Browse MEN1 in pancancer
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05053 Menin |
||||||||||
| Function |
Essential component of a MLL/SET1 histone methyltransferase (HMT) complex, a complex that specifically methylates 'Lys-4' of histone H3 (H3K4). Functions as a transcriptional regulator. Binds to the TERT promoter and represses telomerase expression. Plays a role in TGFB1-mediated inhibition of cell-proliferation, possibly regulating SMAD3 transcriptional activity. Represses JUND-mediated transcriptional activation on AP1 sites, as well as that mediated by NFKB subunit RELA. Positively regulates HOXC8 and HOXC6 gene expression. May be involved in normal hematopoiesis through the activation of HOXA9 expression (By similarity). May be involved in DNA repair. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000079 regulation of cyclin-dependent protein serine/threonine kinase activity GO:0001503 ossification GO:0001649 osteoblast differentiation GO:0001678 cellular glucose homeostasis GO:0001890 placenta development GO:0001893 maternal placenta development GO:0001933 negative regulation of protein phosphorylation GO:0002065 columnar/cuboidal epithelial cell differentiation GO:0002067 glandular epithelial cell differentiation GO:0002076 osteoblast development GO:0003309 type B pancreatic cell differentiation GO:0006469 negative regulation of protein kinase activity GO:0006479 protein methylation GO:0007162 negative regulation of cell adhesion GO:0007178 transmembrane receptor protein serine/threonine kinase signaling pathway GO:0007179 transforming growth factor beta receptor signaling pathway GO:0007254 JNK cascade GO:0007565 female pregnancy GO:0008213 protein alkylation GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0009743 response to carbohydrate GO:0009746 response to hexose GO:0009749 response to glucose GO:0010212 response to ionizing radiation GO:0010332 response to gamma radiation GO:0010810 regulation of cell-substrate adhesion GO:0010812 negative regulation of cell-substrate adhesion GO:0010948 negative regulation of cell cycle process GO:0016570 histone modification GO:0016571 histone methylation GO:0017015 regulation of transforming growth factor beta receptor signaling pathway GO:0018022 peptidyl-lysine methylation GO:0018205 peptidyl-lysine modification GO:0030278 regulation of ossification GO:0030279 negative regulation of ossification GO:0030511 positive regulation of transforming growth factor beta receptor signaling pathway GO:0031016 pancreas development GO:0031018 endocrine pancreas development GO:0031098 stress-activated protein kinase signaling cascade GO:0031589 cell-substrate adhesion GO:0032092 positive regulation of protein binding GO:0032259 methylation GO:0032872 regulation of stress-activated MAPK cascade GO:0032873 negative regulation of stress-activated MAPK cascade GO:0032924 activin receptor signaling pathway GO:0032925 regulation of activin receptor signaling pathway GO:0033500 carbohydrate homeostasis GO:0033673 negative regulation of kinase activity GO:0034284 response to monosaccharide GO:0034968 histone lysine methylation GO:0035270 endocrine system development GO:0035883 enteroendocrine cell differentiation GO:0042326 negative regulation of phosphorylation GO:0042593 glucose homeostasis GO:0043393 regulation of protein binding GO:0043409 negative regulation of MAPK cascade GO:0043414 macromolecule methylation GO:0043433 negative regulation of sequence-specific DNA binding transcription factor activity GO:0043434 response to peptide hormone GO:0044342 type B pancreatic cell proliferation GO:0044706 multi-multicellular organism process GO:0044770 cell cycle phase transition GO:0044843 cell cycle G1/S phase transition GO:0045667 regulation of osteoblast differentiation GO:0045668 negative regulation of osteoblast differentiation GO:0045736 negative regulation of cyclin-dependent protein serine/threonine kinase activity GO:0045786 negative regulation of cell cycle GO:0046328 regulation of JNK cascade GO:0046329 negative regulation of JNK cascade GO:0046697 decidualization GO:0048608 reproductive structure development GO:0050673 epithelial cell proliferation GO:0050678 regulation of epithelial cell proliferation GO:0050680 negative regulation of epithelial cell proliferation GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051348 negative regulation of transferase activity GO:0051403 stress-activated MAPK cascade GO:0051972 regulation of telomerase activity GO:0051974 negative regulation of telomerase activity GO:0060135 maternal process involved in female pregnancy GO:0061458 reproductive system development GO:0061469 regulation of type B pancreatic cell proliferation GO:0070302 regulation of stress-activated protein kinase signaling cascade GO:0070303 negative regulation of stress-activated protein kinase signaling cascade GO:0071322 cellular response to carbohydrate stimulus GO:0071326 cellular response to monosaccharide stimulus GO:0071331 cellular response to hexose stimulus GO:0071333 cellular response to glucose stimulus GO:0071375 cellular response to peptide hormone stimulus GO:0071417 cellular response to organonitrogen compound GO:0071559 response to transforming growth factor beta GO:0071560 cellular response to transforming growth factor beta stimulus GO:0071897 DNA biosynthetic process GO:0071900 regulation of protein serine/threonine kinase activity GO:0071901 negative regulation of protein serine/threonine kinase activity GO:0090092 regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090100 positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway GO:0090287 regulation of cellular response to growth factor stimulus GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1902532 negative regulation of intracellular signal transduction GO:1902806 regulation of cell cycle G1/S phase transition GO:1902807 negative regulation of cell cycle G1/S phase transition GO:1903844 regulation of cellular response to transforming growth factor beta stimulus GO:1903846 positive regulation of cellular response to transforming growth factor beta stimulus GO:1904029 regulation of cyclin-dependent protein kinase activity GO:1904030 negative regulation of cyclin-dependent protein kinase activity GO:1904837 beta-catenin-TCF complex assembly GO:2000278 regulation of DNA biosynthetic process GO:2000279 negative regulation of DNA biosynthetic process |
| Molecular Function |
GO:0000217 DNA secondary structure binding GO:0000400 four-way junction DNA binding GO:0000403 Y-form DNA binding GO:0003682 chromatin binding GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018024 histone-lysine N-methyltransferase activity GO:0030674 protein binding, bridging GO:0042054 histone methyltransferase activity GO:0043566 structure-specific DNA binding GO:0046332 SMAD binding GO:0047485 protein N-terminus binding GO:0060090 binding, bridging GO:0070412 R-SMAD binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016363 nuclear matrix GO:0032153 cell division site GO:0032154 cleavage furrow GO:0032155 cell division site part GO:0034399 nuclear periphery GO:0034708 methyltransferase complex GO:0035097 histone methyltransferase complex GO:0044454 nuclear chromosome part GO:0097610 cell surface furrow GO:0098687 chromosomal region |
| KEGG | - |
| Reactome |
R-HSA-3769402: Deactivation of the beta-catenin transactivating complex R-HSA-201722: Formation of the beta-catenin R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-195258: RHO GTPase Effectors R-HSA-5626467: RHO GTPases activate IQGAPs R-HSA-2173796: SMAD2/SMAD3 R-HSA-162582: Signal Transduction R-HSA-194315: Signaling by Rho GTPases R-HSA-170834: Signaling by TGF-beta Receptor Complex R-HSA-195721: Signaling by Wnt R-HSA-201681: TCF dependent signaling in response to WNT R-HSA-2173793: Transcriptional activity of SMAD2/SMAD3 |
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
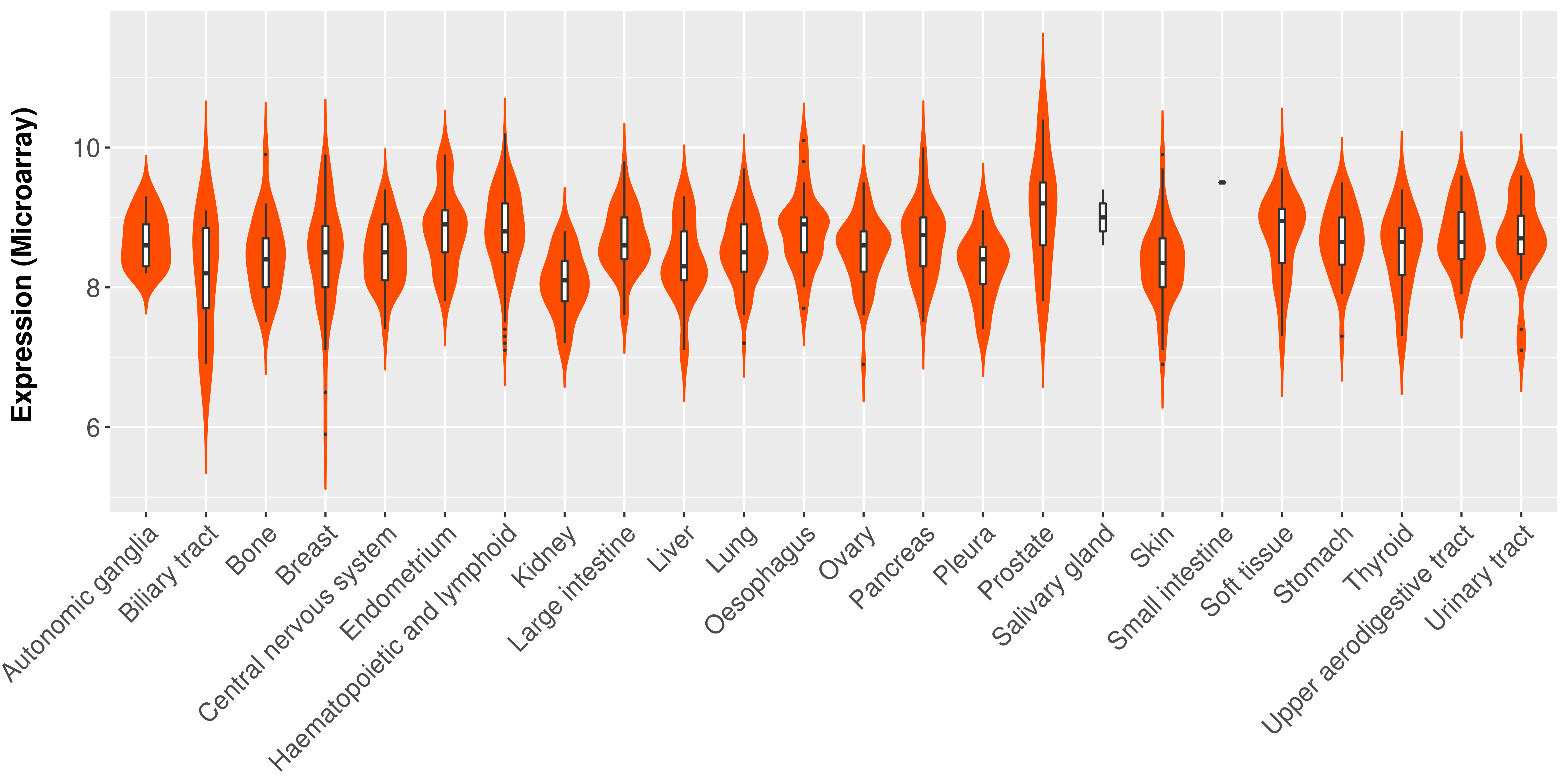
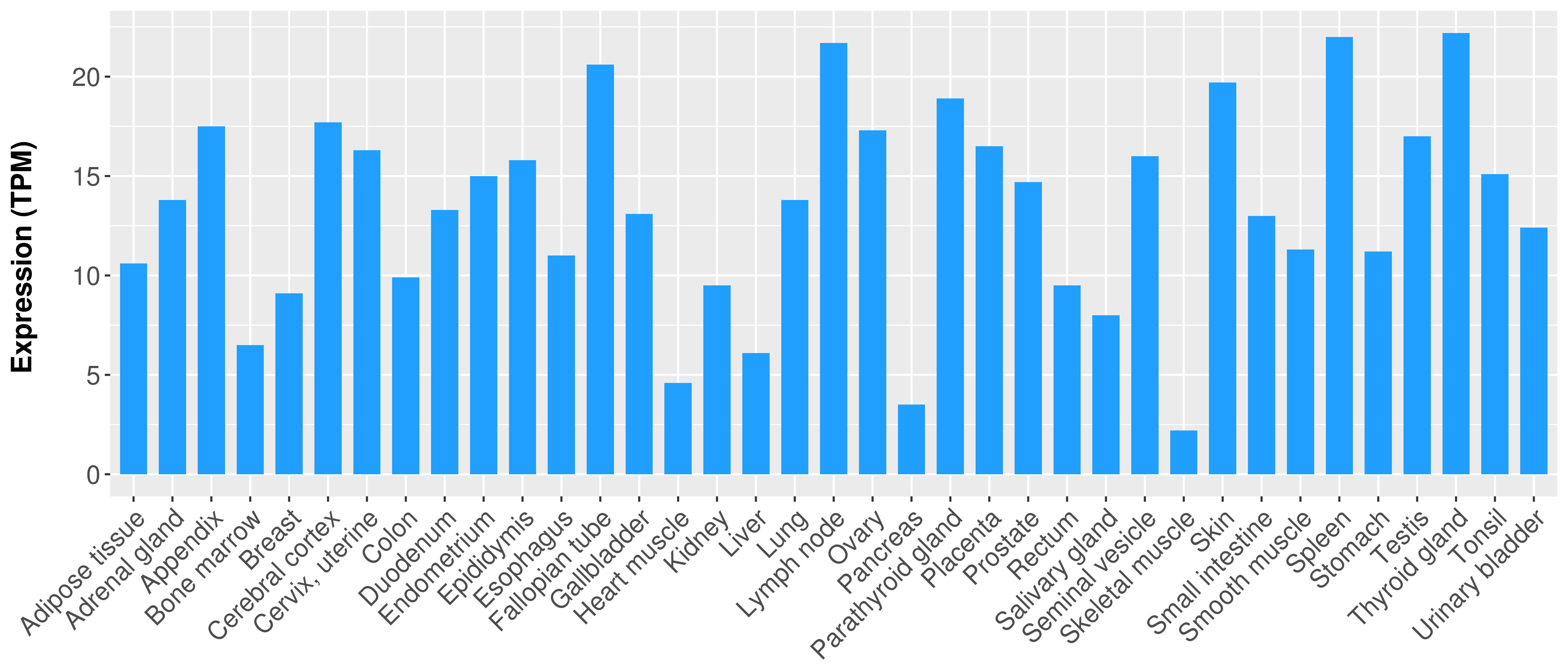
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
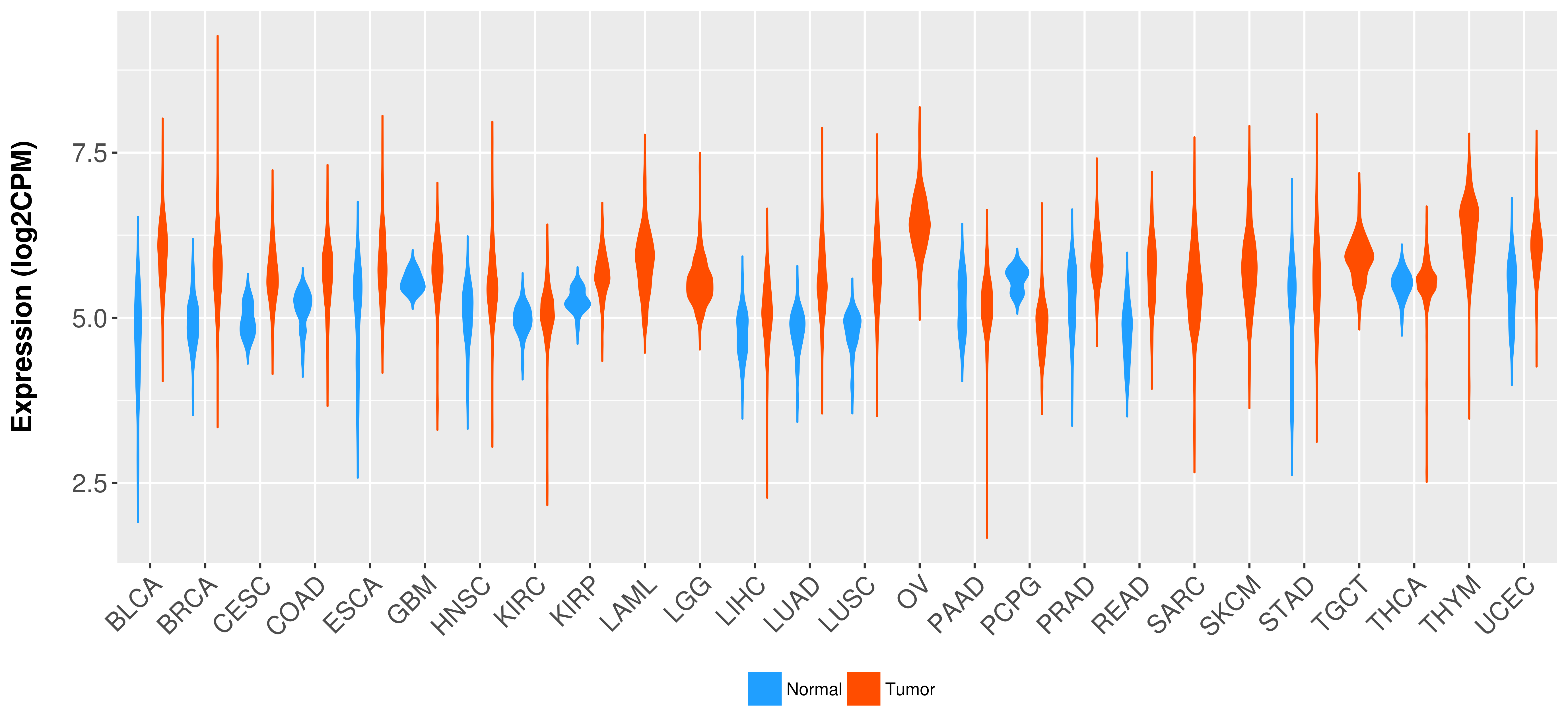
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
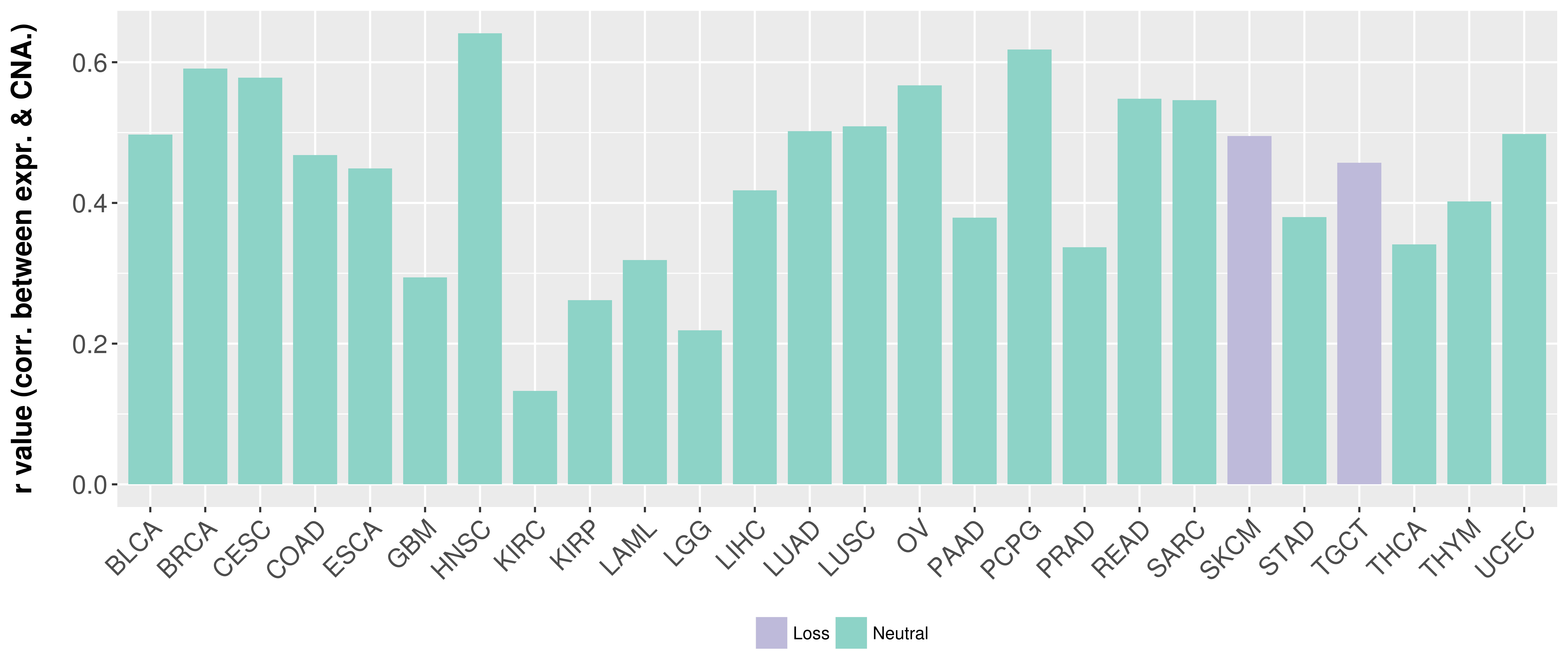
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
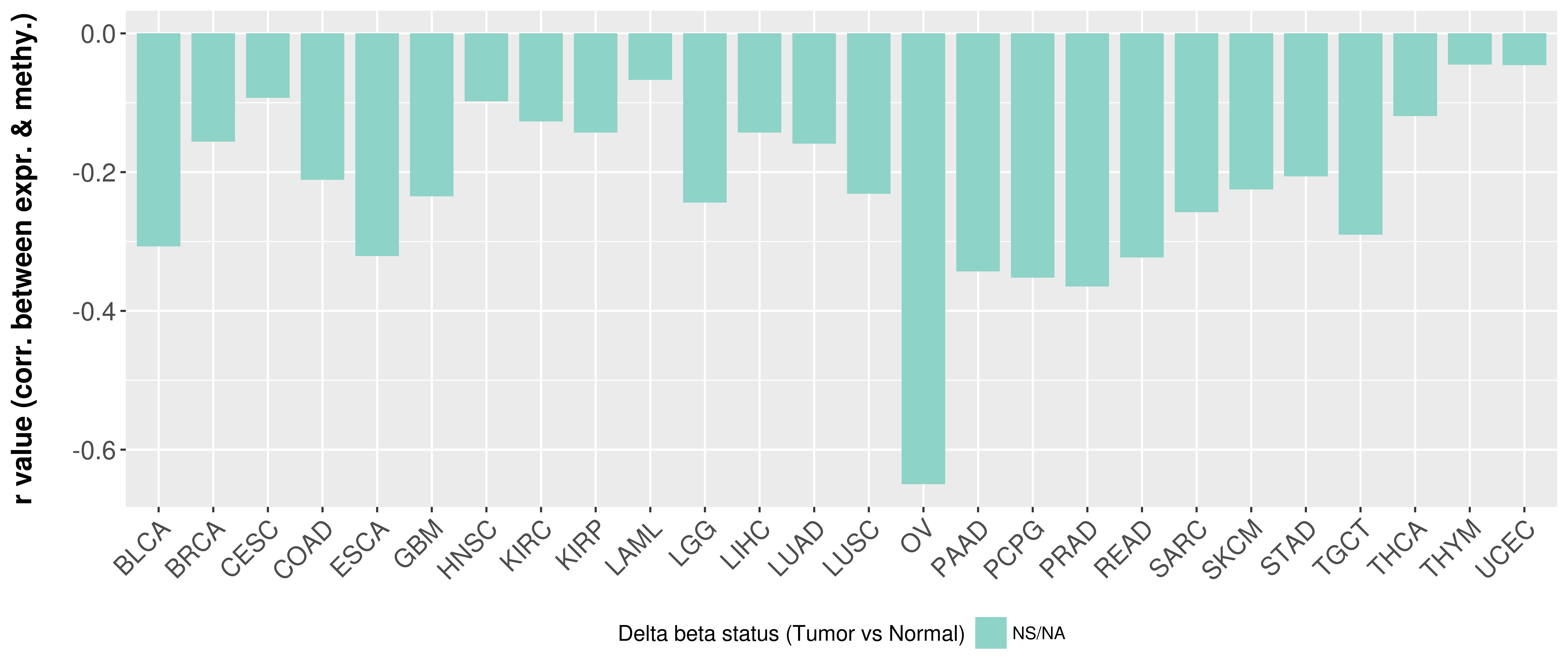
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
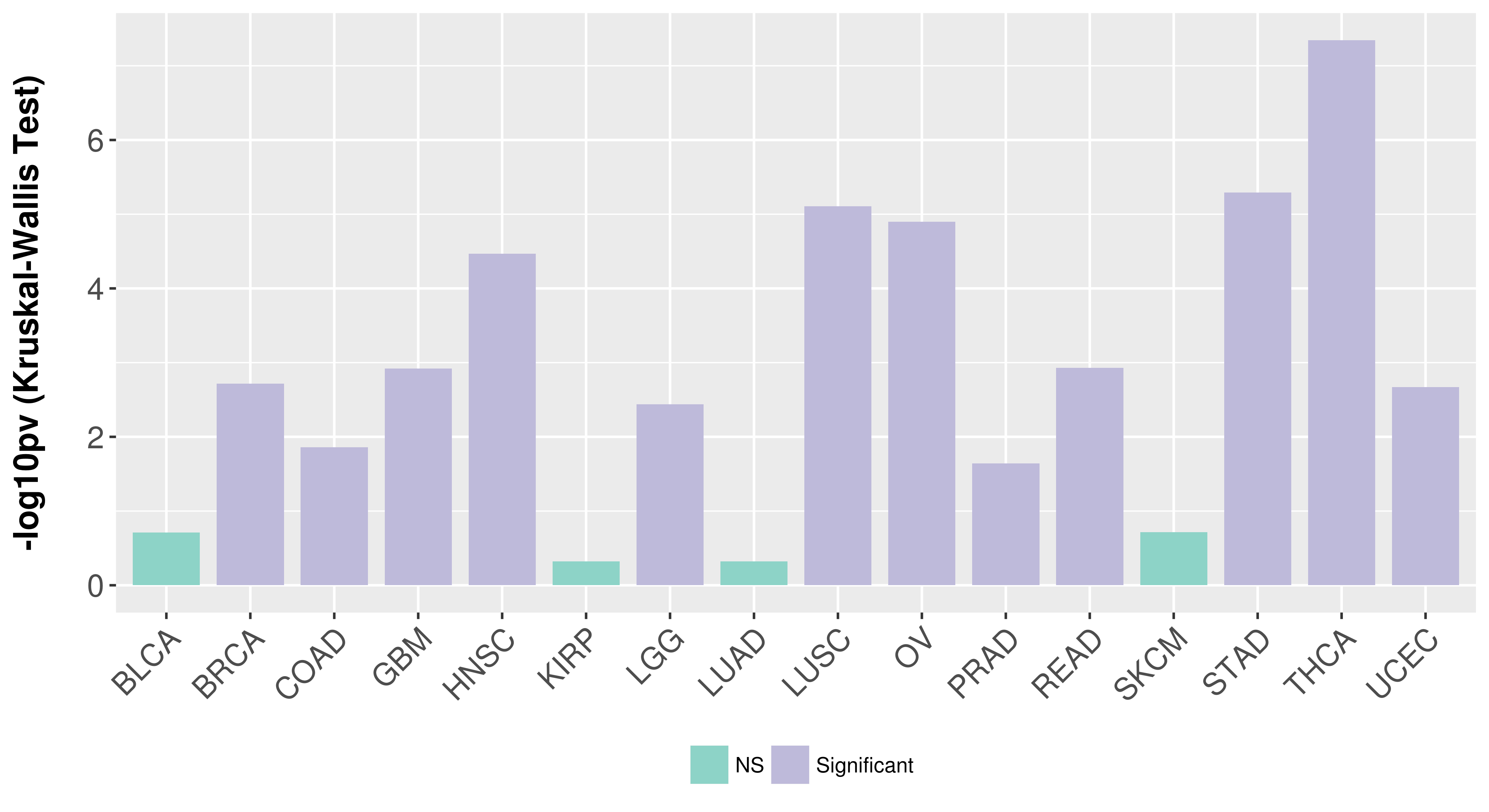
Association between expresson and subtype.
|
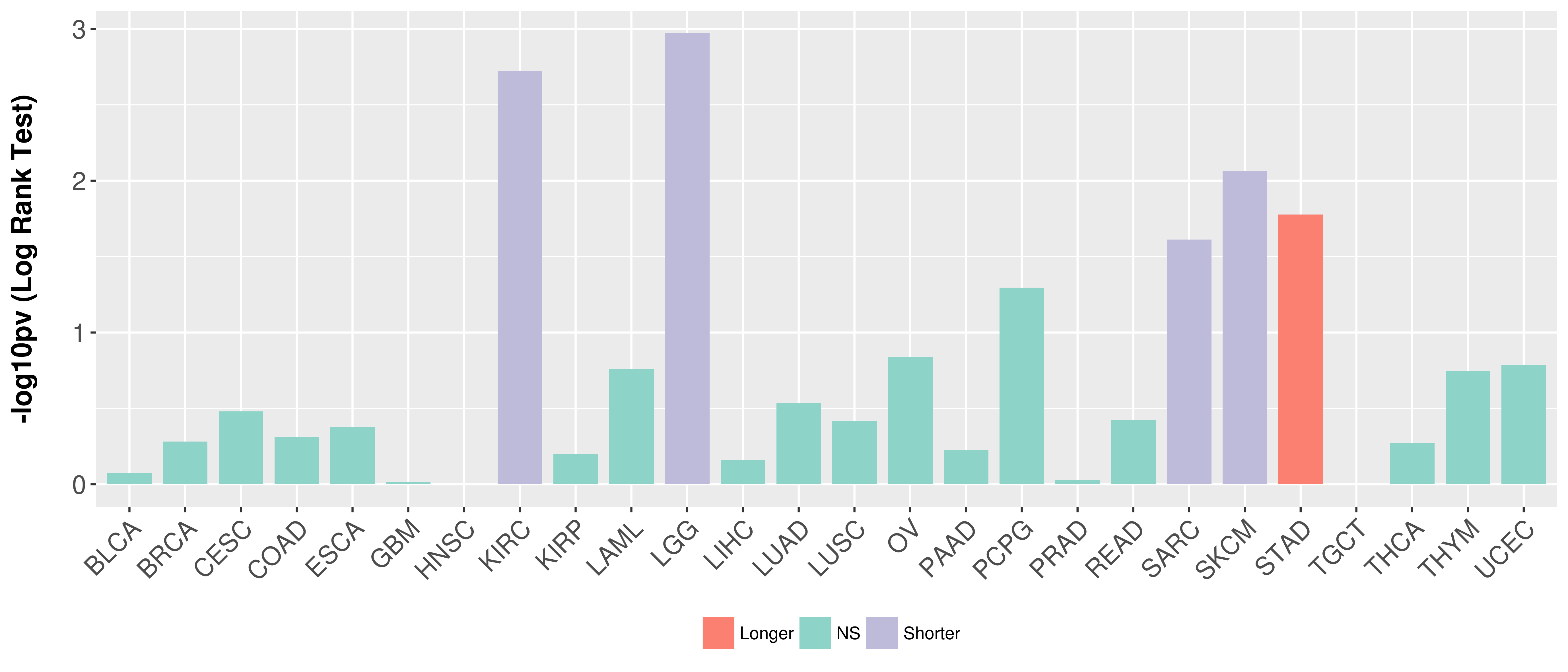
Overall survival analysis based on expression.
|
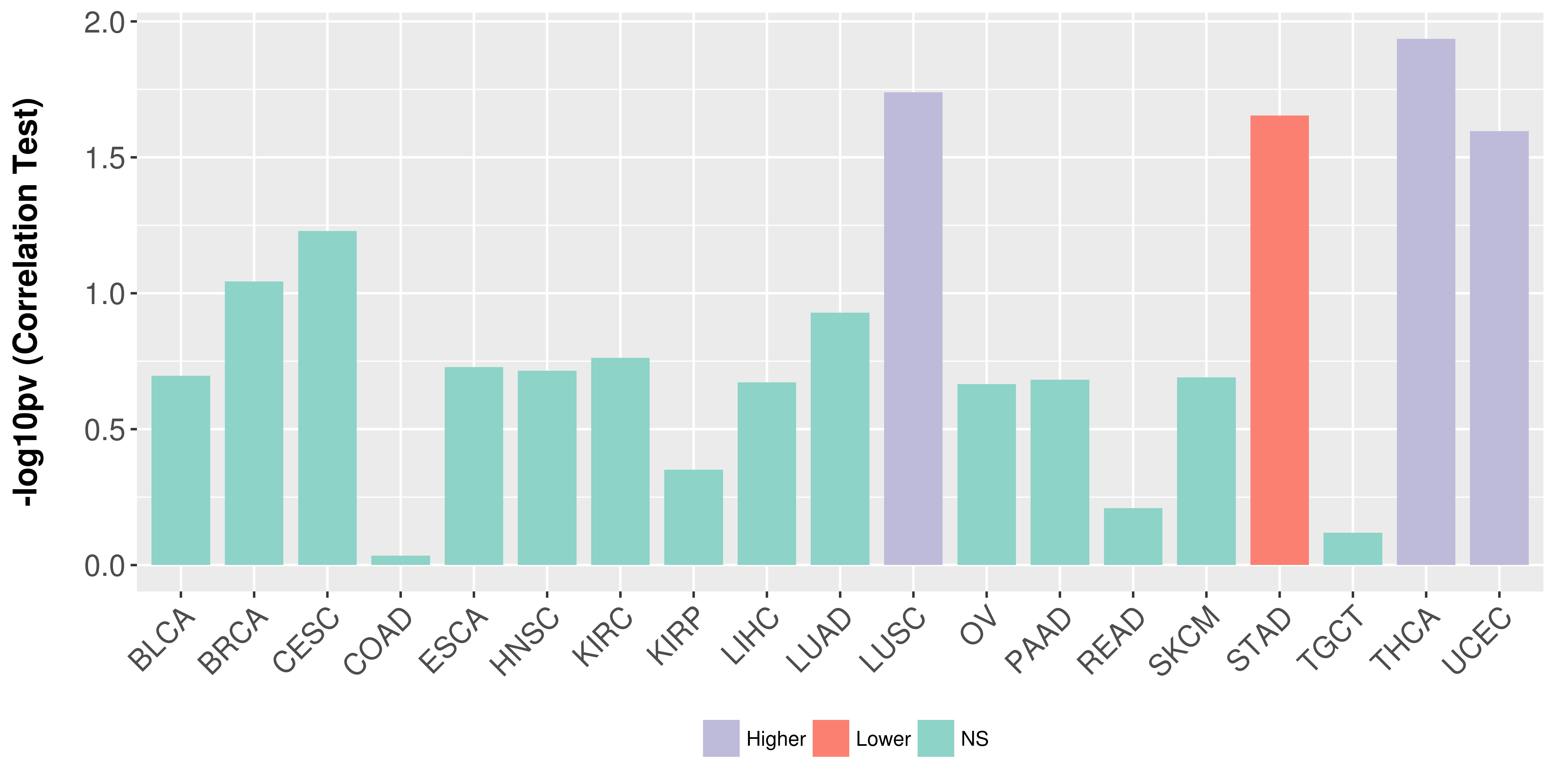
Association between expresson and stage.
|
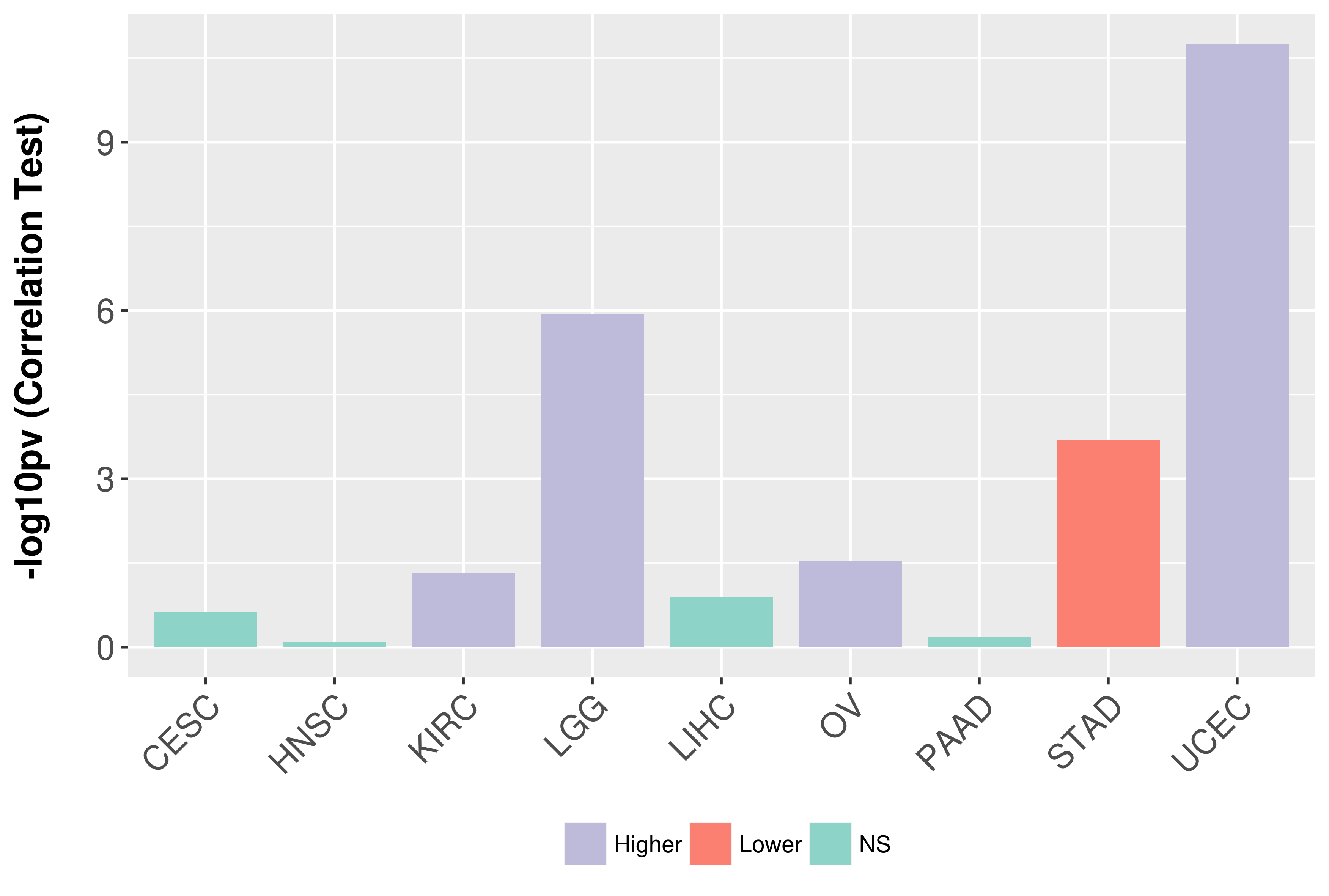
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for MEN1. |
| Summary | |
|---|---|
| Symbol | MEN1 |
| Name | menin 1 |
| Aliases | menin; MEAI |
| Location | 11q13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for MEN1. |
|