:0007613 memory GO:0007616 long-term memory GO:0007626 locomotory behavior GO:0007632 visual behavior GO:0008015 blood circulation GO:0008202 steroid metabolic process GO:0008211 glucocorticoid metabolic process GO:0008213 protein alkylation GO:0008217 regulation of blood pressure GO:0008306 associative learning GO:0008344 adult locomotory behavior GO:0008542 visual learning GO:0009064 glutamine family amino acid metabolic process GO:0009116 nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009123 nucleoside monophosphate metabolic process GO:0009126 purine nucleoside monophosphate metabolic process GO:0009141 nucleoside triphosphate metabolic process GO:0009144 purine nucleoside triphosphate metabolic process GO:0009150 purine ribonucleotide metabolic process GO:0009161 ribonucleoside monophosphate metabolic process GO:0009167 purine ribonucleoside monophosphate metabolic process GO:0009199 ribonucleoside triphosphate metabolic process GO:0009205 purine ribonucleoside triphosphate metabolic process GO:0009308 amine metabolic process GO:0009314 response to radiation GO:0009405 pathogenesis GO:0009416 response to light stimulus GO:0009791 post-embryonic development GO:0015844 monoamine transport GO:0015850 organic hydroxy compound transport GO:0015980 energy derivation by oxidation of organic compounds GO:0016358 dendrite development GO:0016458 gene silencing GO:0016570 histone modification GO:0016571 histone methylation GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019230 proprioception GO:0019233 sensory perception of pain GO:0019751 polyol metabolic process GO:0021549 cerebellum development GO:0021591 ventricular system development GO:0021700 developmental maturation GO:0022037 metencephalon development GO:0022900 electron transport chain GO:0022904 respiratory electron transport chain GO:0030534 adult behavior GO:0030902 hindbrain development GO:0031056 regulation of histone modification GO:0031057 negative regulation of histone modification GO:0031060 regulation of histone methylation GO:0031061 negative regulation of histone methylation GO:0032048 cardiolipin metabolic process GO:0032259 methylation GO:0033555 multicellular organismal response to stress GO:0035065 regulation of histone acetylation GO:0035067 negative regulation of histone acetylation GO:0035176 social behavior GO:0036293 response to decreased oxygen levels GO:0040029 regulation of gene expression, epigenetic GO:0042278 purine nucleoside metabolic process GO:0042391 regulation of membrane potential GO:0042551 neuron maturation GO:0042596 fear response GO:0042773 ATP synthesis coupled electron transport GO:0042775 mitochondrial ATP synthesis coupled electron transport GO:0043414 macromolecule methylation GO:0043523 regulation of neuron apoptotic process GO:0043524 negative regulation of neuron apoptotic process GO:0043543 protein acylation GO:0043576 regulation of respiratory gaseous exchange GO:0044057 regulation of system process GO:0044065 regulation of respiratory system process GO:0044089 positive regulation of cellular component biogenesis GO:0044106 cellular amine metabolic process GO:0044262 cellular carbohydrate metabolic process GO:0044708 single-organism behavior GO:0044723 single-organism carbohydrate metabolic process GO:0045333 cellular respiration GO:0045814 negative regulation of gene expression, epigenetic GO:0046034 ATP metabolic process GO:0046128 purine ribonucleoside metabolic process GO:0046470 phosphatidylcholine metabolic process GO:0046471 phosphatidylglycerol metabolic process GO:0046486 glycerolipid metabolic process GO:0048167 regulation of synaptic plasticity GO:0048469 cell maturation GO:0050432 catecholamine secretion GO:0050803 regulation of synapse structure or activity GO:0050804 modulation of synaptic transmission GO:0050806 positive regulation of synaptic transmission GO:0050807 regulation of synapse organization GO:0050808 synapse organization GO:0050884 neuromuscular process controlling posture GO:0050890 cognition GO:0050905 neuromuscular process GO:0051402 neuron apoptotic process GO:0051703 intraspecies interaction between organisms GO:0051705 multi-organism behavior GO:0051937 catecholamine transport GO:0051962 positive regulation of nervous system development GO:0051963 regulation of synapse assembly GO:0051965 positive regulation of synapse assembly GO:0060078 regulation of postsynaptic membrane potential GO:0060079 excitatory postsynaptic potential GO:0060291 long-term synaptic potentiation GO:0070482 response to oxygen levels GO:0070997 neuron death GO:0071514 genetic imprinting GO:0097164 ammonium ion metabolic process GO:0099565 chemical synaptic transmission, postsynaptic GO:1901214 regulation of neuron death GO:1901215 negative regulation of neuron death GO:1901605 alpha-amino acid metabolic process GO:1901615 organic hydroxy compound metabolic process GO:1901657 glycosyl compound metabolic process GO:1901983 regulation of protein acetylation GO:1901984 negative regulation of protein acetylation GO:1902275 regulation of chromatin organization GO:1905268 negative regulation of chromatin organization GO:2000756 regulation of peptidyl-lysine acetylation GO:2000757 negative regulation of peptidyl-lysine acetylation |
| Molecular Function |
GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0003729 mRNA binding GO:0008134 transcription factor binding GO:0008327 methyl-CpG binding GO:0010385 double-stranded methylated DNA binding GO:0035197 siRNA binding GO:0042826 histone deacetylase binding GO:0047485 protein N-terminus binding |
| Cellular Component |
GO:0000785 chromatin GO:0000792 heterochromatin GO:0098794 postsynapse |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
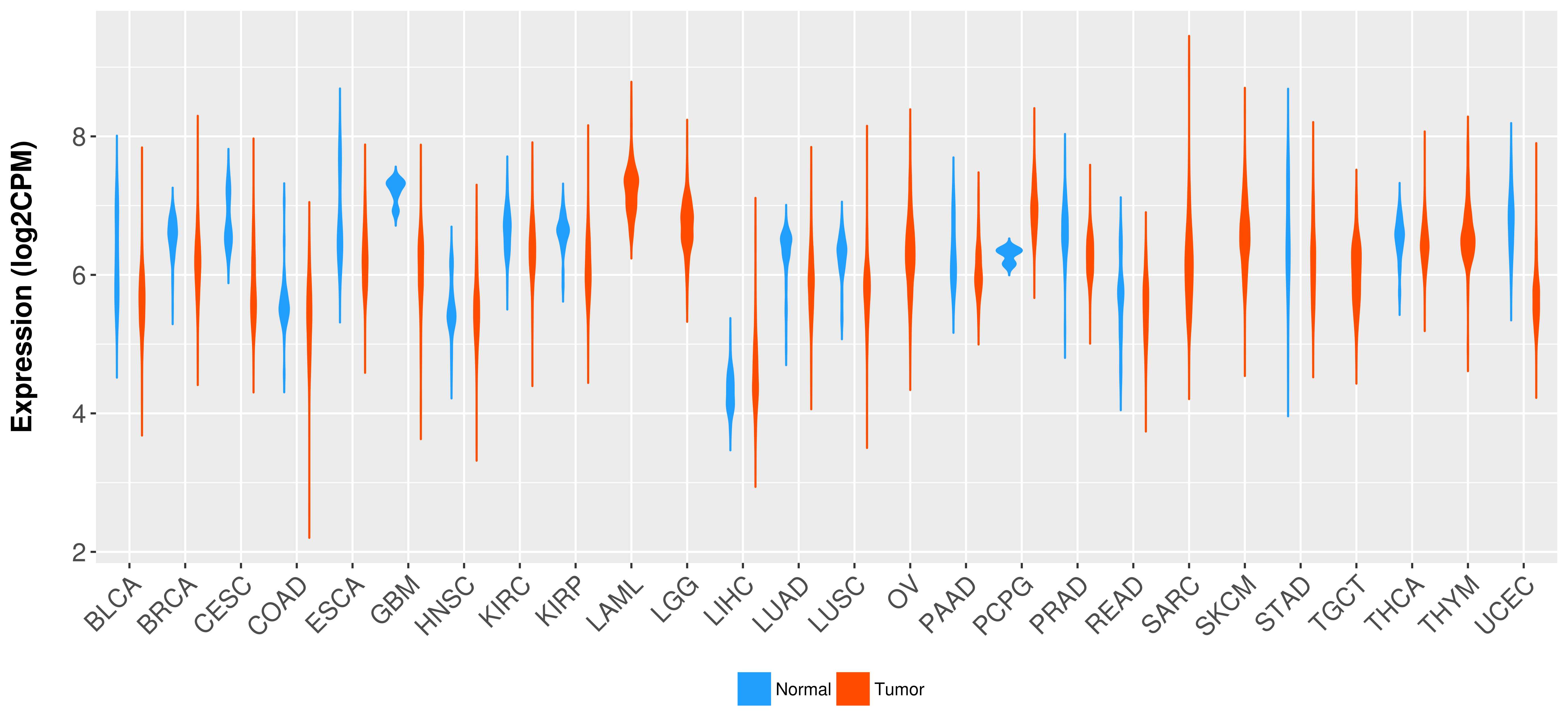
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
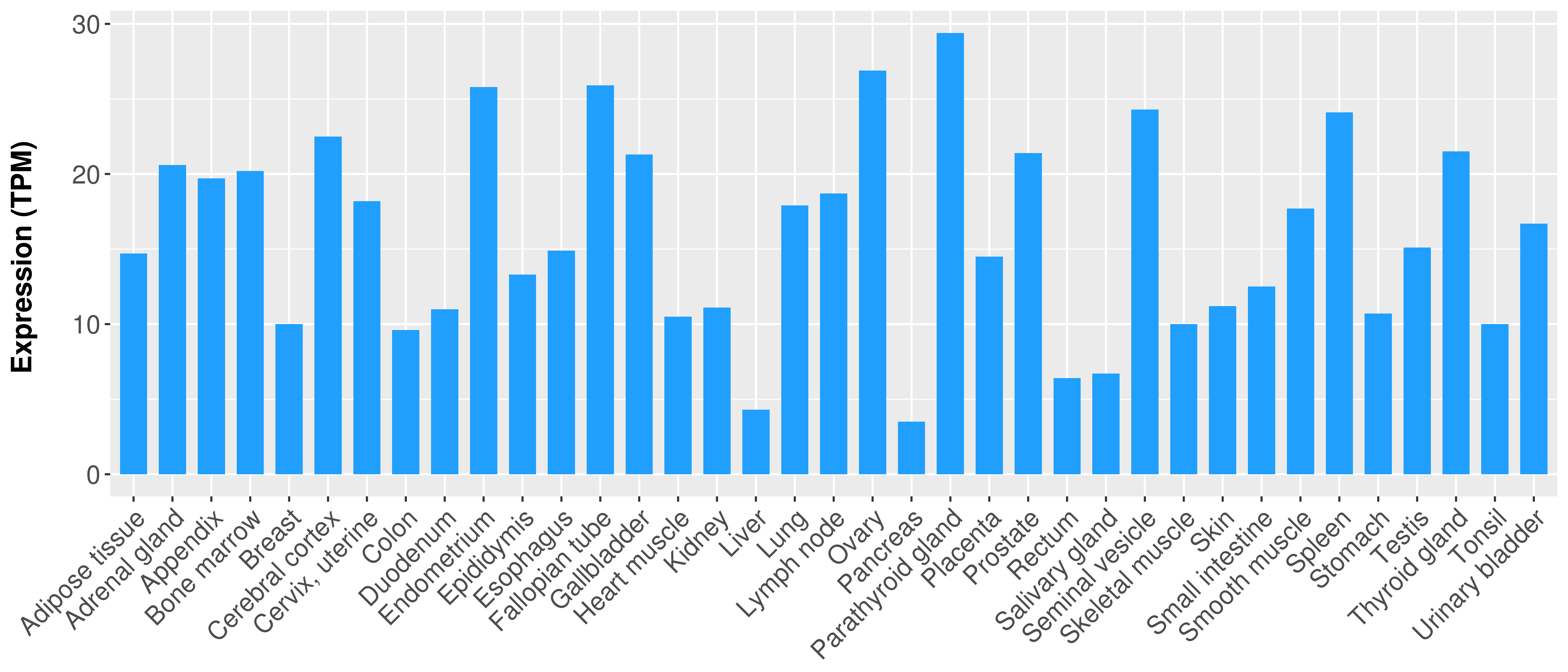
|
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
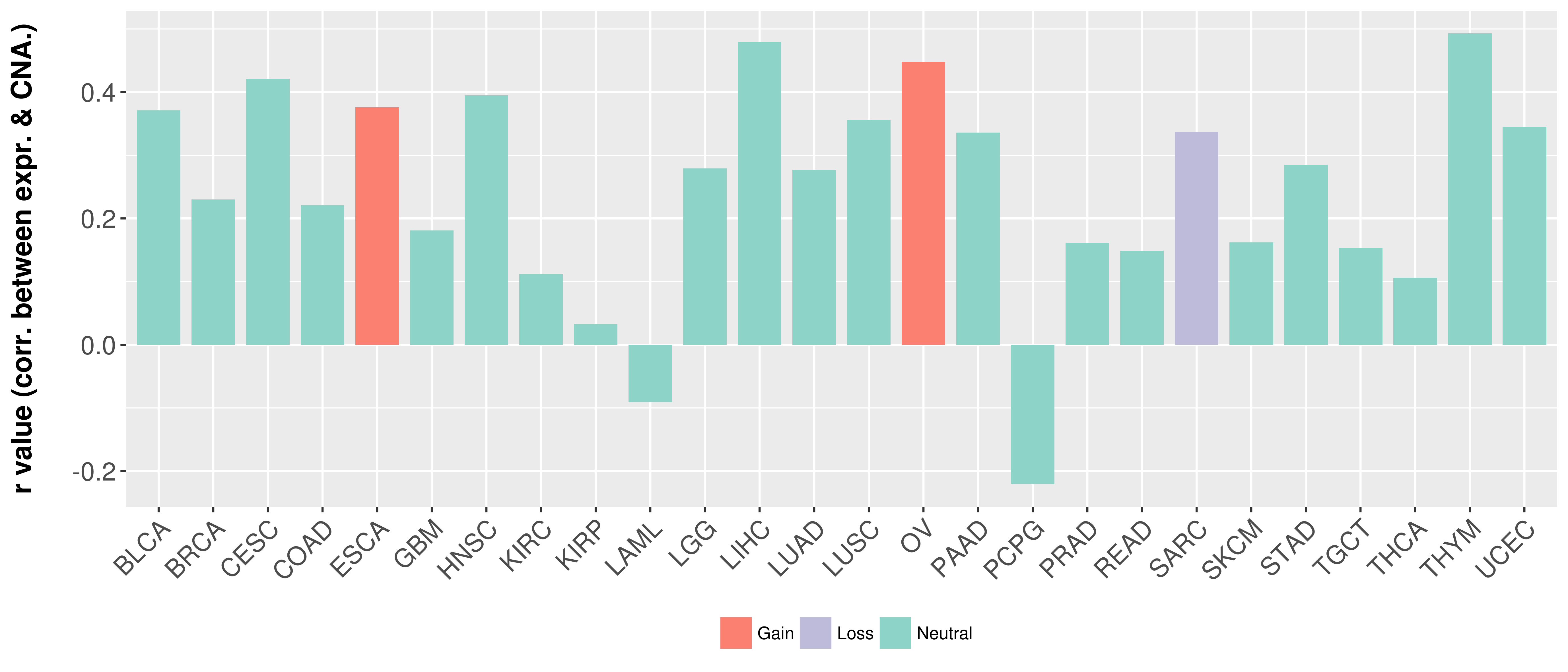
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
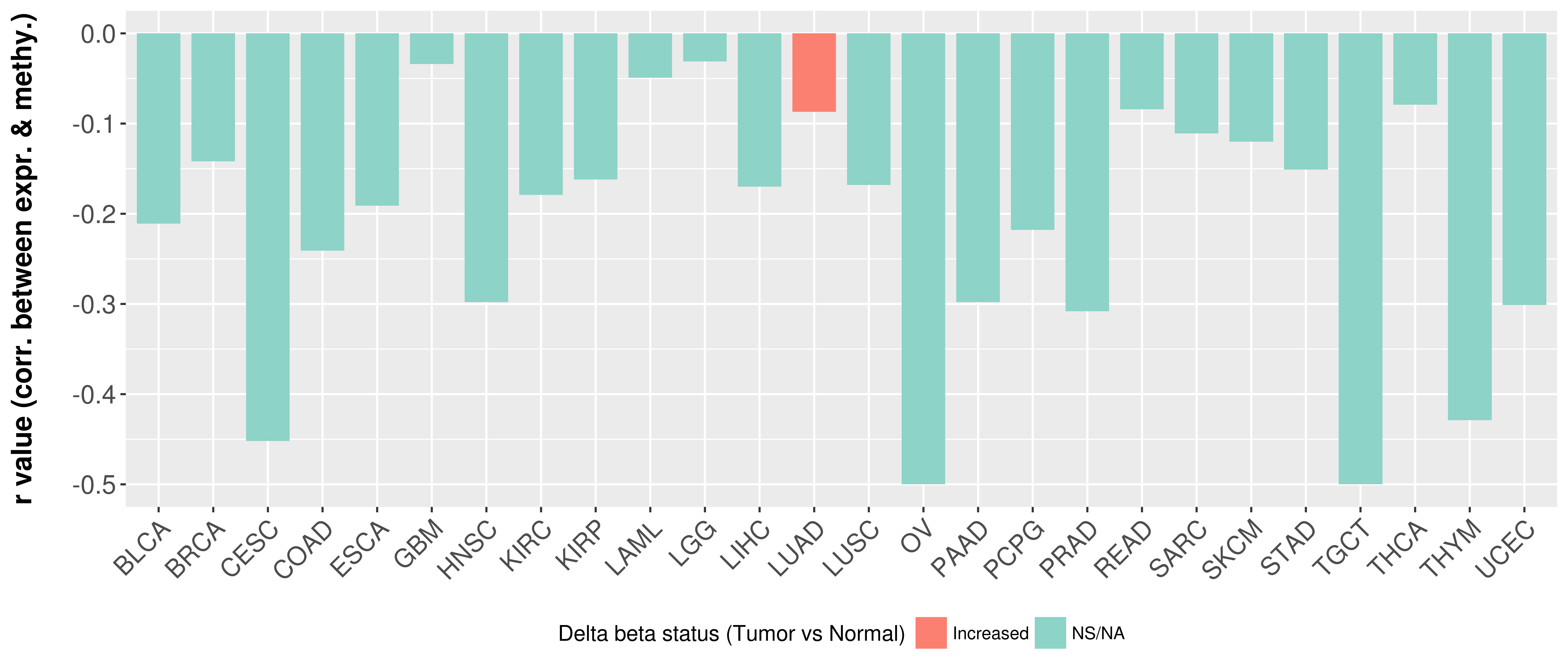
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
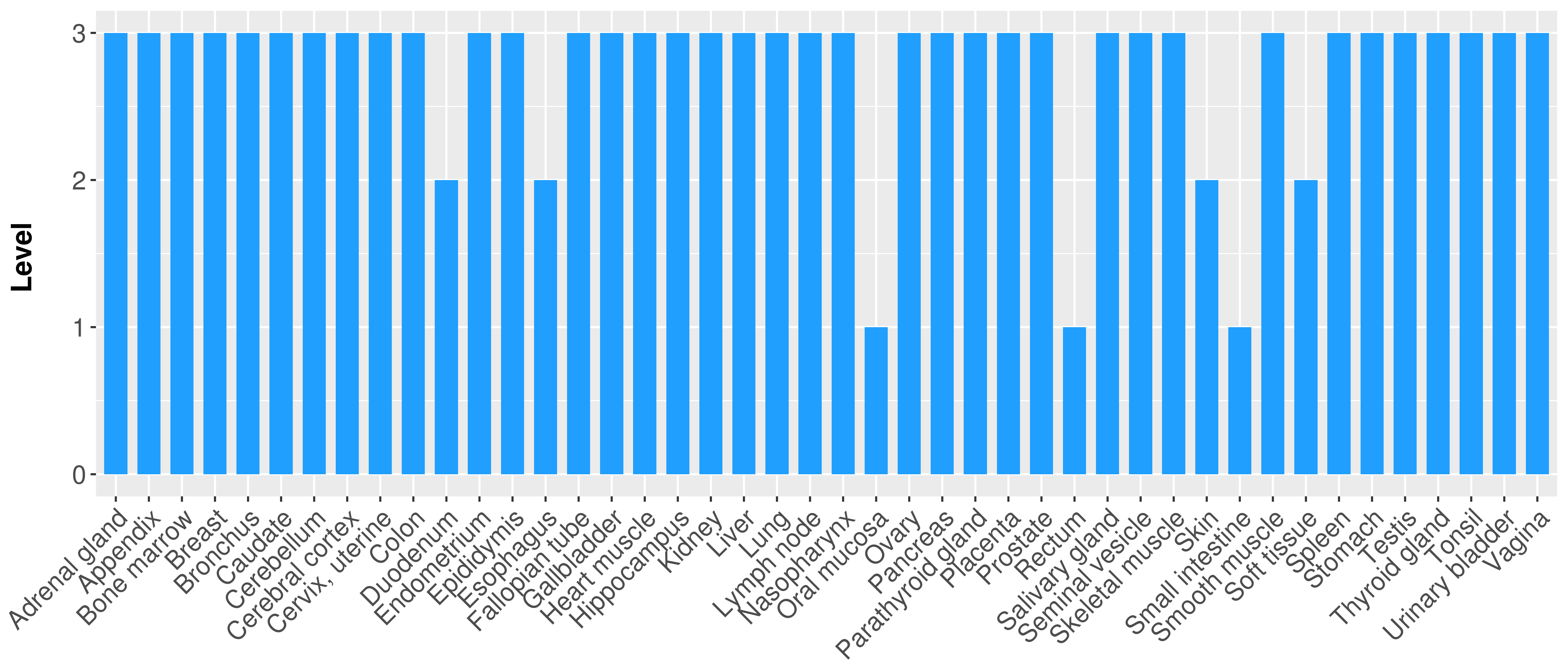
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
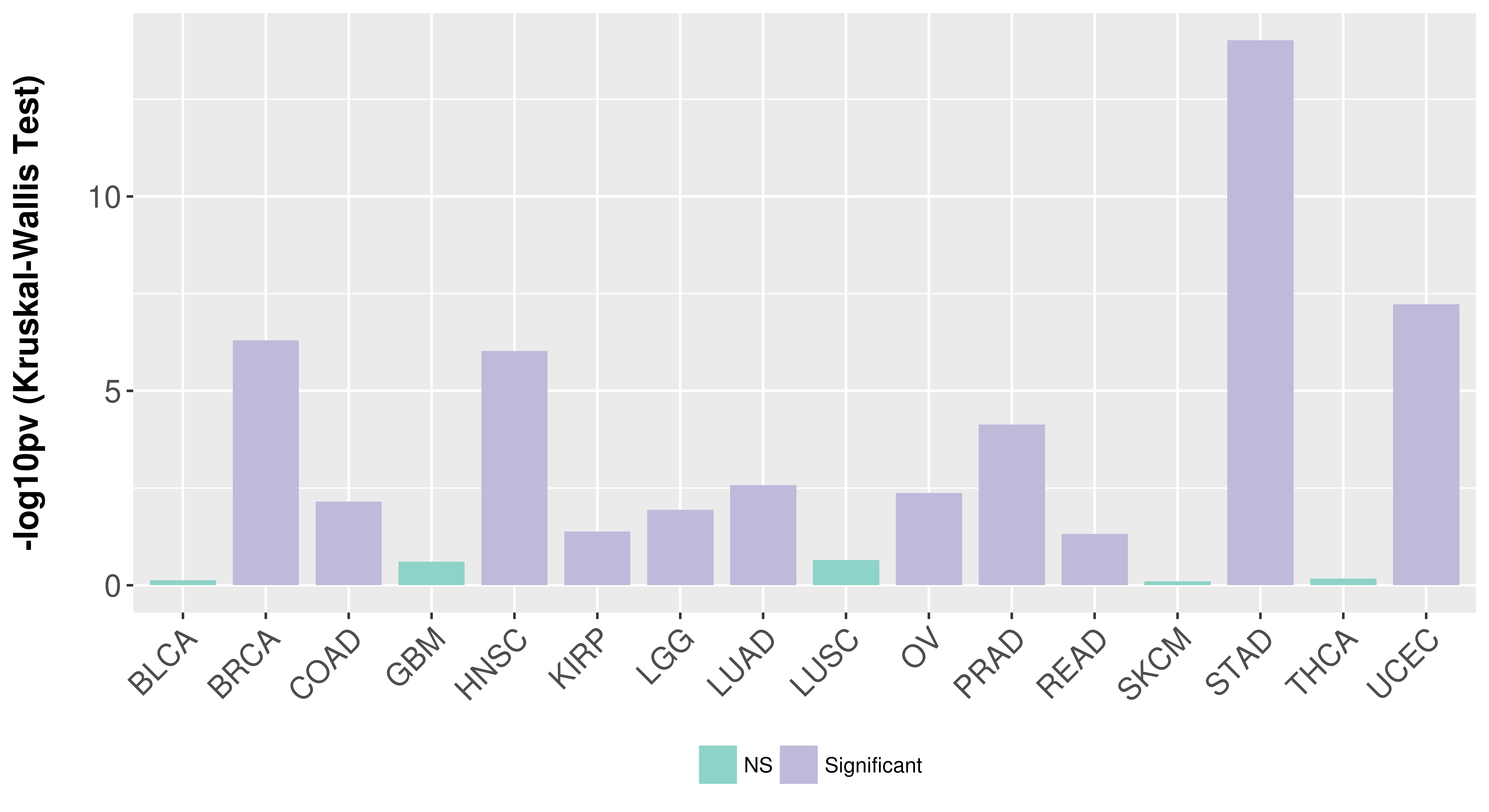
Association between expresson and subtype.
|
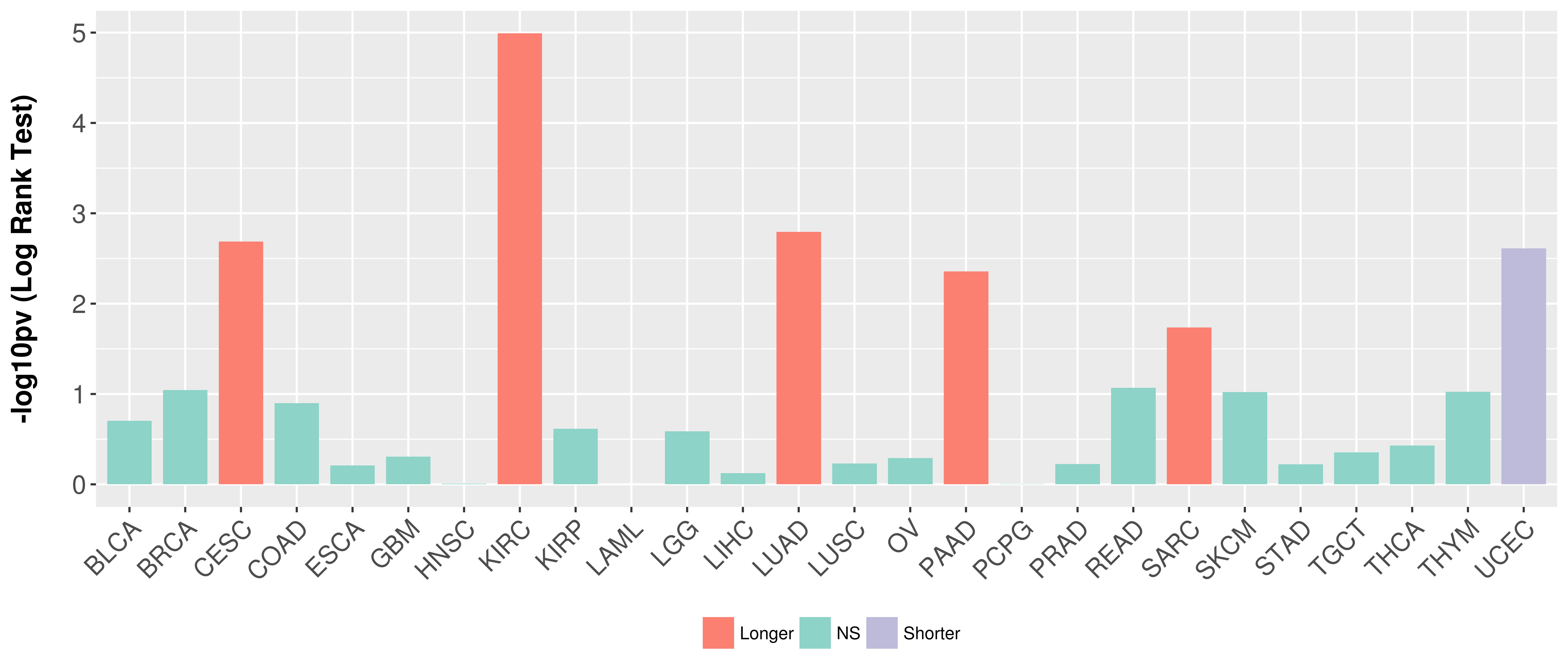
Overall survival analysis based on expression.
|
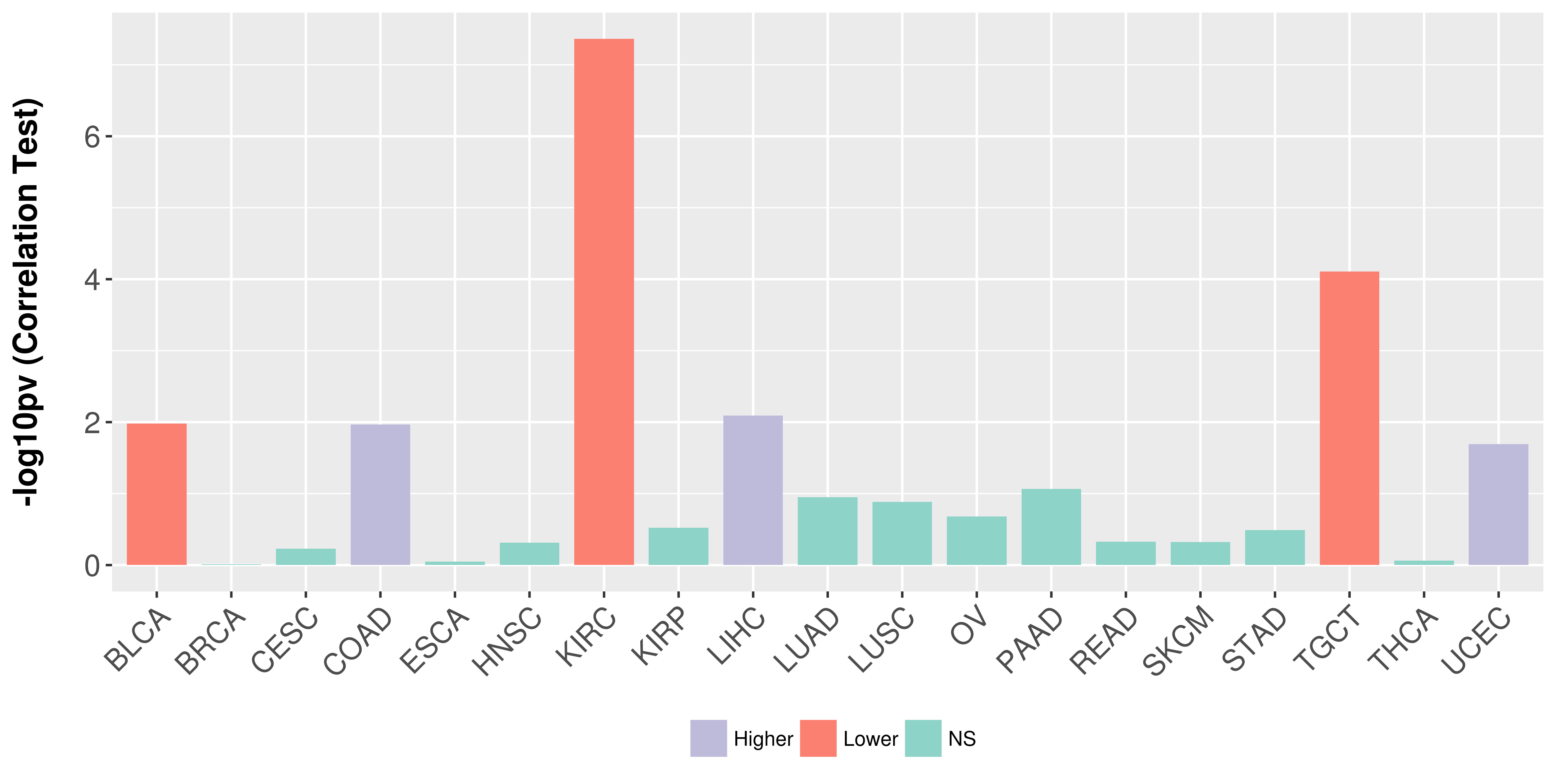
Association between expresson and stage.
|
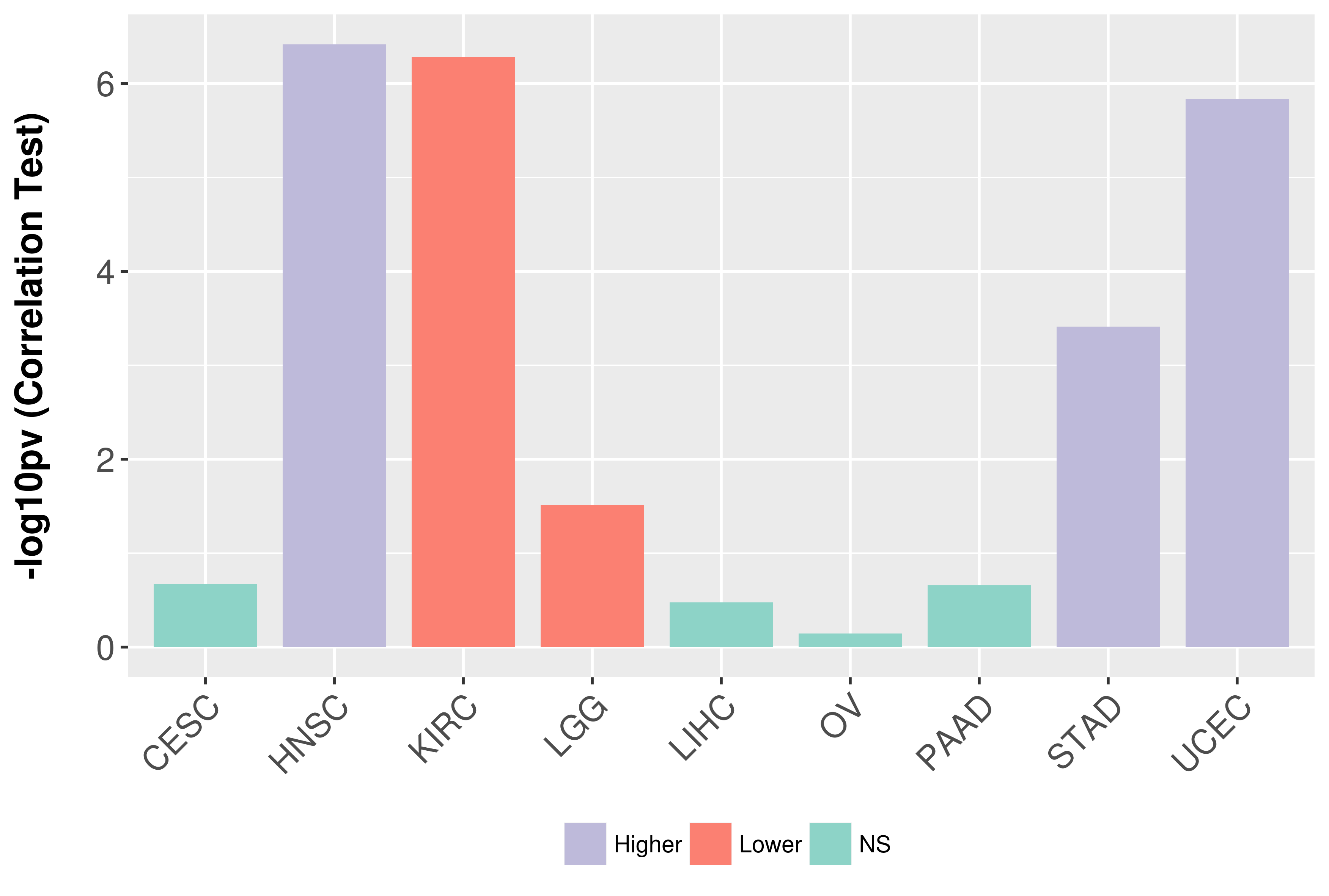
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for MECP2. |
| Summary | |
|---|---|
| Symbol | MECP2 |
| Name | methyl-CpG binding protein 2 |
| Aliases | RTT; MRX16; MRX79; mental retardation, X-linked 16; mental retardation, X-linked 79; Rett syndrome; methyl C ...... |
| Location | Xq28 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|