Browse MORF4L2 in pancancer
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05712 MRG |
||||||||||
| Function |
Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histone H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when directly recruited to sites of DNA damage. Also component of the MSIN3A complex which acts to repress transcription by deacetylation of nucleosomal histones. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006338 chromatin remodeling GO:0006342 chromatin silencing GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006476 protein deacetylation GO:0016458 gene silencing GO:0016570 histone modification GO:0016573 histone acetylation GO:0016575 histone deacetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0035601 protein deacylation GO:0040029 regulation of gene expression, epigenetic GO:0042692 muscle cell differentiation GO:0043543 protein acylation GO:0043967 histone H4 acetylation GO:0043968 histone H2A acetylation GO:0045814 negative regulation of gene expression, epigenetic GO:0051146 striated muscle cell differentiation GO:0051147 regulation of muscle cell differentiation GO:0051149 positive regulation of muscle cell differentiation GO:0051153 regulation of striated muscle cell differentiation GO:0051155 positive regulation of striated muscle cell differentiation GO:0098732 macromolecule deacylation |
| Molecular Function | - |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0031248 protein acetyltransferase complex GO:0035267 NuA4 histone acetyltransferase complex GO:0043189 H4/H2A histone acetyltransferase complex GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214847: HATs acetylate histones |
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for MORF4L2. |
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
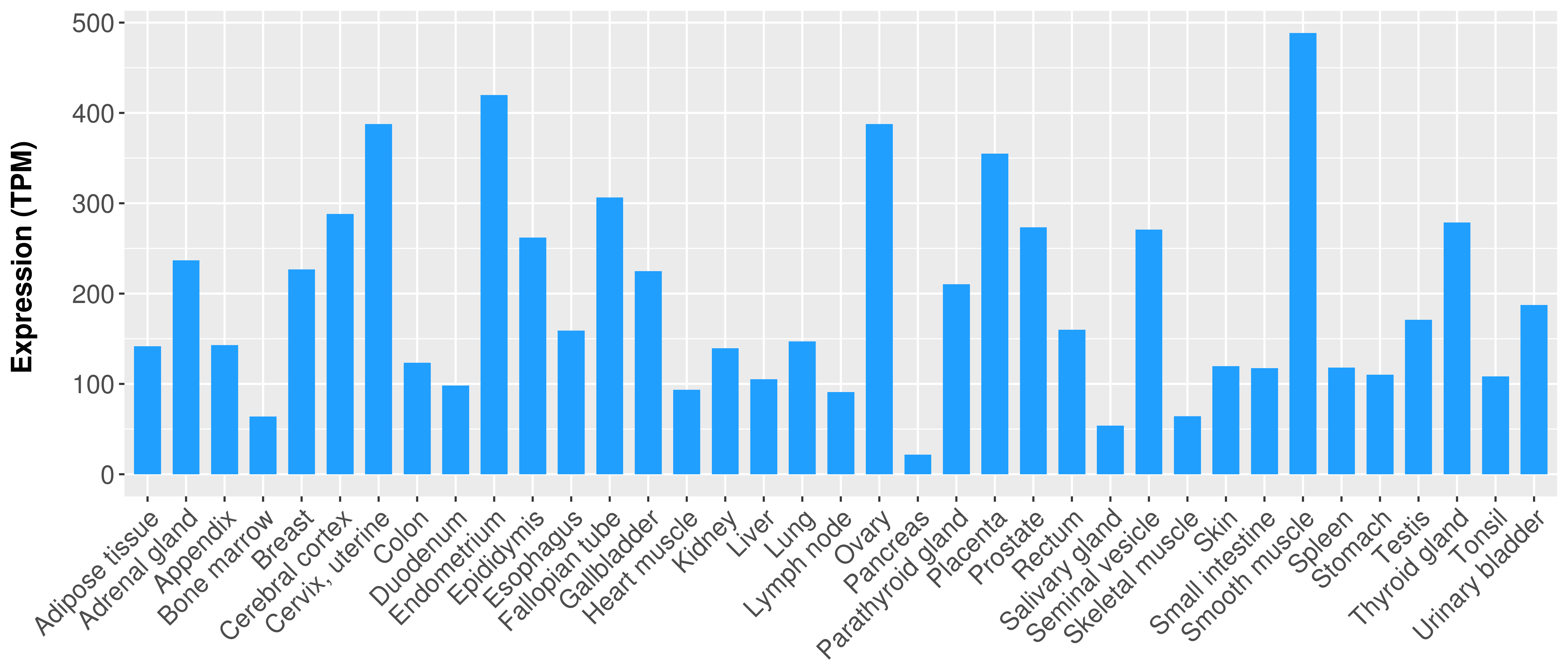
|
|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
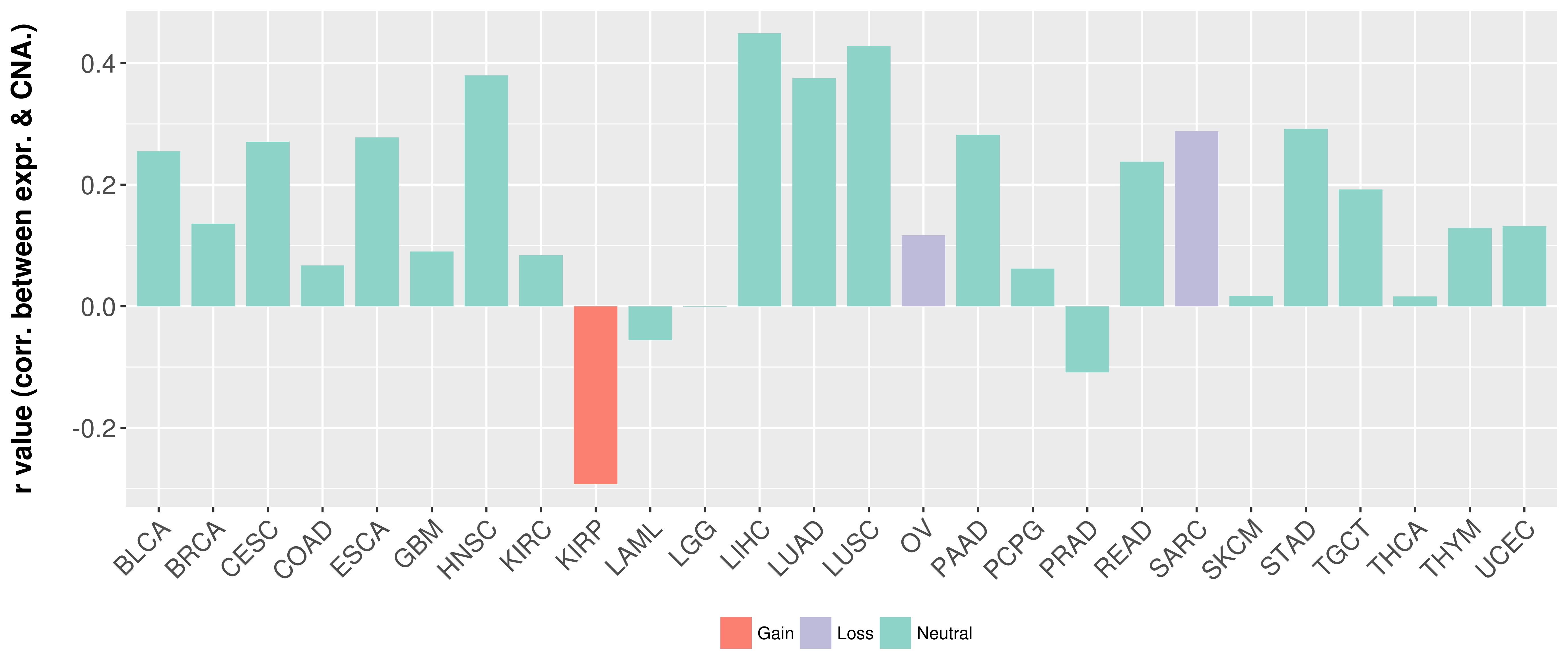
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
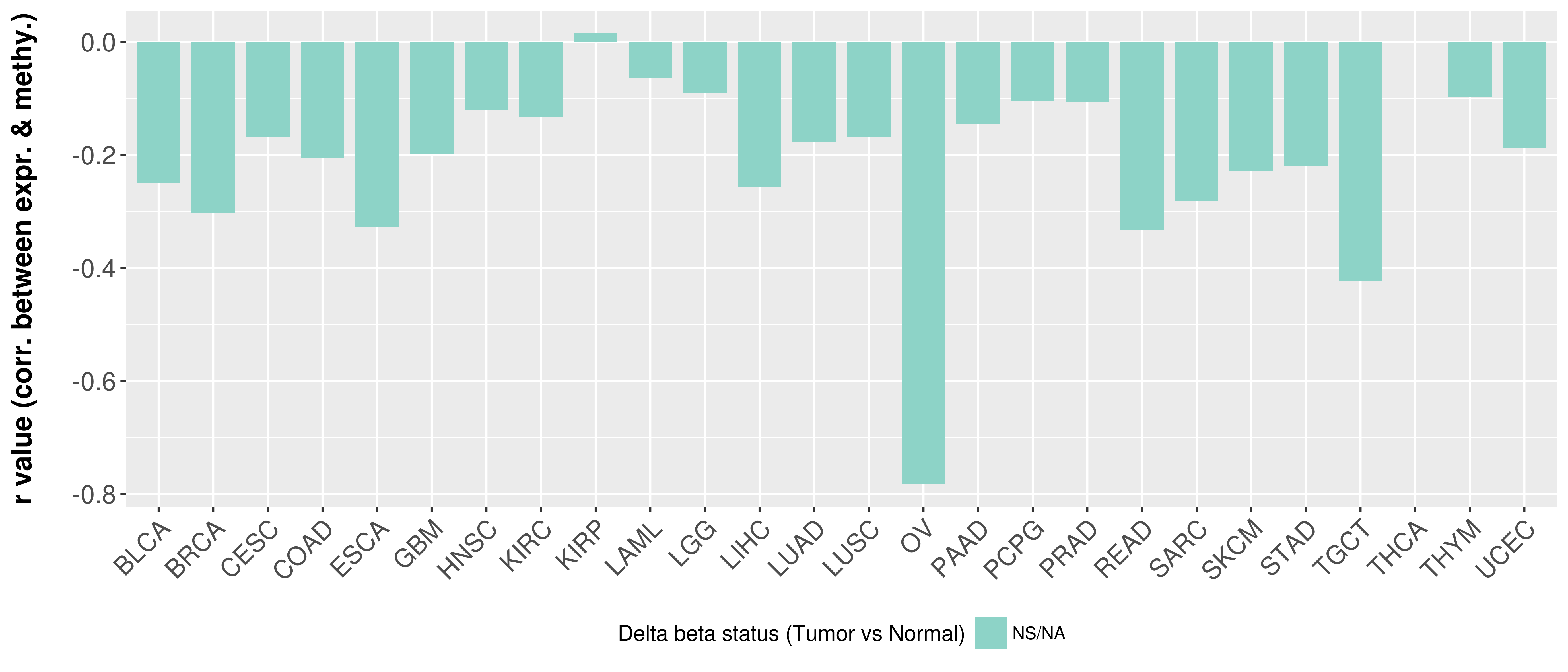
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |

|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
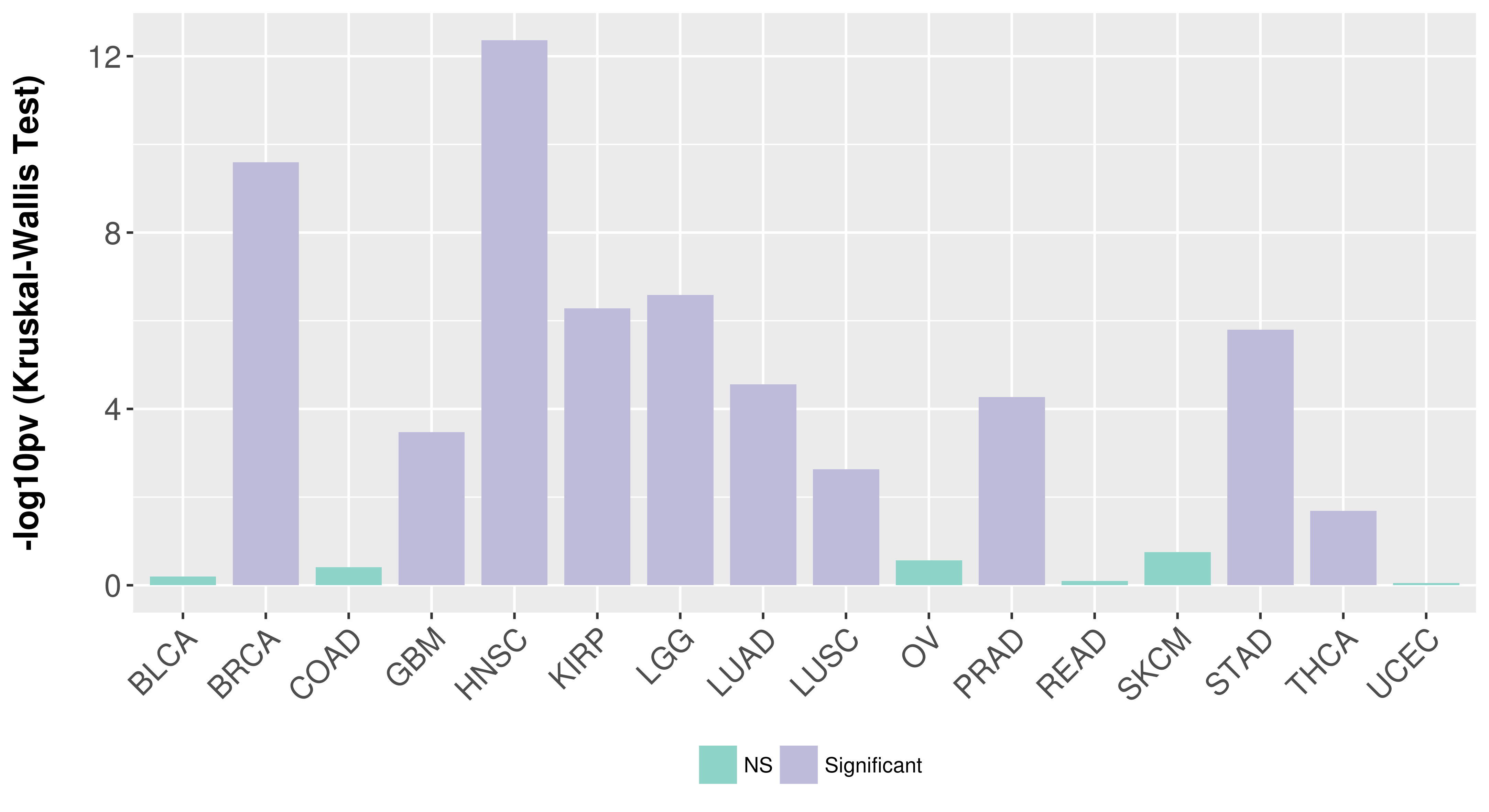
Association between expresson and subtype.
|
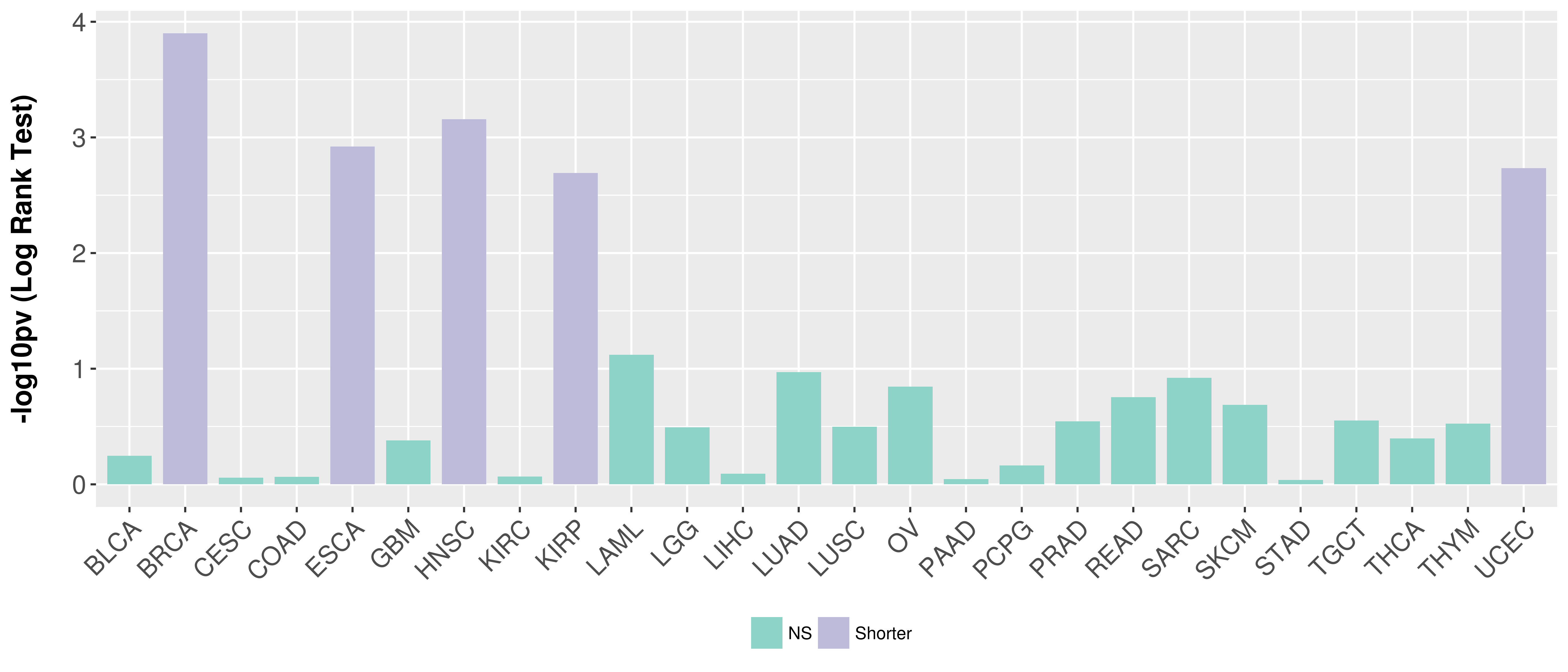
Overall survival analysis based on expression.
|
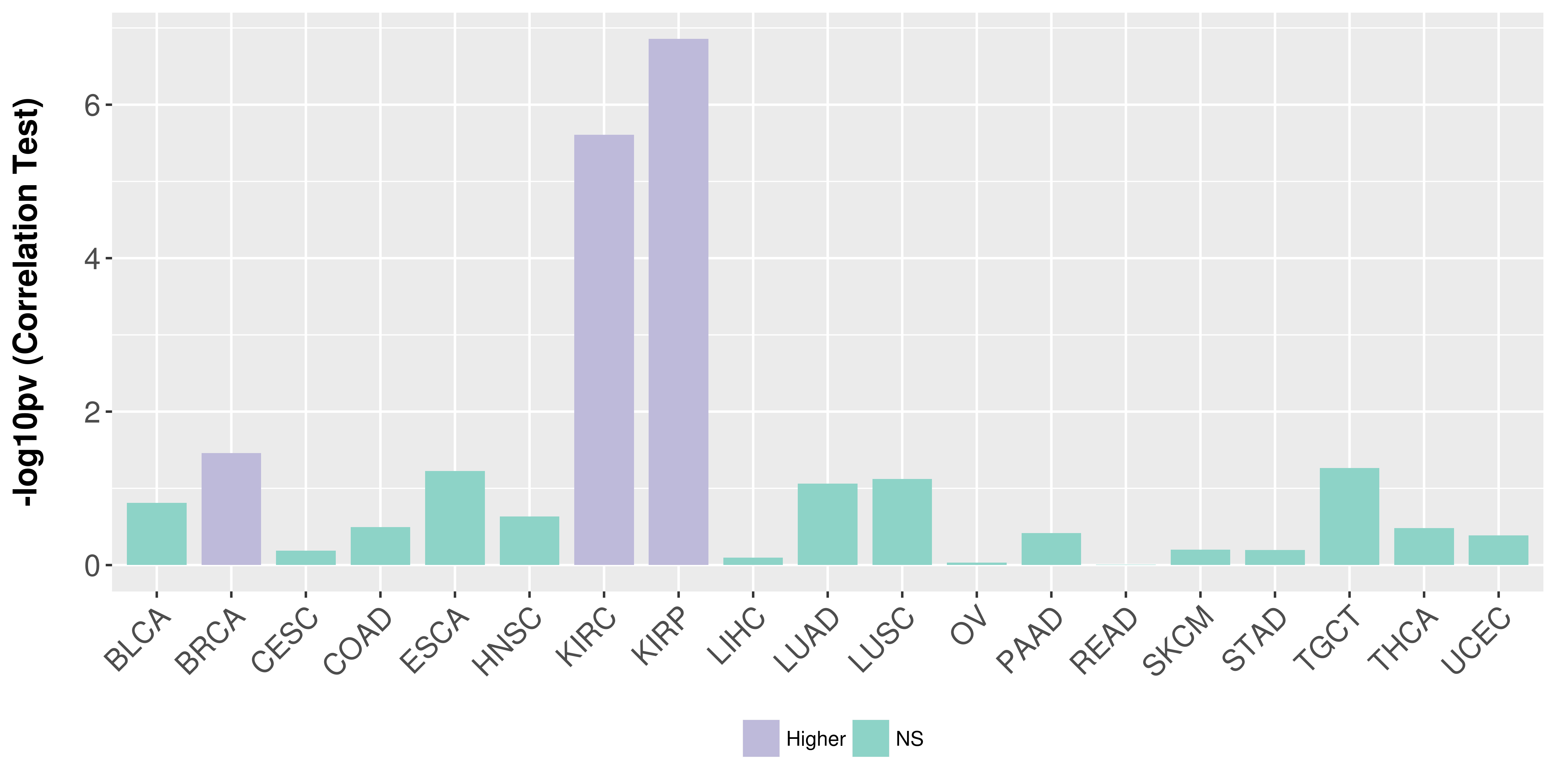
Association between expresson and stage.
|
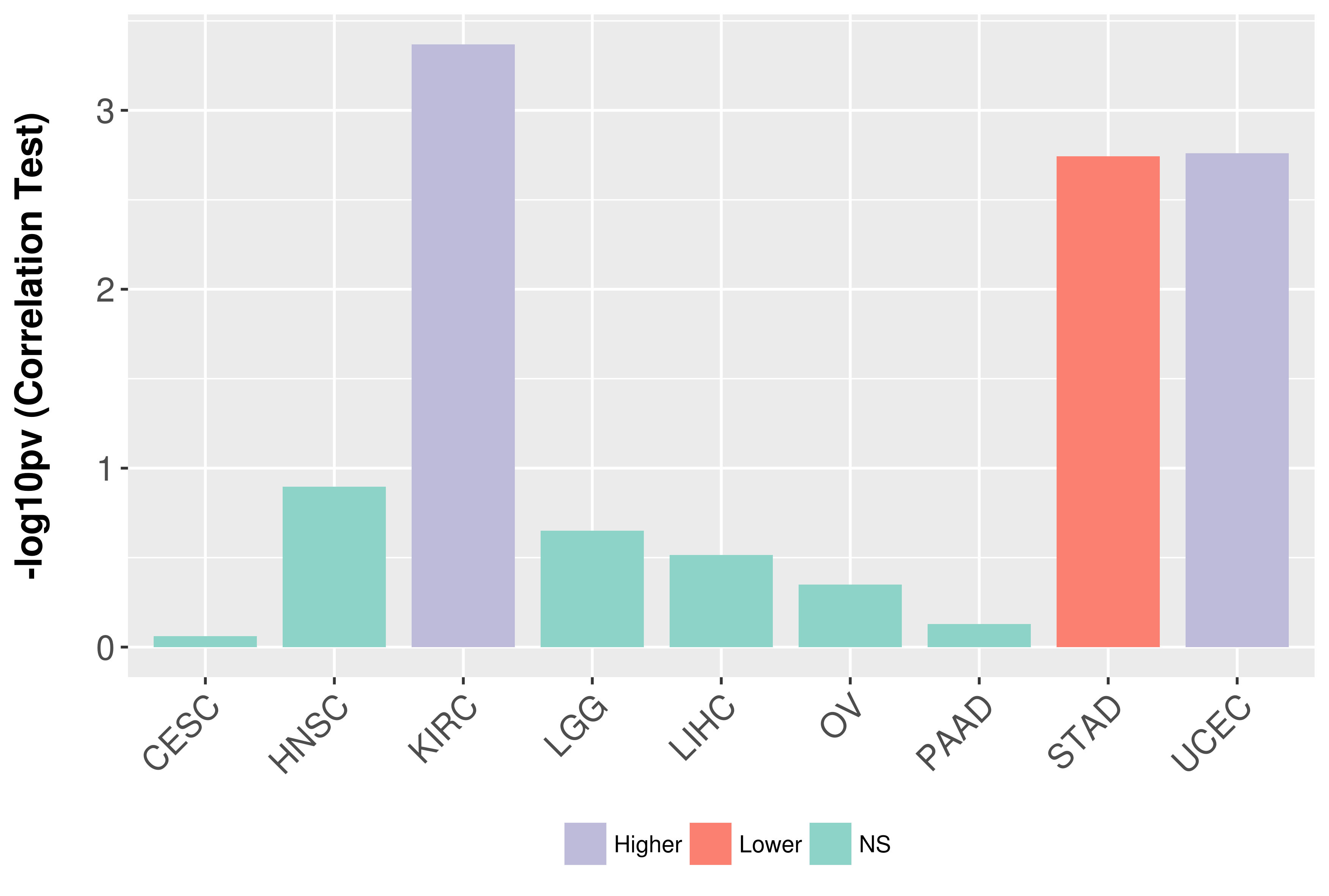
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for MORF4L2. |
| Summary | |
|---|---|
| Symbol | MORF4L2 |
| Name | mortality factor 4 like 2 |
| Aliases | KIAA0026; MRGX; MORF-related gene X; MORFL2; MORF-related gene X protein; MSL3-2 protein; protein MSL3-2; tr ...... |
| Location | Xq22.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for MORF4L2. |