Browse MSH6 in pancancer
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF01624 MutS domain I PF05188 MutS domain II PF05192 MutS domain III PF05190 MutS family domain IV PF00488 MutS domain V PF00855 PWWP domain |
||||||||||
| Function |
Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MSH2 to form MutS alpha, which binds to DNA mismatches thereby initiating DNA repair. When bound, MutS alpha bends the DNA helix and shields approximately 20 base pairs, and recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. After mismatch binding, forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. Recruited on chromatin in G1 and early S phase via its PWWP domain that specifically binds trimethylated 'Lys-36' of histone H3 (H3K36me3): early recruitment to chromatin to be replicated allowing a quick identification of mismatch repair to initiate the DNA mismatch repair reaction. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000018 regulation of DNA recombination GO:0000710 meiotic mismatch repair GO:0002200 somatic diversification of immune receptors GO:0002204 somatic recombination of immunoglobulin genes involved in immune response GO:0002208 somatic diversification of immunoglobulins involved in immune response GO:0002250 adaptive immune response GO:0002263 cell activation involved in immune response GO:0002285 lymphocyte activation involved in immune response GO:0002312 B cell activation involved in immune response GO:0002366 leukocyte activation involved in immune response GO:0002377 immunoglobulin production GO:0002381 immunoglobulin production involved in immunoglobulin mediated immune response GO:0002440 production of molecular mediator of immune response GO:0002443 leukocyte mediated immunity GO:0002449 lymphocyte mediated immunity GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002562 somatic diversification of immune receptors via germline recombination within a single locus GO:0002566 somatic diversification of immune receptors via somatic mutation GO:0002637 regulation of immunoglobulin production GO:0002639 positive regulation of immunoglobulin production GO:0002694 regulation of leukocyte activation GO:0002696 positive regulation of leukocyte activation GO:0002697 regulation of immune effector process GO:0002699 positive regulation of immune effector process GO:0002700 regulation of production of molecular mediator of immune response GO:0002702 positive regulation of production of molecular mediator of immune response GO:0002703 regulation of leukocyte mediated immunity GO:0002705 positive regulation of leukocyte mediated immunity GO:0002706 regulation of lymphocyte mediated immunity GO:0002708 positive regulation of lymphocyte mediated immunity GO:0002712 regulation of B cell mediated immunity GO:0002714 positive regulation of B cell mediated immunity GO:0002819 regulation of adaptive immune response GO:0002821 positive regulation of adaptive immune response GO:0002822 regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002824 positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002889 regulation of immunoglobulin mediated immune response GO:0002891 positive regulation of immunoglobulin mediated immune response GO:0006298 mismatch repair GO:0006310 DNA recombination GO:0007126 meiotic nuclear division GO:0007127 meiosis I GO:0007131 reciprocal meiotic recombination GO:0007568 aging GO:0008340 determination of adult lifespan GO:0008630 intrinsic apoptotic signaling pathway in response to DNA damage GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0010259 multicellular organism aging GO:0016064 immunoglobulin mediated immune response GO:0016444 somatic cell DNA recombination GO:0016445 somatic diversification of immunoglobulins GO:0016446 somatic hypermutation of immunoglobulin genes GO:0016447 somatic recombination of immunoglobulin gene segments GO:0019724 B cell mediated immunity GO:0035825 reciprocal DNA recombination GO:0042113 B cell activation GO:0045190 isotype switching GO:0045191 regulation of isotype switching GO:0045830 positive regulation of isotype switching GO:0045910 negative regulation of DNA recombination GO:0045911 positive regulation of DNA recombination GO:0050864 regulation of B cell activation GO:0050865 regulation of cell activation GO:0050867 positive regulation of cell activation GO:0050871 positive regulation of B cell activation GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051095 regulation of helicase activity GO:0051096 positive regulation of helicase activity GO:0051249 regulation of lymphocyte activation GO:0051251 positive regulation of lymphocyte activation GO:0051321 meiotic cell cycle GO:0097193 intrinsic apoptotic signaling pathway GO:1903046 meiotic cell cycle process |
| Molecular Function |
GO:0000217 DNA secondary structure binding GO:0000287 magnesium ion binding GO:0000400 four-way junction DNA binding GO:0003682 chromatin binding GO:0003684 damaged DNA binding GO:0016887 ATPase activity GO:0030983 mismatched DNA binding GO:0032135 DNA insertion or deletion binding GO:0032137 guanine/thymine mispair binding GO:0032138 single base insertion or deletion binding GO:0032142 single guanine insertion binding GO:0032143 single thymine insertion binding GO:0032356 oxidized DNA binding GO:0032357 oxidized purine DNA binding GO:0032404 mismatch repair complex binding GO:0032405 MutLalpha complex binding GO:0035064 methylated histone binding GO:0042393 histone binding GO:0043531 ADP binding GO:0043566 structure-specific DNA binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0032300 mismatch repair complex GO:0032301 MutSalpha complex GO:0044454 nuclear chromosome part GO:1990391 DNA repair complex |
| KEGG |
hsa03430 Mismatch repair |
| Reactome |
R-HSA-73894: DNA Repair R-HSA-5358508: Mismatch Repair R-HSA-5358565: Mismatch repair (MMR) directed by MSH2 |
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
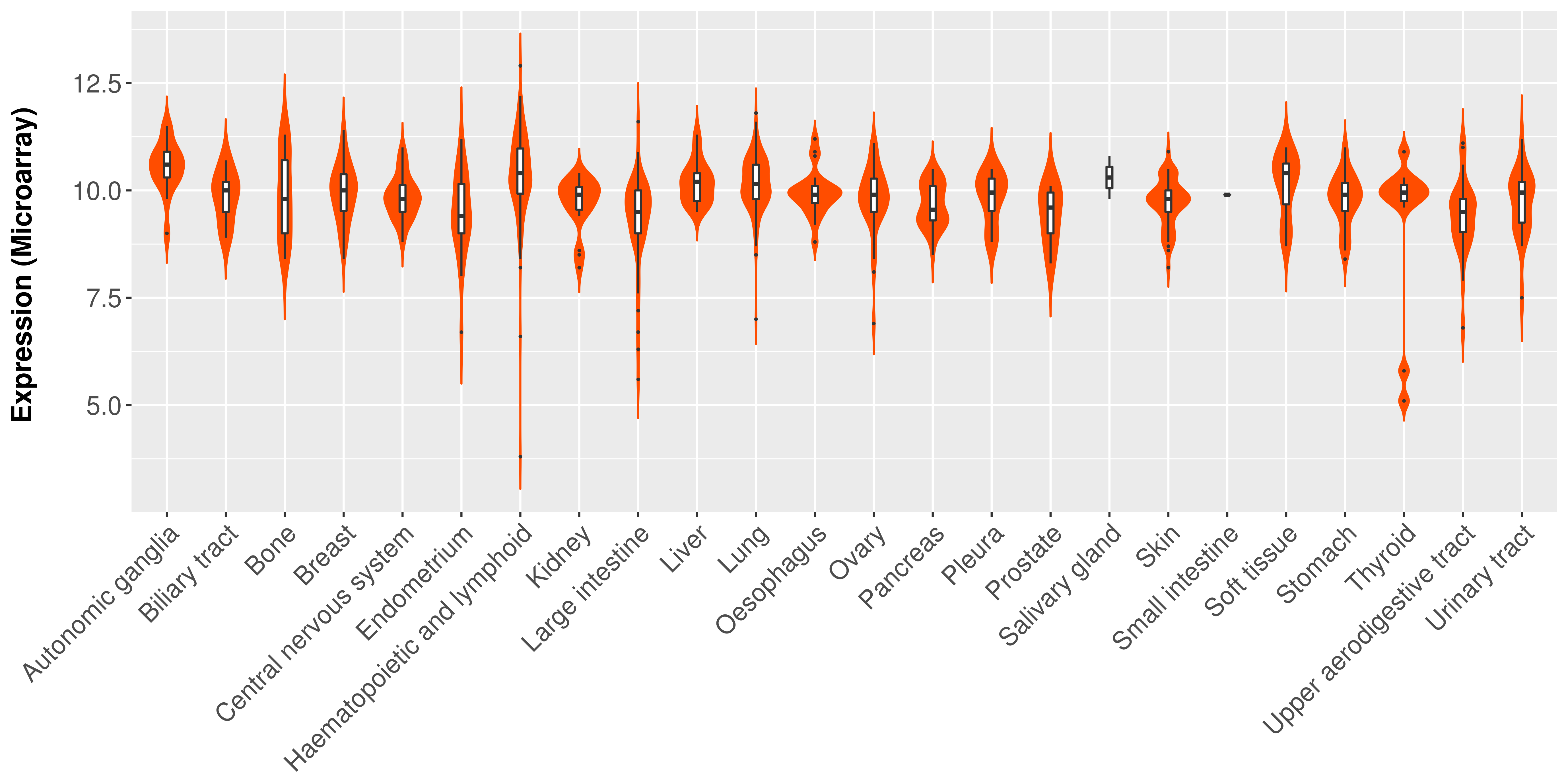
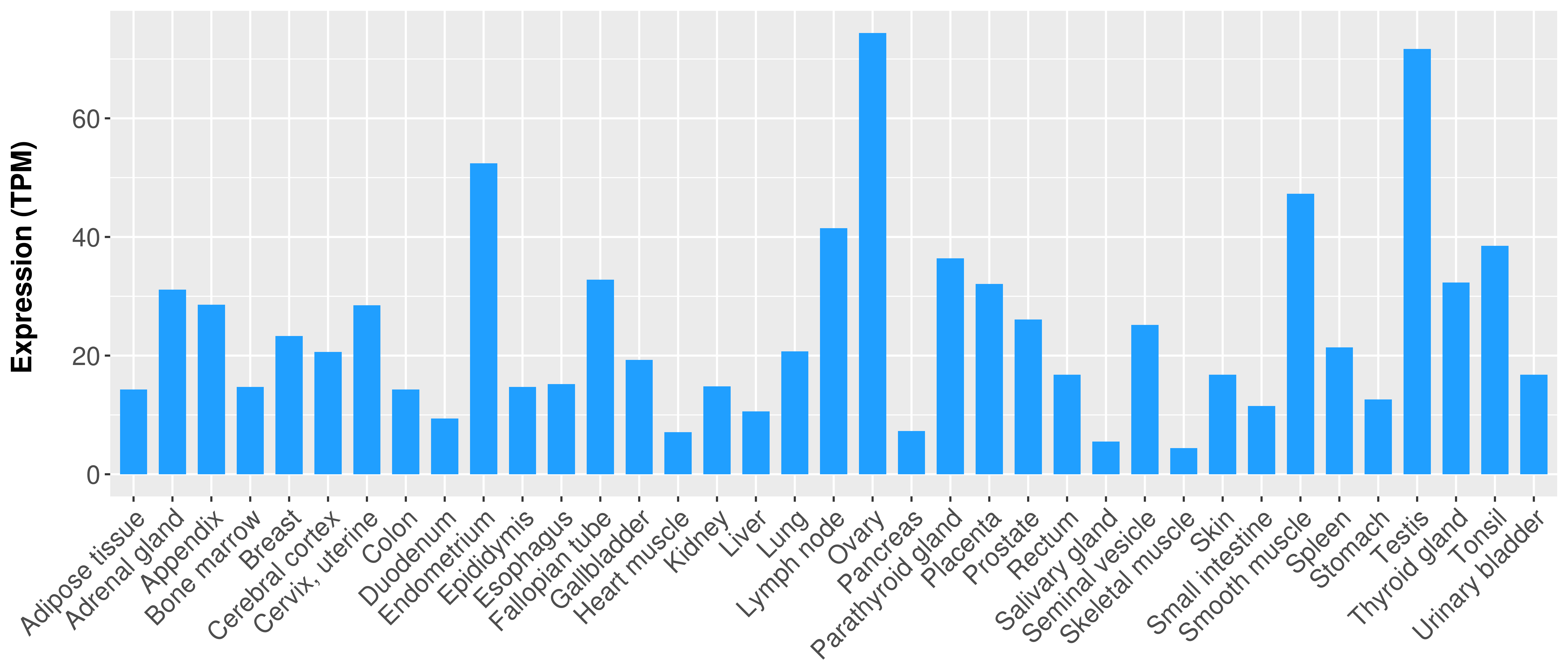
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
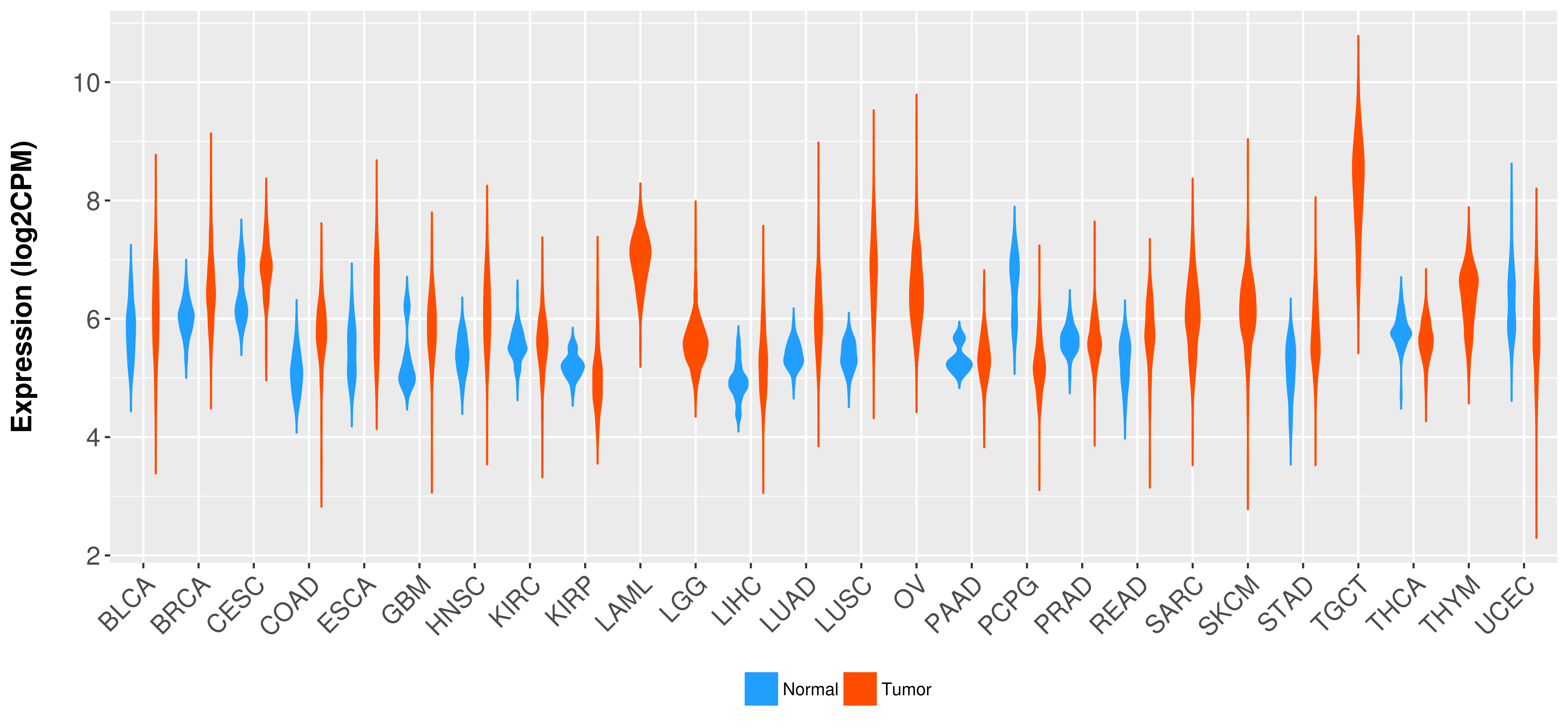
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
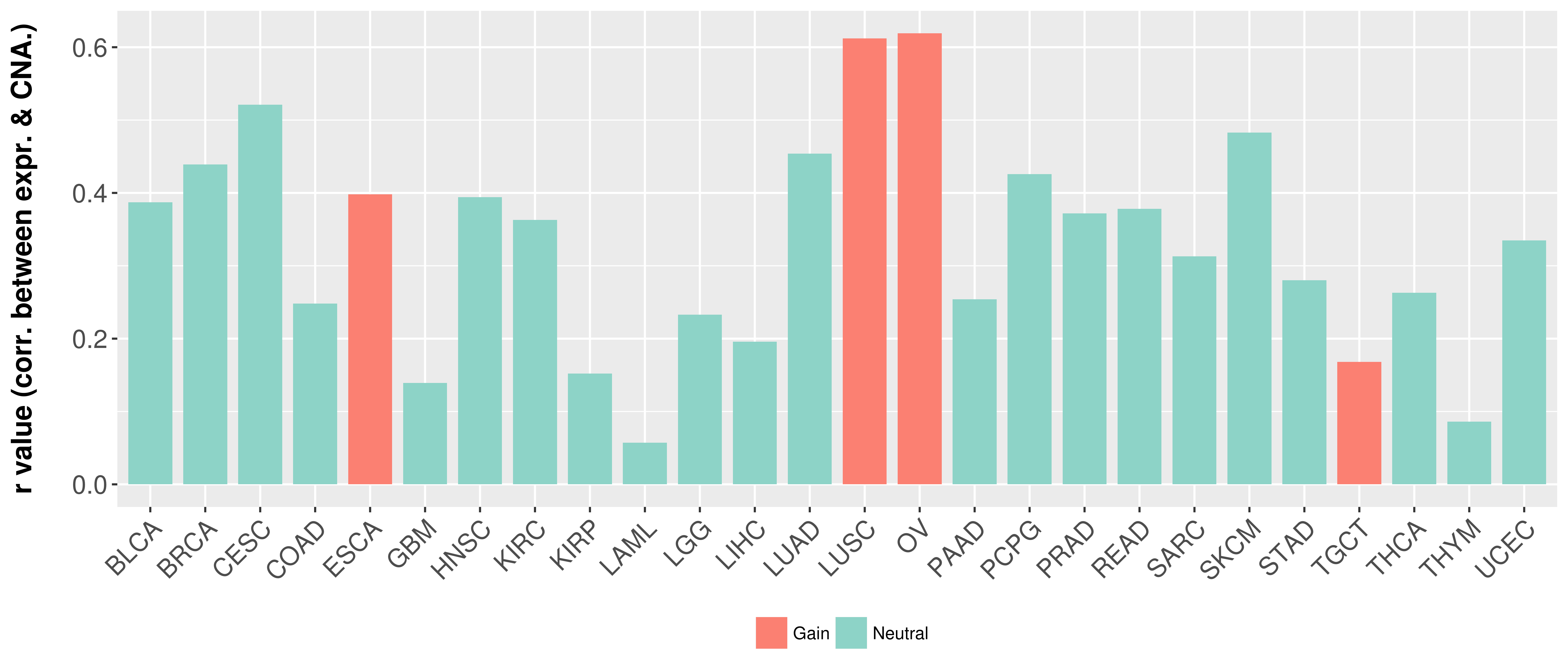
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
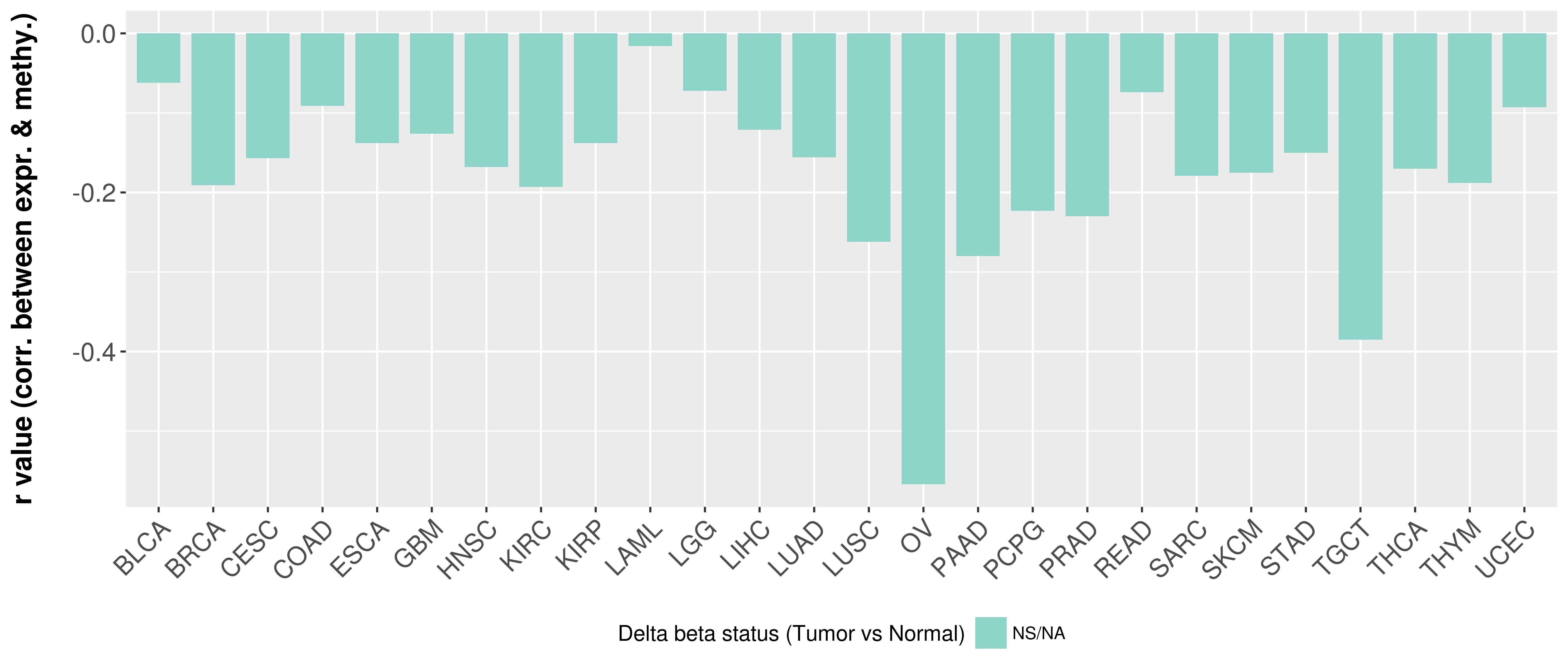
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
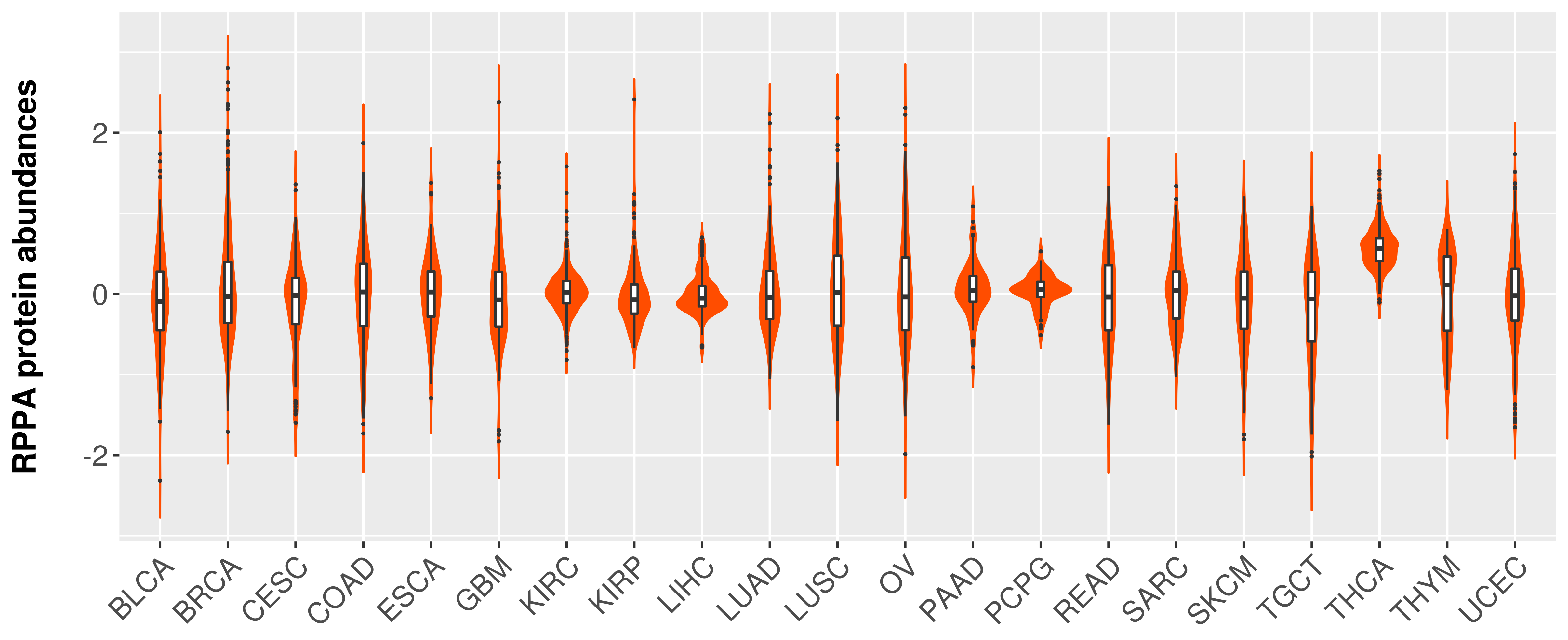
|
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
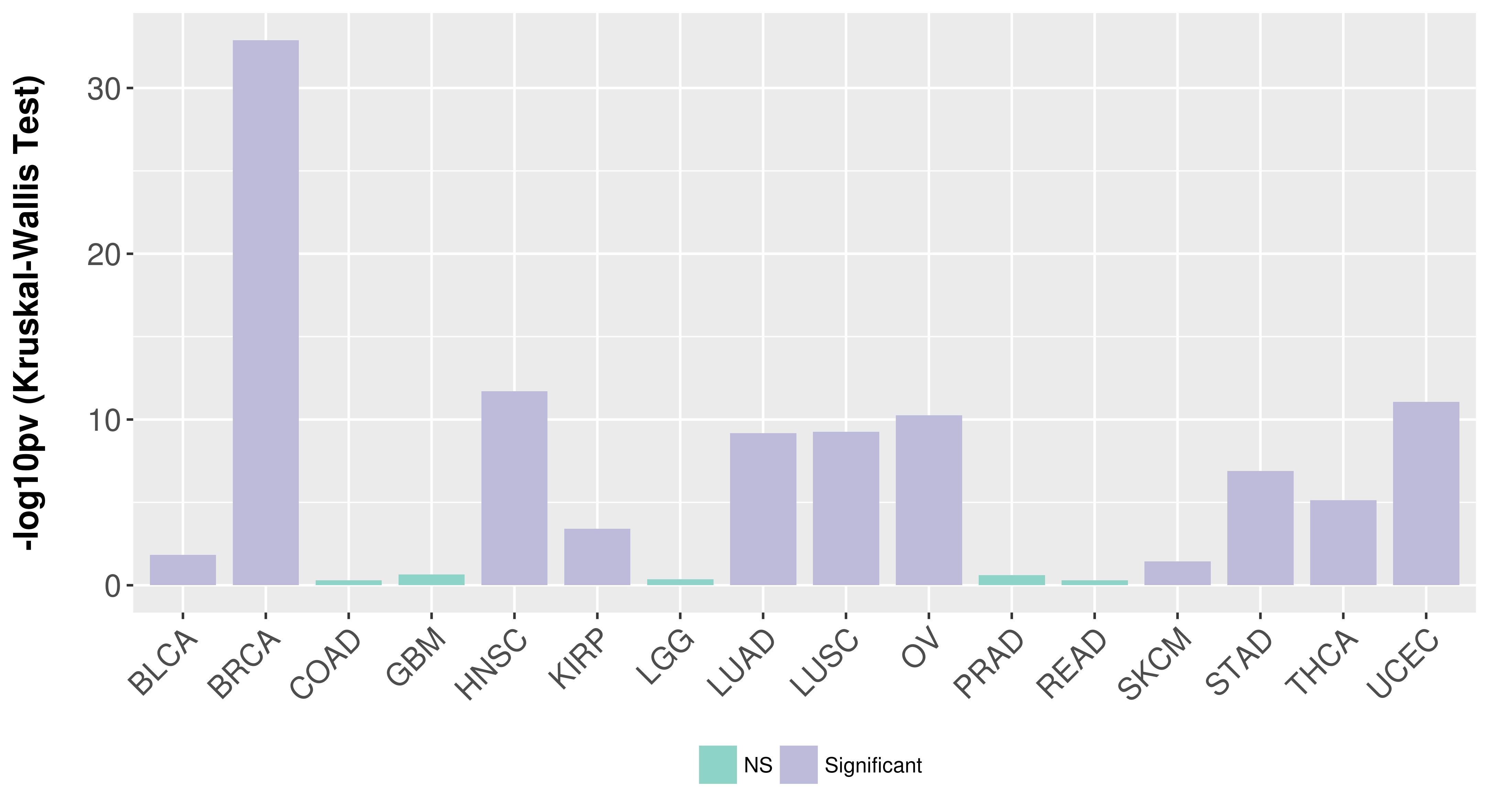
Association between expresson and subtype.
|
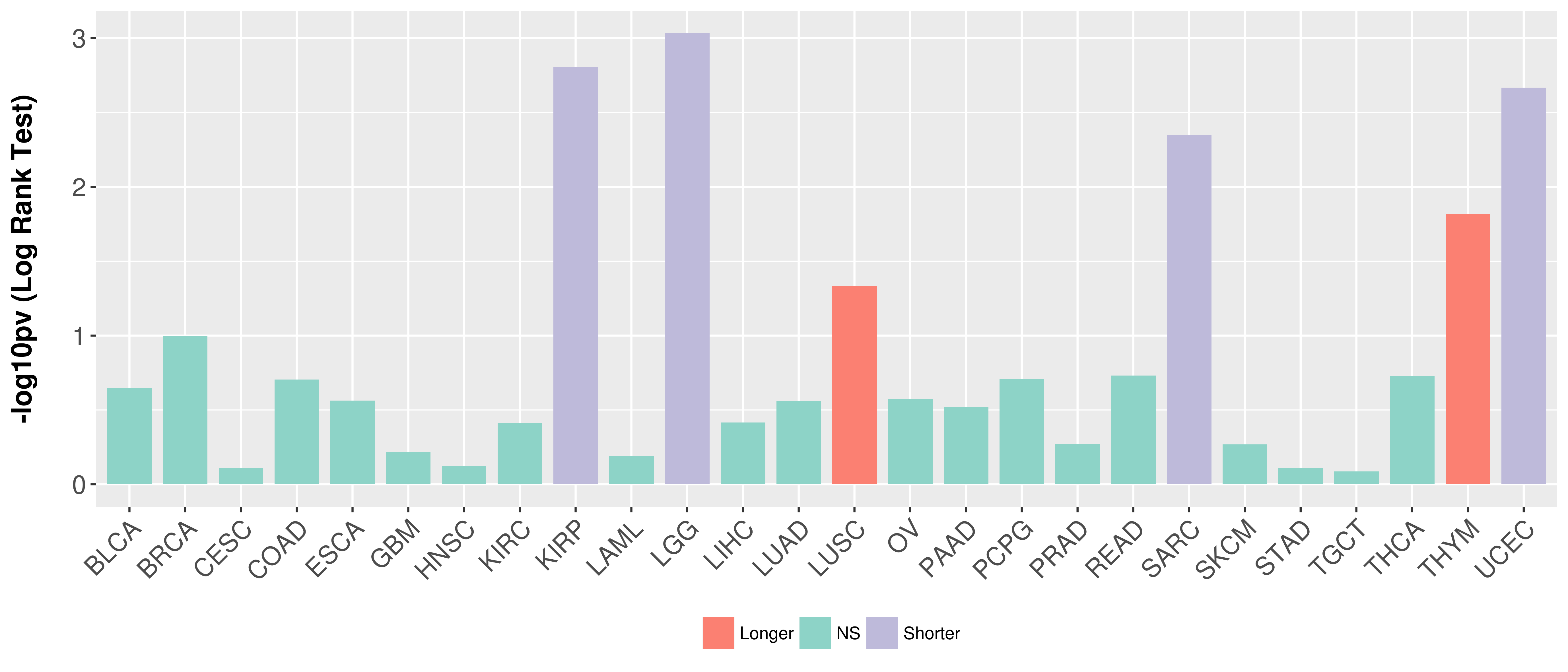
Overall survival analysis based on expression.
|
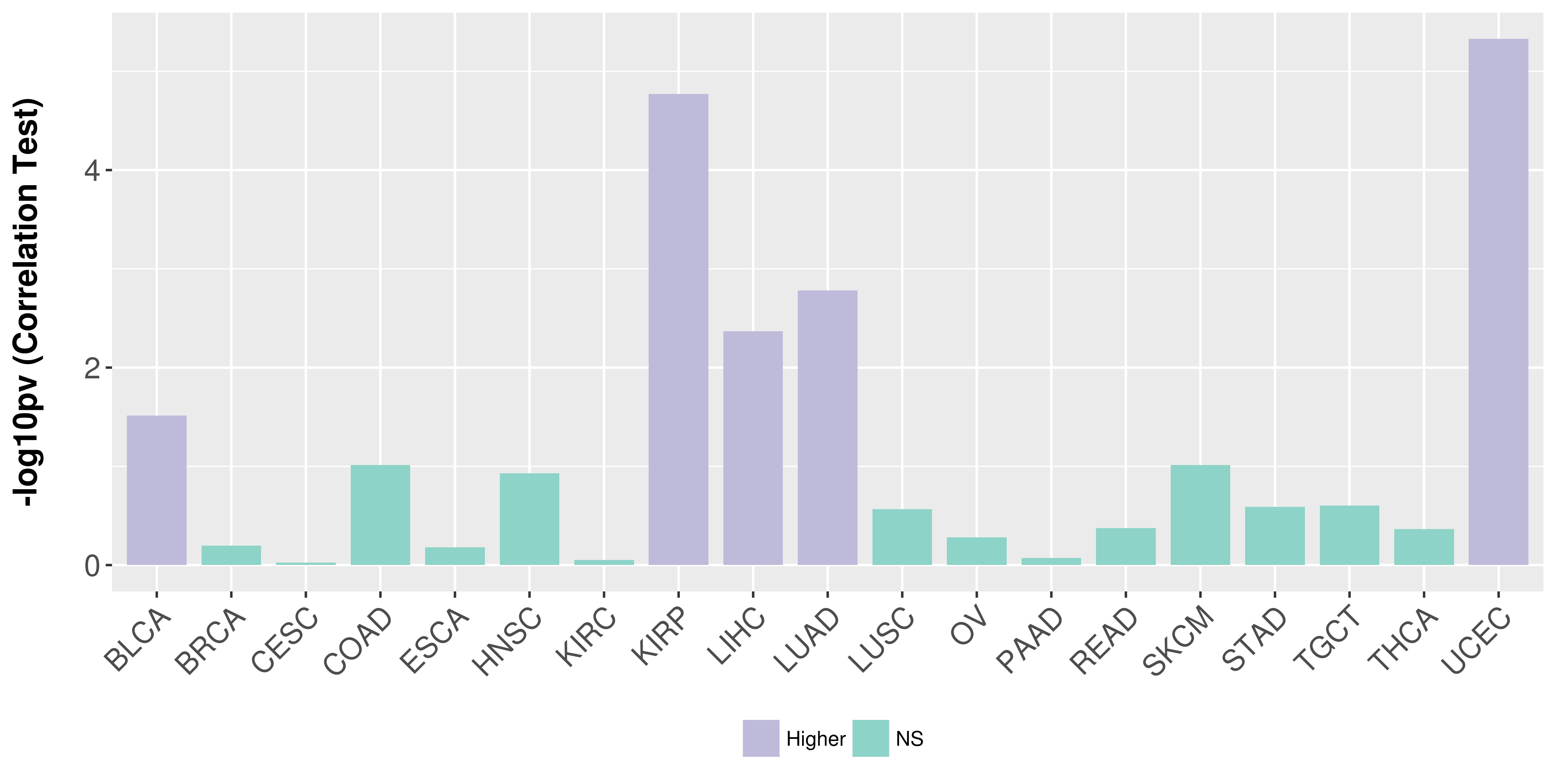
Association between expresson and stage.
|
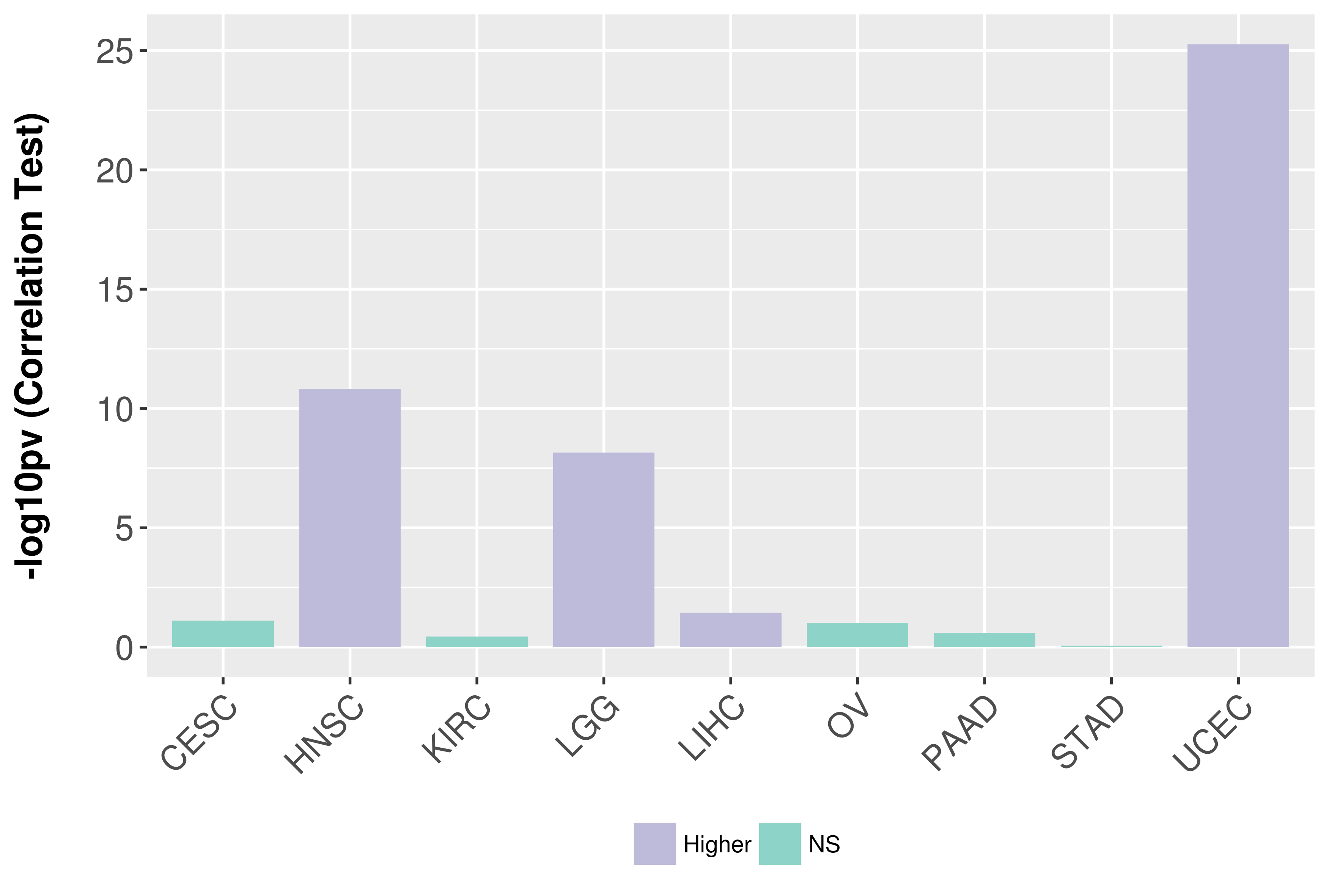
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for MSH6. |
| Summary | |
|---|---|
| Symbol | MSH6 |
| Name | mutS homolog 6 |
| Aliases | GTBP; mutS (E. coli) homolog 6; mutS homolog 6 (E. coli); GTMBP; HNPCC5; HSAP; G/T mismatch-binding protein; ...... |
| Location | 2p16.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|