Browse NCOA1 in pancancer
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF07469 Domain of unknown function (DUF1518) PF00010 Helix-loop-helix DNA-binding domain PF08815 Nuclear receptor coactivator PF00989 PAS fold PF08832 Steroid receptor coactivator |
||||||||||
| Function |
Nuclear receptor coactivator that directly binds nuclear receptors and stimulates the transcriptional activities in a hormone-dependent fashion. Involved in the coactivation of different nuclear receptors, such as for steroids (PGR, GR and ER), retinoids (RXRs), thyroid hormone (TRs) and prostanoids (PPARs). Also involved in coactivation mediated by STAT3, STAT5A, STAT5B and STAT6 transcription factors. Displays histone acetyltransferase activity toward H3 and H4; the relevance of such activity remains however unclear. Plays a central role in creating multisubunit coactivator complexes that act via remodeling of chromatin, and possibly acts by participating in both chromatin remodeling and recruitment of general transcription factors. Required with NCOA2 to control energy balance between white and brown adipose tissues. Required for mediating steroid hormone response. Isoform 2 has a higher thyroid hormone-dependent transactivation activity than isoform 1 and isoform 3. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000409 regulation of transcription by galactose GO:0000411 positive regulation of transcription by galactose GO:0000429 carbon catabolite regulation of transcription from RNA polymerase II promoter GO:0000431 regulation of transcription from RNA polymerase II promoter by galactose GO:0000435 positive regulation of transcription from RNA polymerase II promoter by galactose GO:0000436 carbon catabolite activation of transcription from RNA polymerase II promoter GO:0001101 response to acid chemical GO:0001701 in utero embryonic development GO:0001890 placenta development GO:0001892 embryonic placenta development GO:0002154 thyroid hormone mediated signaling pathway GO:0002155 regulation of thyroid hormone mediated signaling pathway GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006820 anion transport GO:0006869 lipid transport GO:0007548 sex differentiation GO:0007584 response to nutrient GO:0007589 body fluid secretion GO:0007595 lactation GO:0007617 mating behavior GO:0007618 mating GO:0008406 gonad development GO:0008584 male gonad development GO:0009755 hormone-mediated signaling pathway GO:0009991 response to extracellular stimulus GO:0010720 positive regulation of cell development GO:0010876 lipid localization GO:0015711 organic anion transport GO:0015718 monocarboxylic acid transport GO:0015721 bile acid and bile salt transport GO:0015849 organic acid transport GO:0015850 organic hydroxy compound transport GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0019098 reproductive behavior GO:0021536 diencephalon development GO:0021537 telencephalon development GO:0021543 pallium development GO:0021549 cerebellum development GO:0021761 limbic system development GO:0021766 hippocampus development GO:0021854 hypothalamus development GO:0021987 cerebral cortex development GO:0022037 metencephalon development GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0030879 mammary gland development GO:0030900 forebrain development GO:0030902 hindbrain development GO:0031667 response to nutrient levels GO:0031668 cellular response to extracellular stimulus GO:0031669 cellular response to nutrient levels GO:0031670 cellular response to nutrient GO:0032355 response to estradiol GO:0032526 response to retinoic acid GO:0032570 response to progesterone GO:0032941 secretion by tissue GO:0035690 cellular response to drug GO:0042493 response to drug GO:0042698 ovulation cycle GO:0043401 steroid hormone mediated signaling pathway GO:0043543 protein acylation GO:0043967 histone H4 acetylation GO:0044849 estrous cycle GO:0045137 development of primary sexual characteristics GO:0045666 positive regulation of neuron differentiation GO:0045924 regulation of female receptivity GO:0045925 positive regulation of female receptivity GO:0045990 carbon catabolite regulation of transcription GO:0045991 carbon catabolite activation of transcription GO:0046546 development of primary male sexual characteristics GO:0046661 male sex differentiation GO:0046942 carboxylic acid transport GO:0048511 rhythmic process GO:0048545 response to steroid hormone GO:0048568 embryonic organ development GO:0048608 reproductive structure development GO:0048732 gland development GO:0050769 positive regulation of neurogenesis GO:0050878 regulation of body fluid levels GO:0051962 positive regulation of nervous system development GO:0060179 male mating behavior GO:0060180 female mating behavior GO:0060669 embryonic placenta morphogenesis GO:0060711 labyrinthine layer development GO:0060713 labyrinthine layer morphogenesis GO:0061458 reproductive system development GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071496 cellular response to external stimulus GO:1901522 positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus GO:1901654 response to ketone GO:1904016 response to Thyroglobulin triiodothyronine GO:1904017 cellular response to Thyroglobulin triiodothyronine GO:2001023 regulation of response to drug GO:2001038 regulation of cellular response to drug |
| Molecular Function |
GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001104 RNA polymerase II transcription cofactor activity GO:0001105 RNA polymerase II transcription coactivator activity GO:0001190 transcriptional activator activity, RNA polymerase II transcription factor binding GO:0001191 transcriptional repressor activity, RNA polymerase II transcription factor binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0008134 transcription factor binding GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0016922 ligand-dependent nuclear receptor binding GO:0030331 estrogen receptor binding GO:0030374 ligand-dependent nuclear receptor transcription coactivator activity GO:0033142 progesterone receptor binding GO:0034212 peptide N-acetyltransferase activity GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0042974 retinoic acid receptor binding GO:0046965 retinoid X receptor binding GO:0047485 protein N-terminus binding GO:0050681 androgen receptor binding GO:0051427 hormone receptor binding GO:0061733 peptide-lysine-N-acetyltransferase activity GO:0098811 transcriptional repressor activity, RNA polymerase II activating transcription factor binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0044454 nuclear chromosome part |
| KEGG |
hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-2426168: Activation of gene expression by SREBF (SREBP) R-HSA-1368108: BMAL1 R-HSA-194068: Bile acid and bile salt metabolism R-HSA-211859: Biological oxidations R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-211897: Cytochrome P450 - arranged by substrate type R-HSA-1266738: Developmental Biology R-HSA-211976: Endogenous sterols R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214847: HATs acetylate histones R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-1592230: Mitochondrial biogenesis R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-1989781: PPARA activates gene expression R-HSA-211945: Phase 1 - Functionalization of compounds R-HSA-1368082: RORA activates gene expression R-HSA-159418: Recycling of bile acids and salts R-HSA-1655829: Regulation of cholesterol biosynthesis by SREBP (SREBF) R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) R-HSA-192105: Synthesis of bile acids and bile salts R-HSA-193807: Synthesis of bile acids and bile salts via 27-hydroxycholesterol R-HSA-193368: Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis R-HSA-381340: Transcriptional regulation of white adipocyte differentiation R-HSA-2032785: YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
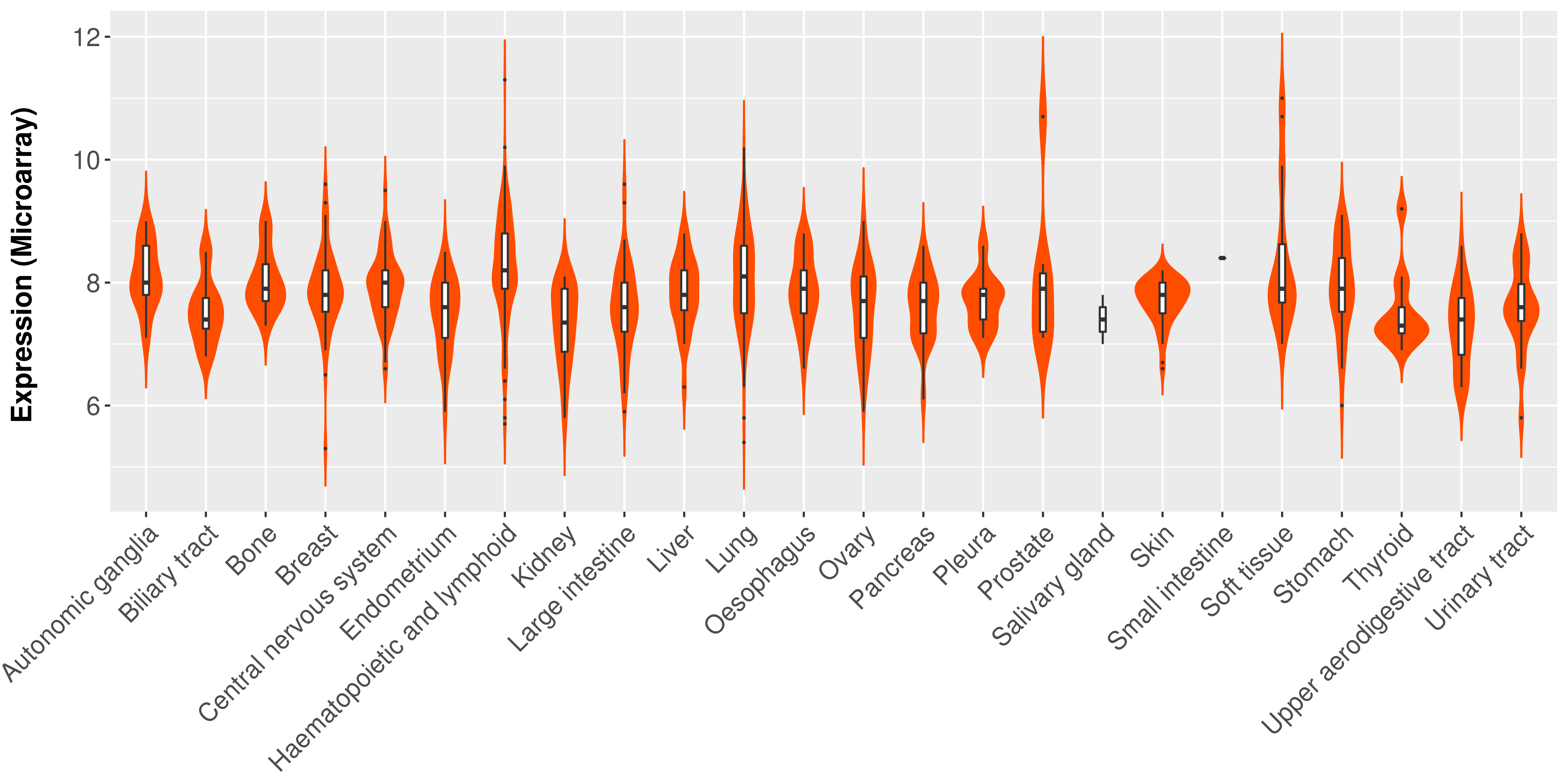
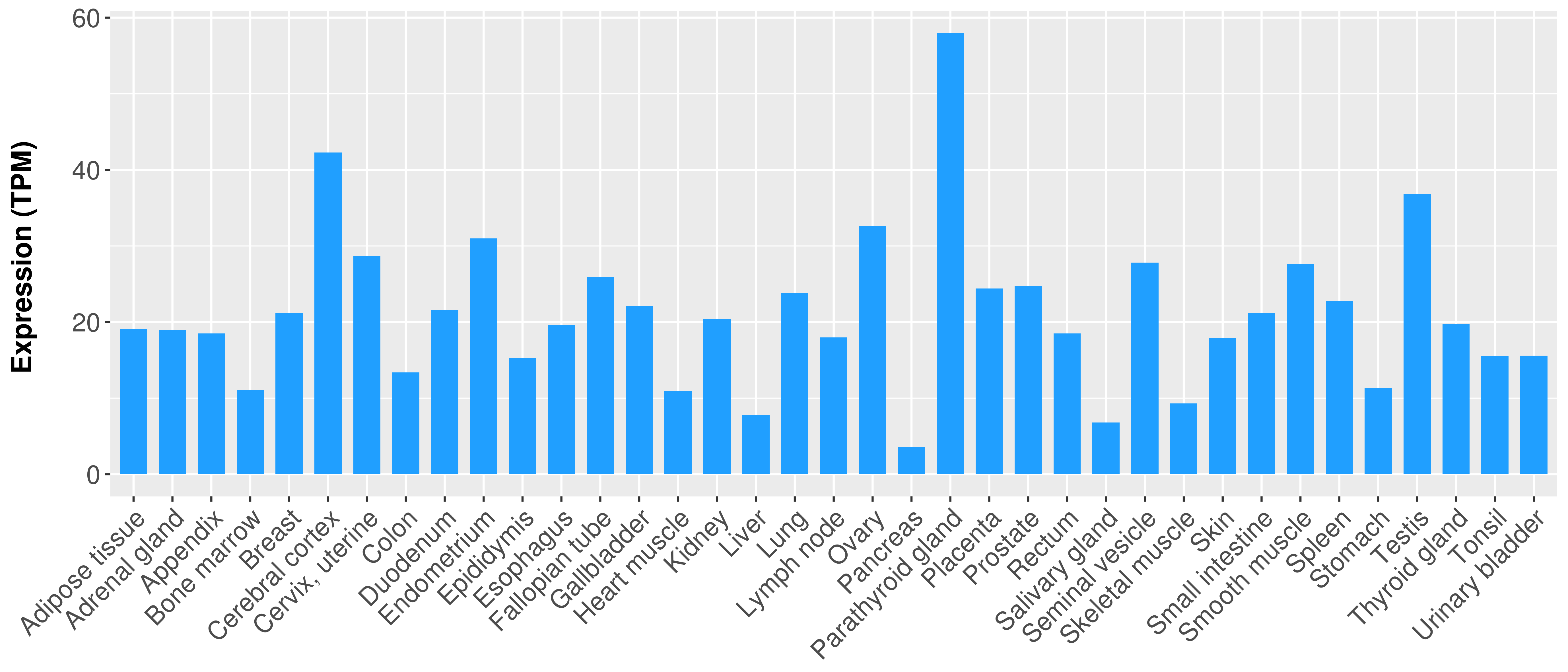
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
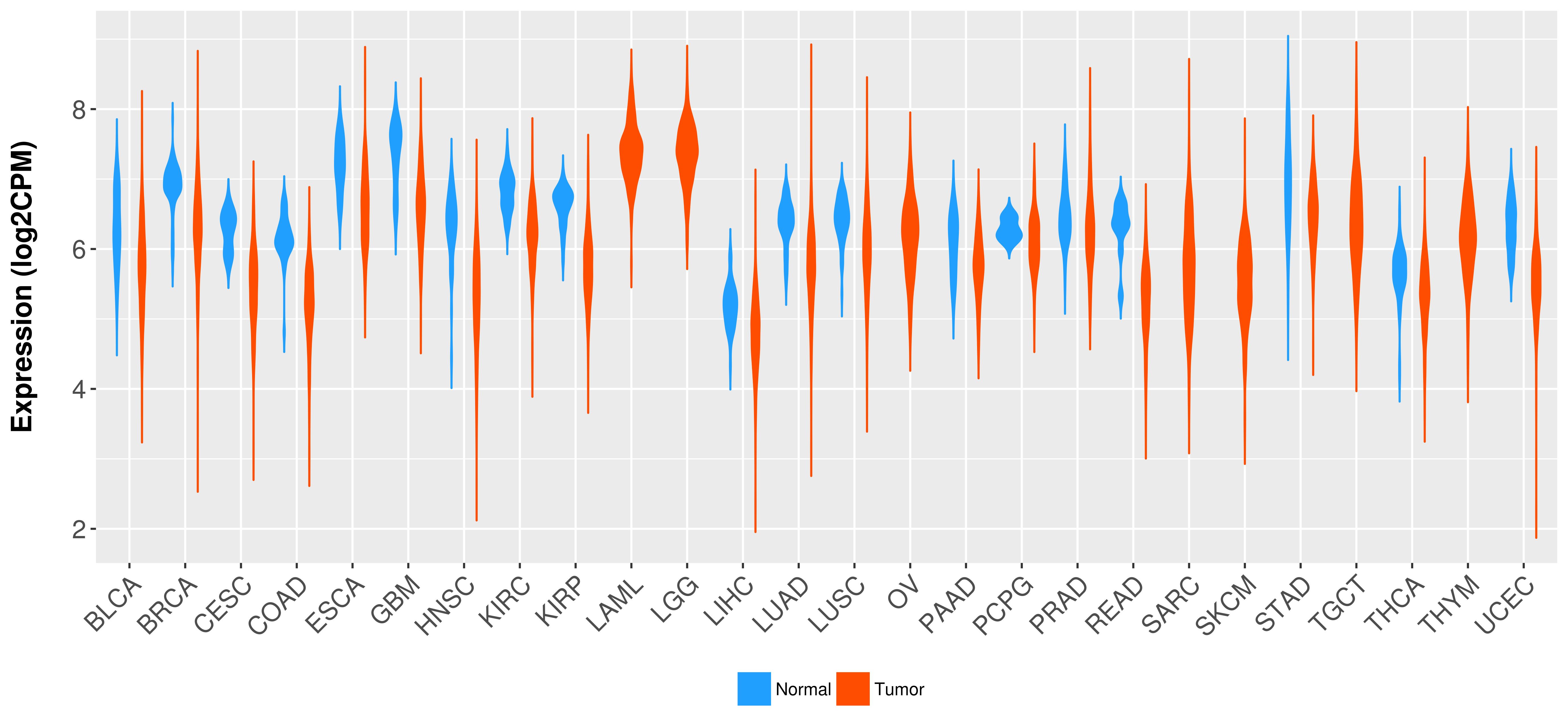
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
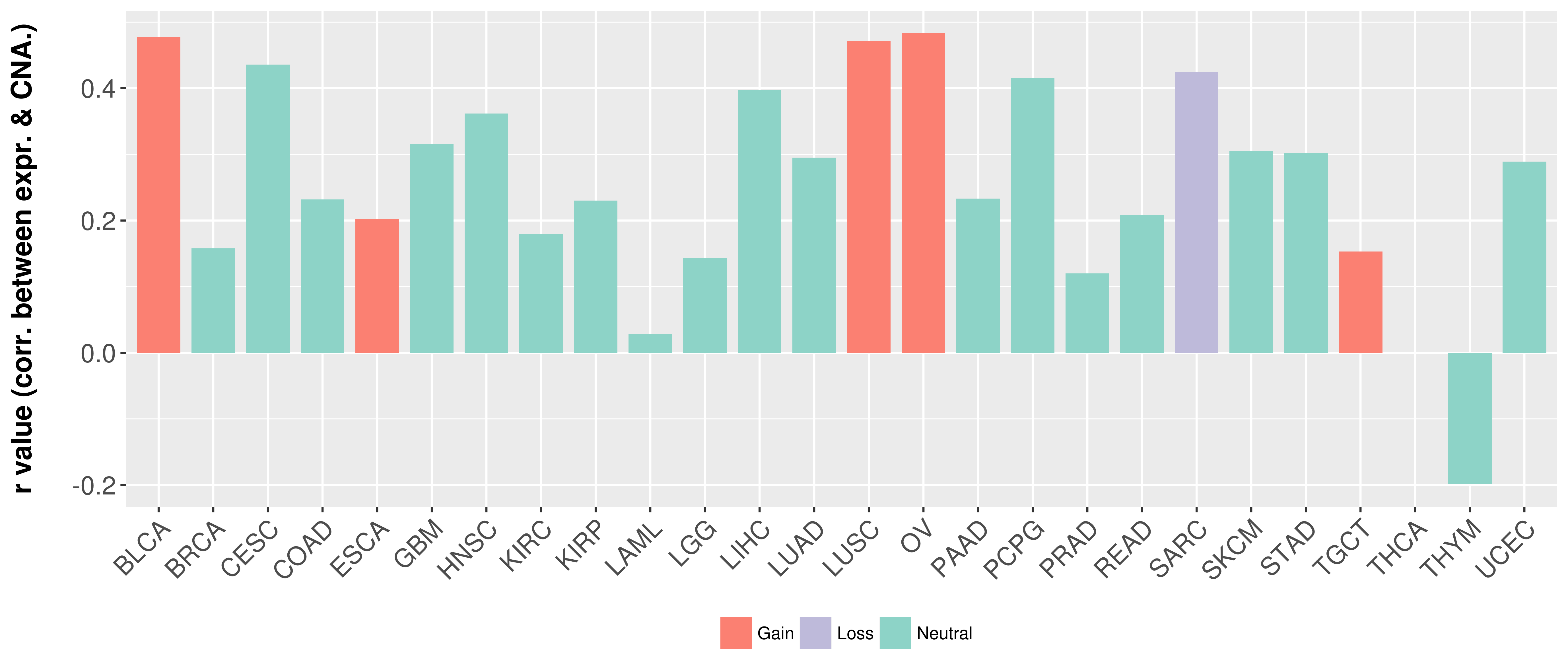
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
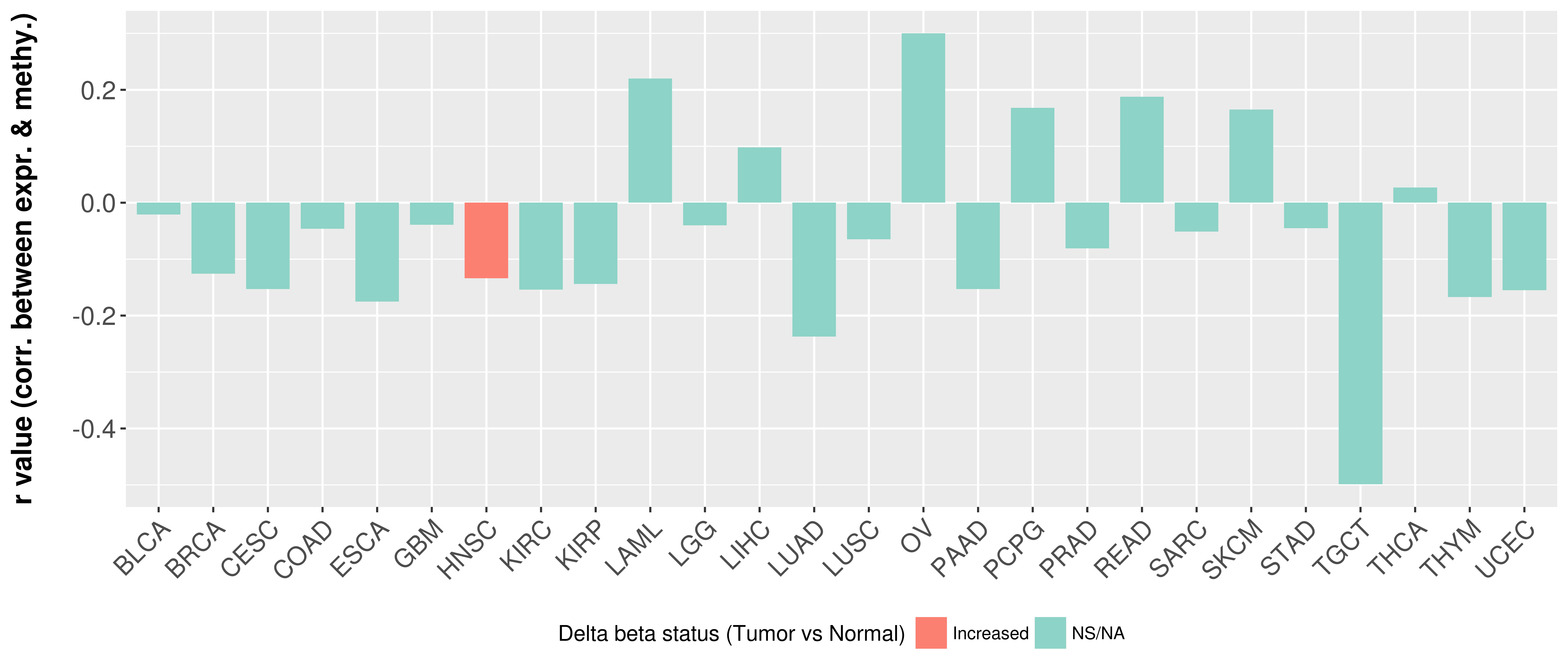
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |

|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
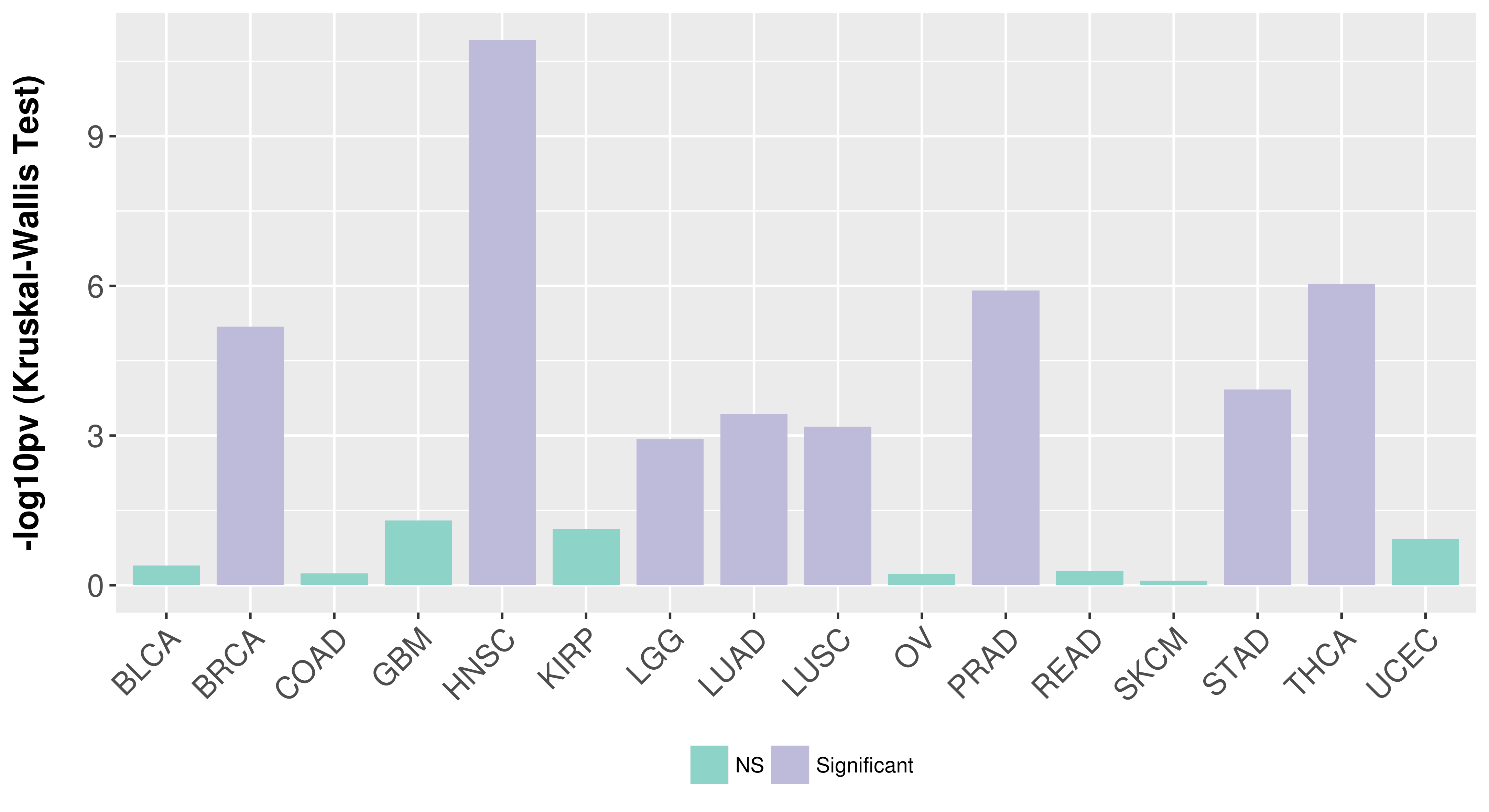
Association between expresson and subtype.
|
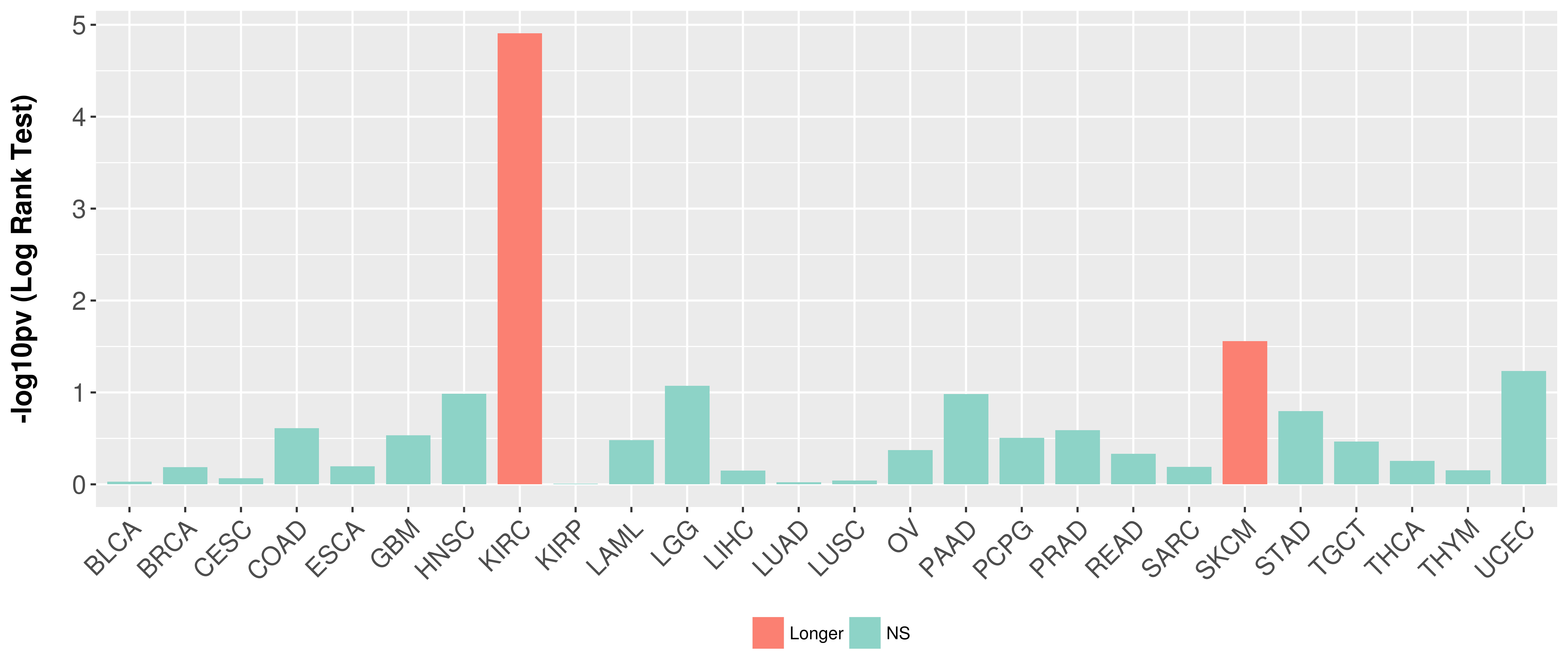
Overall survival analysis based on expression.
|
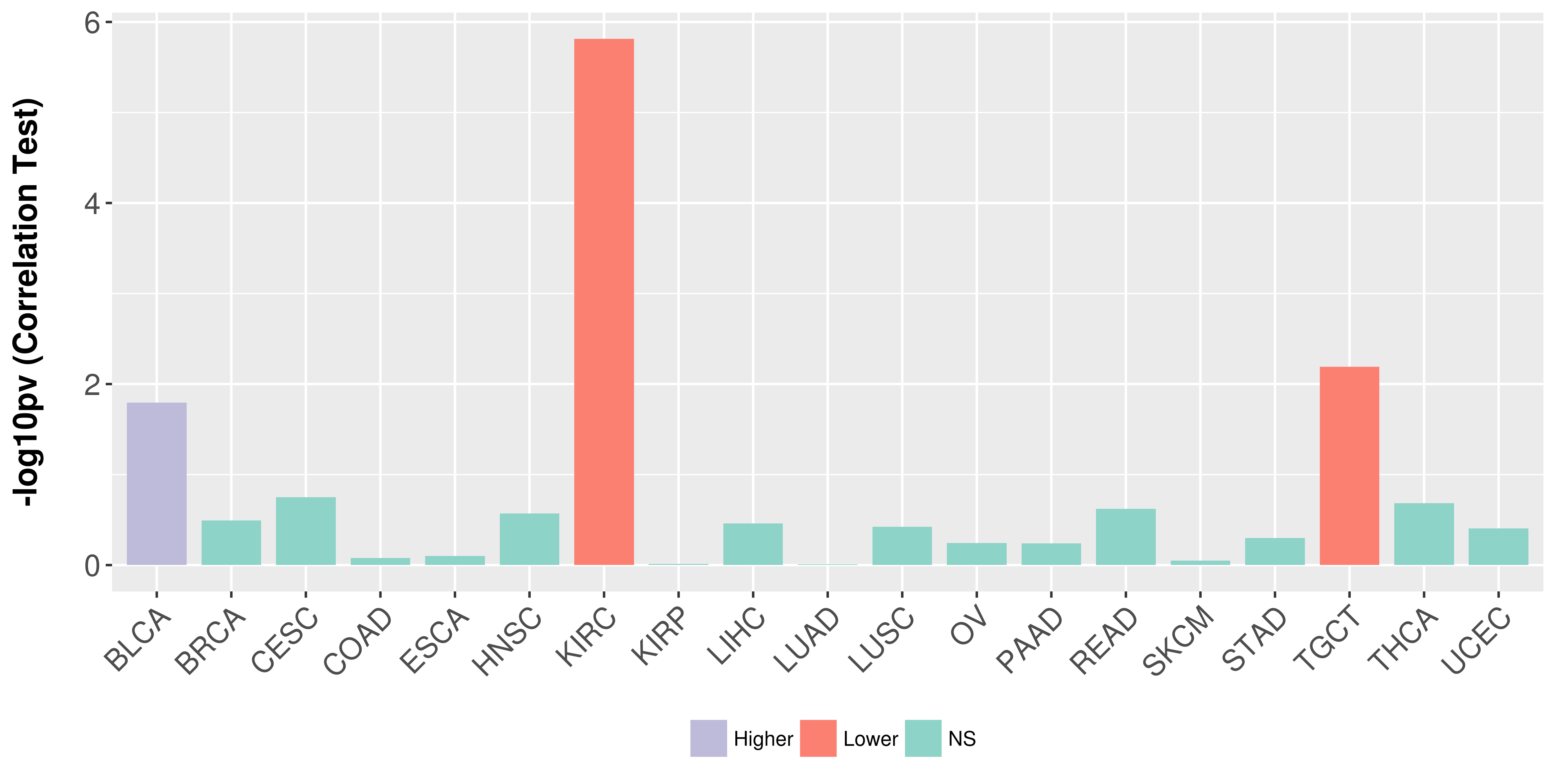
Association between expresson and stage.
|
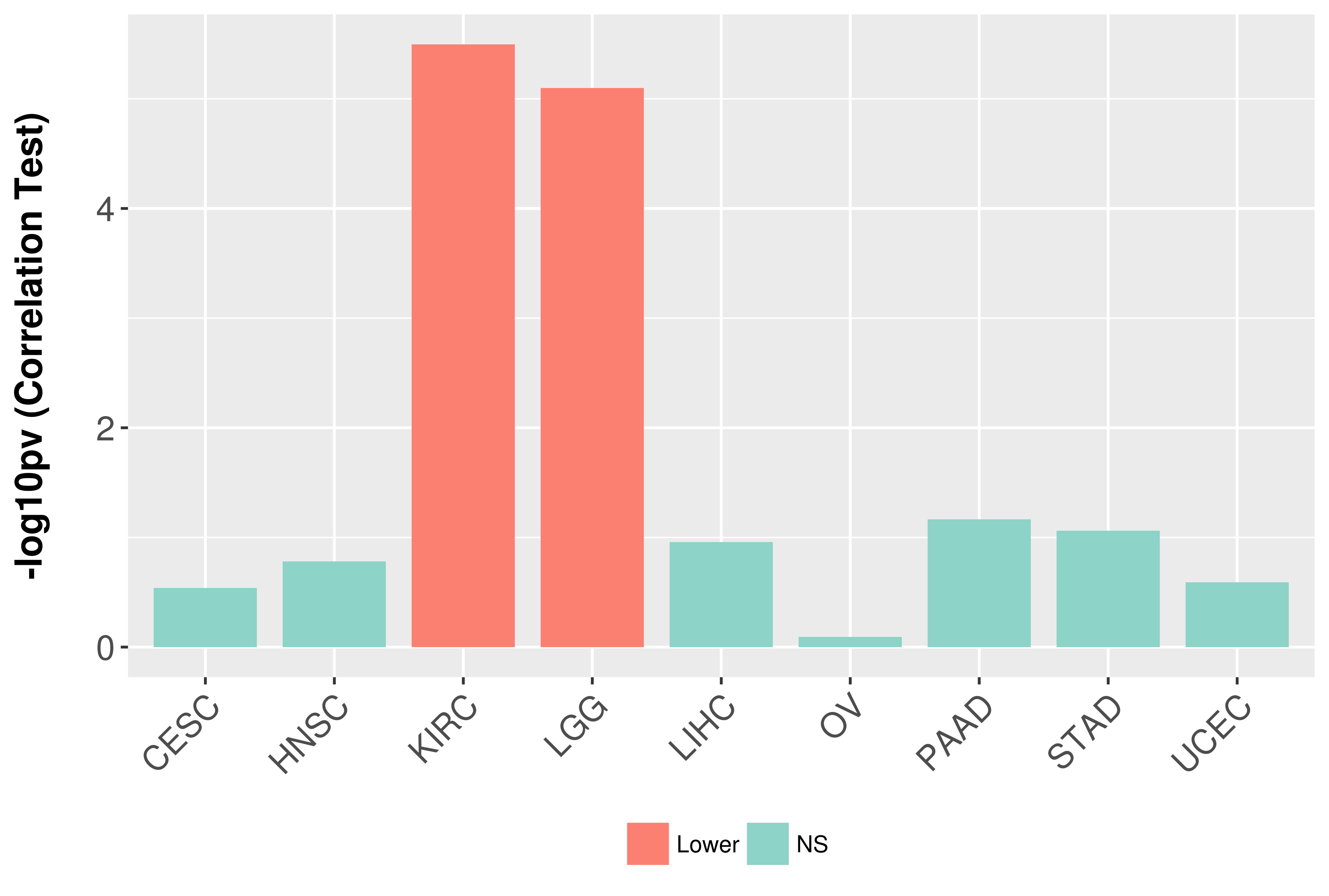
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
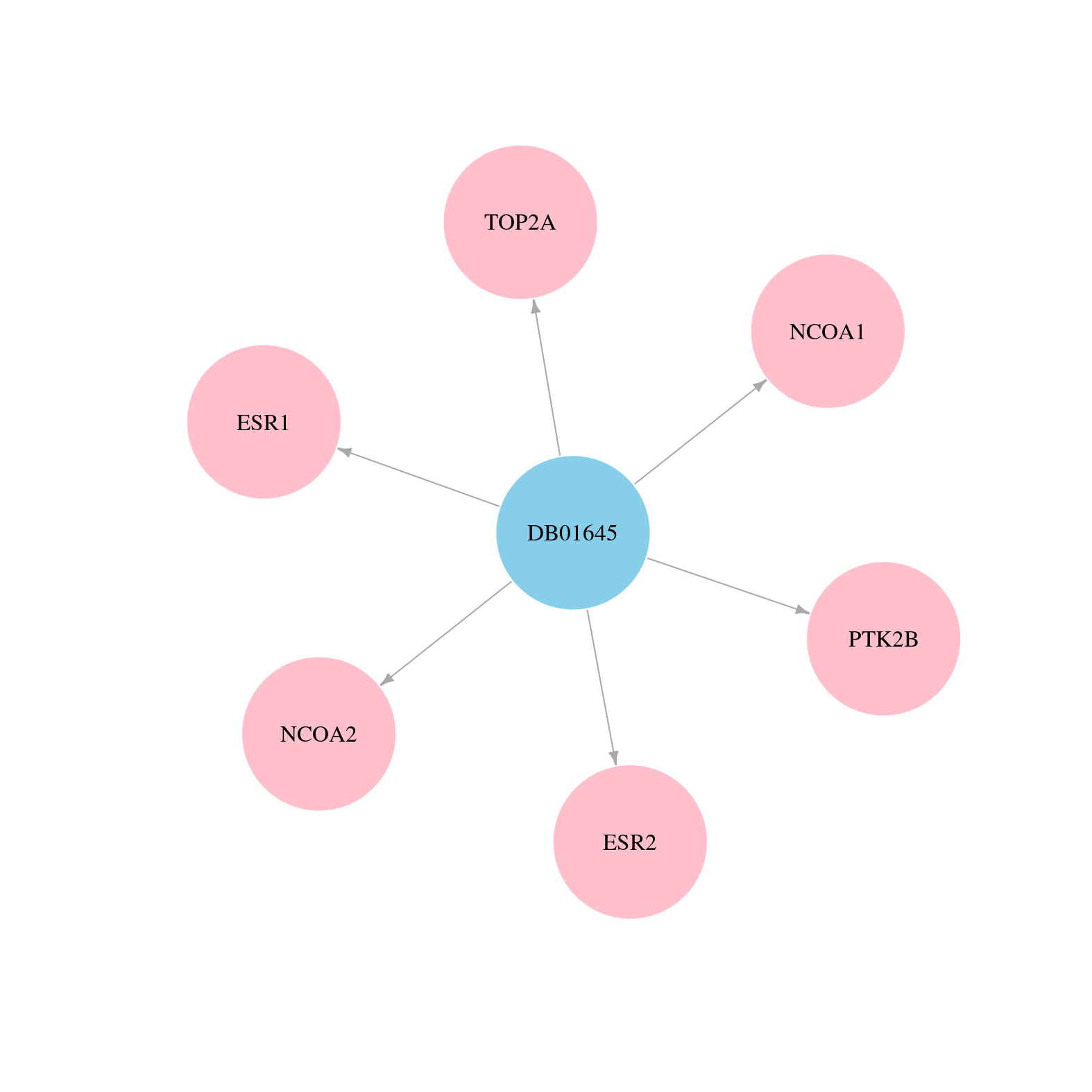
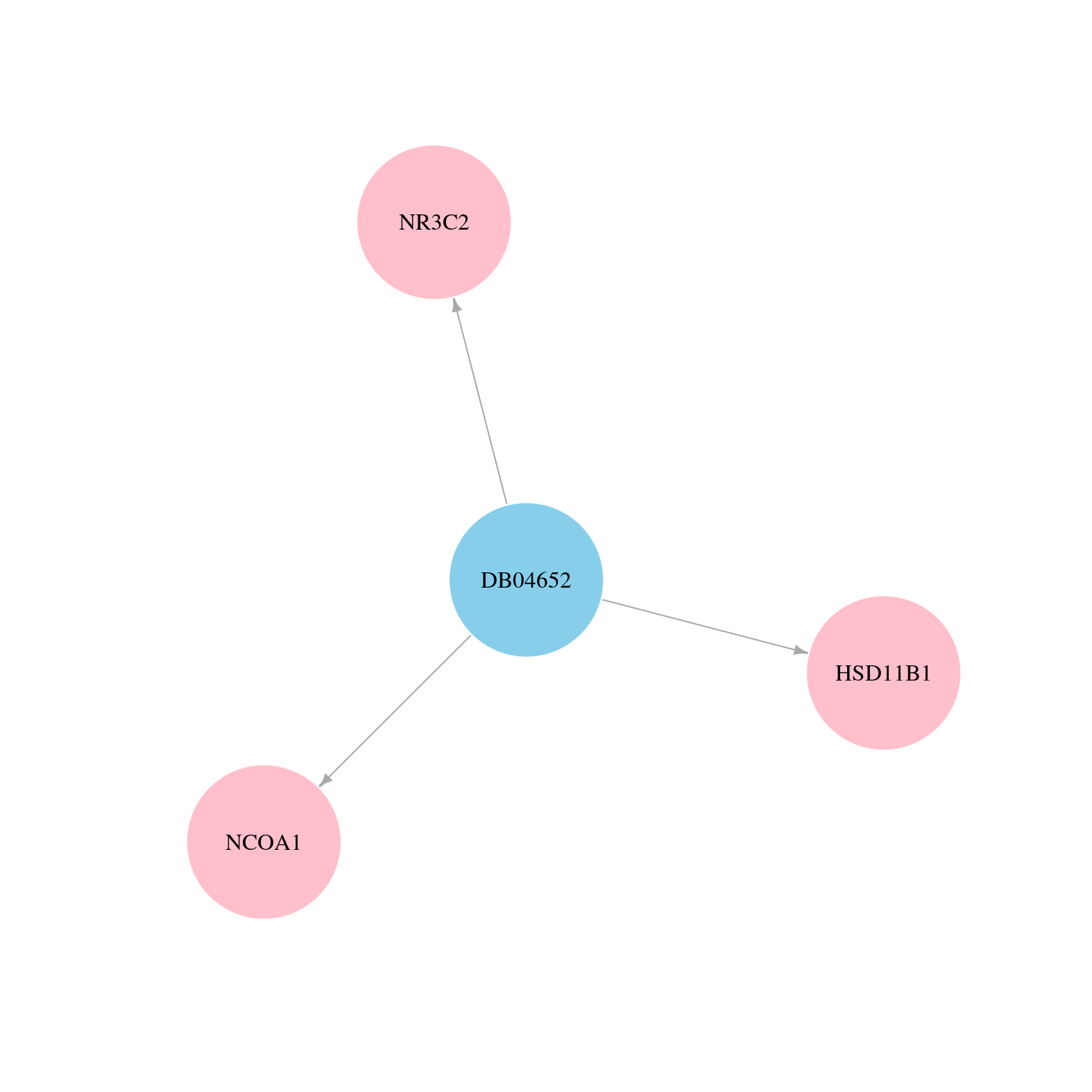
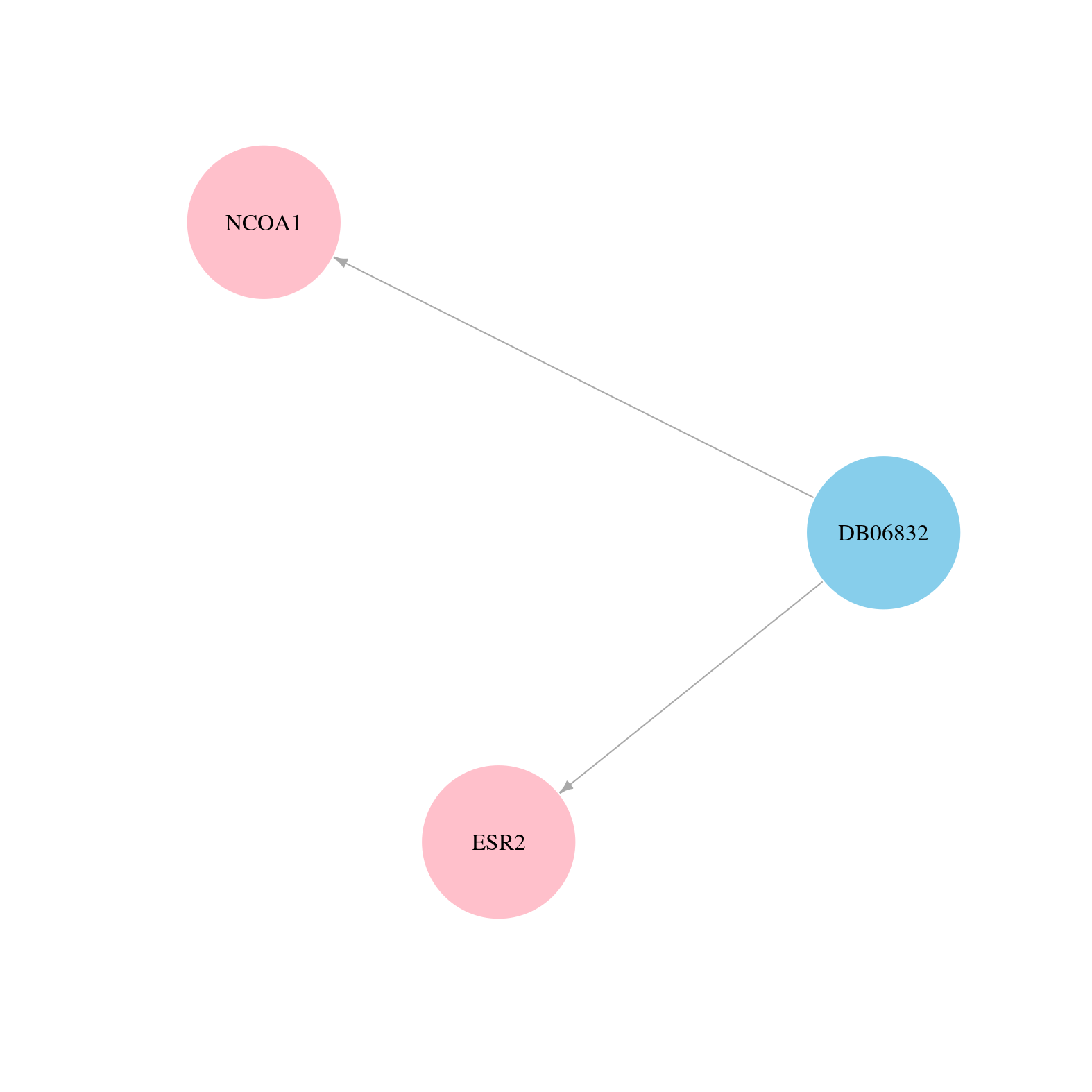
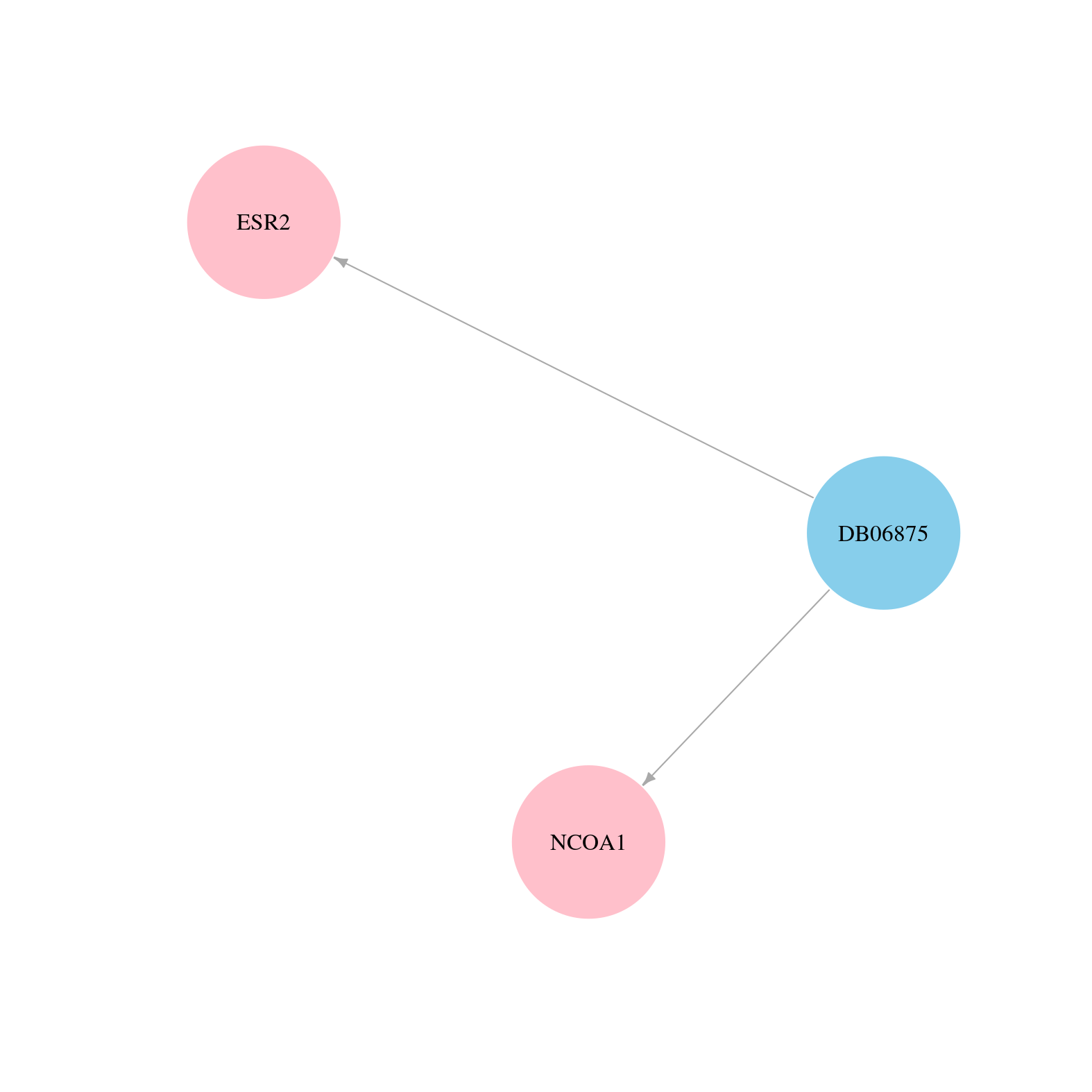
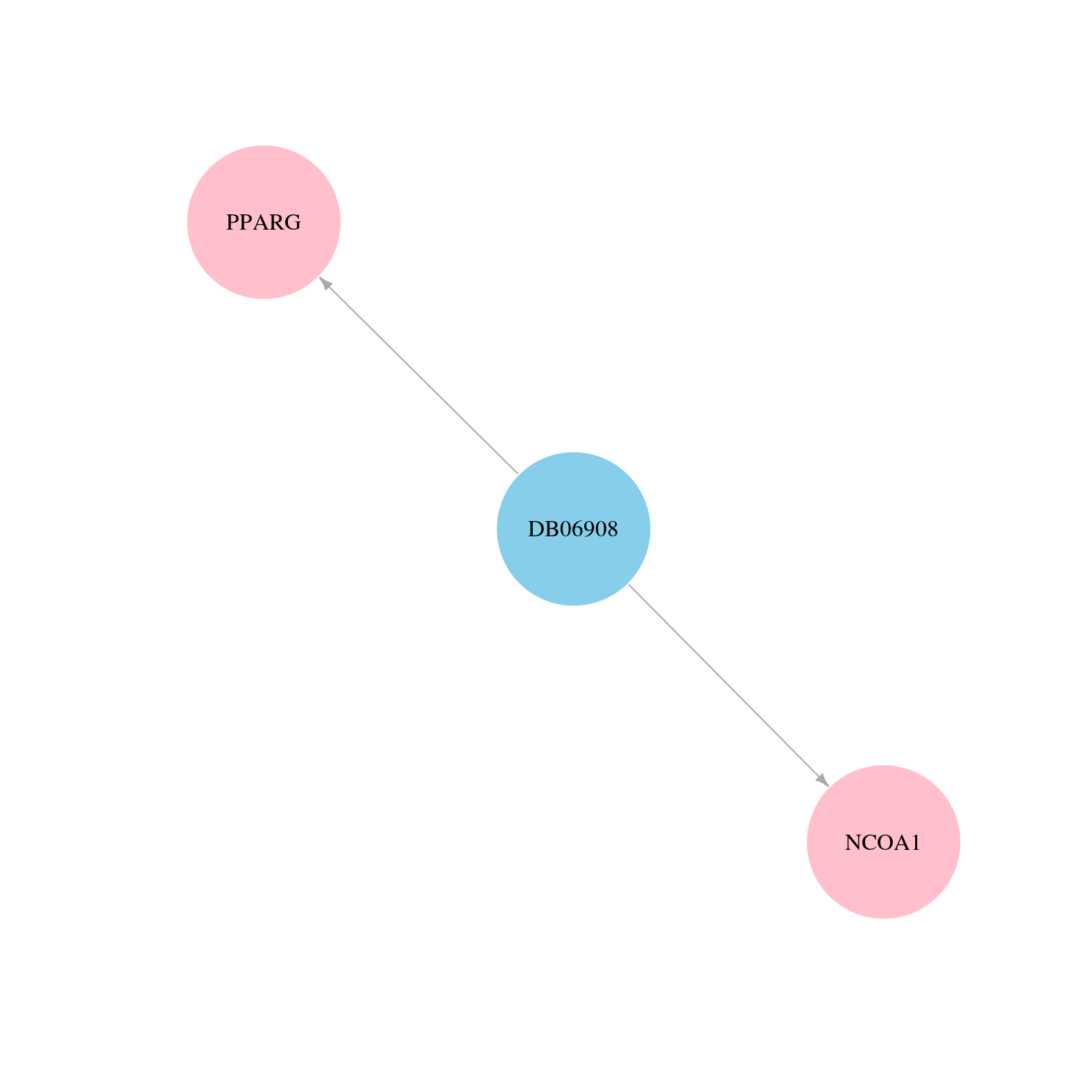
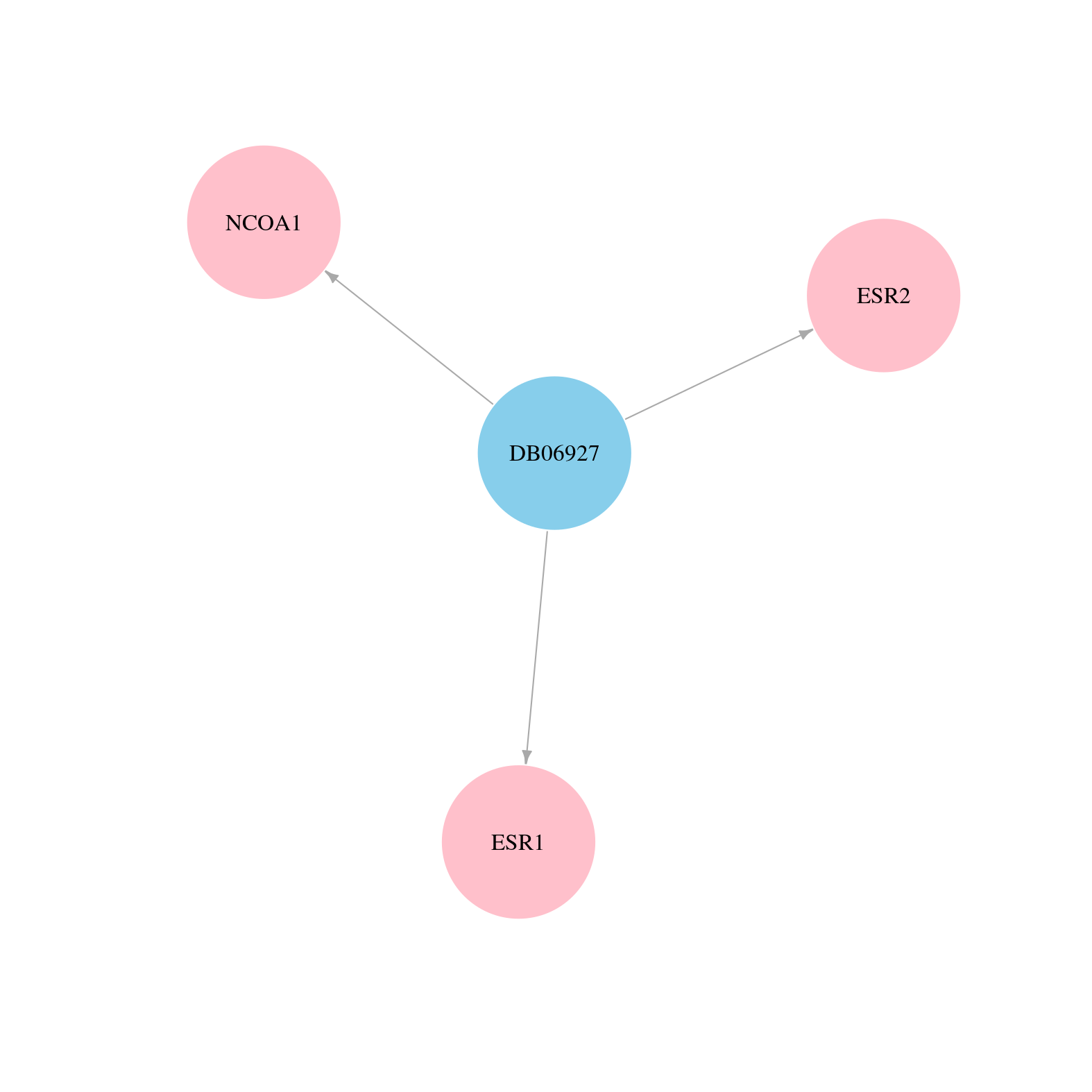
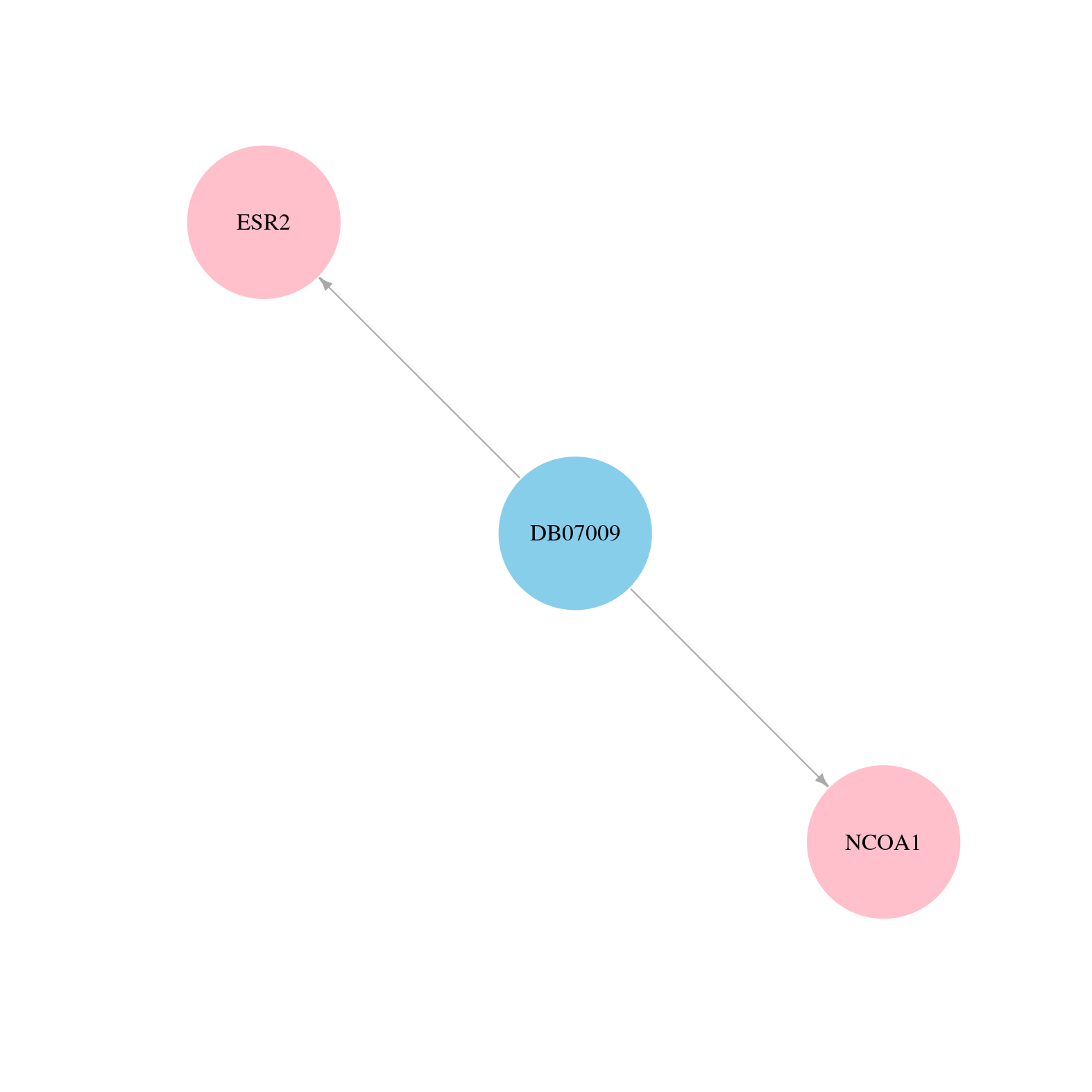
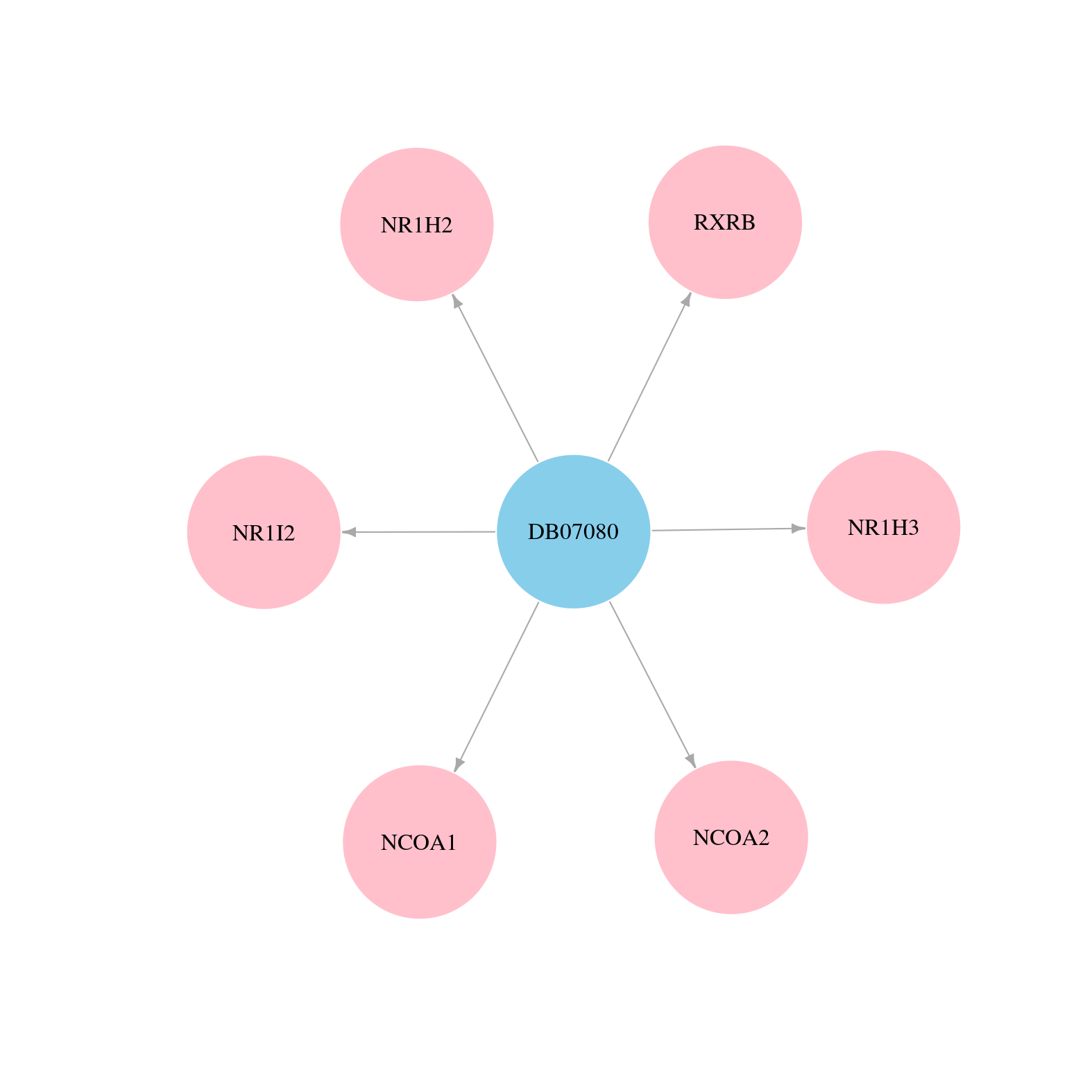
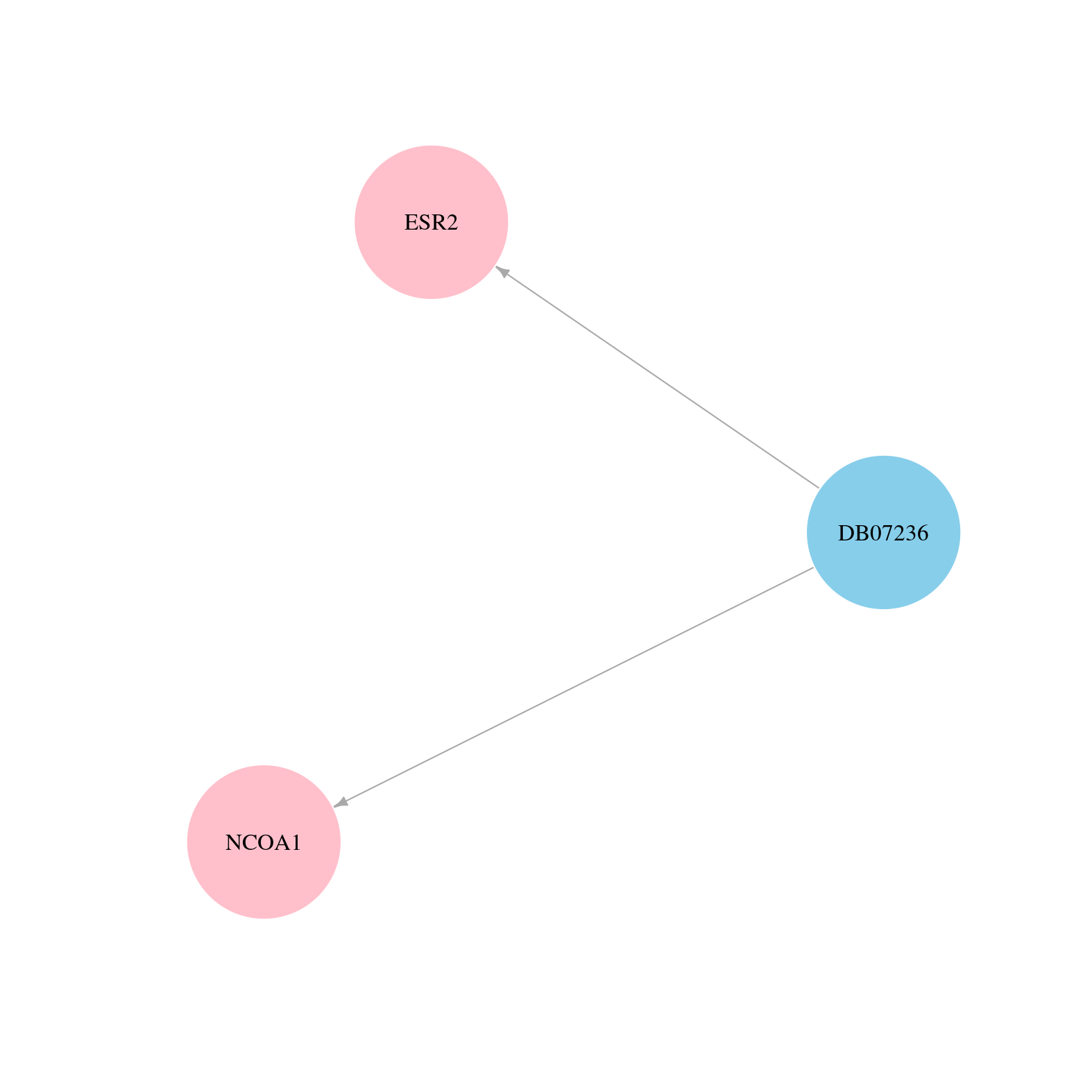
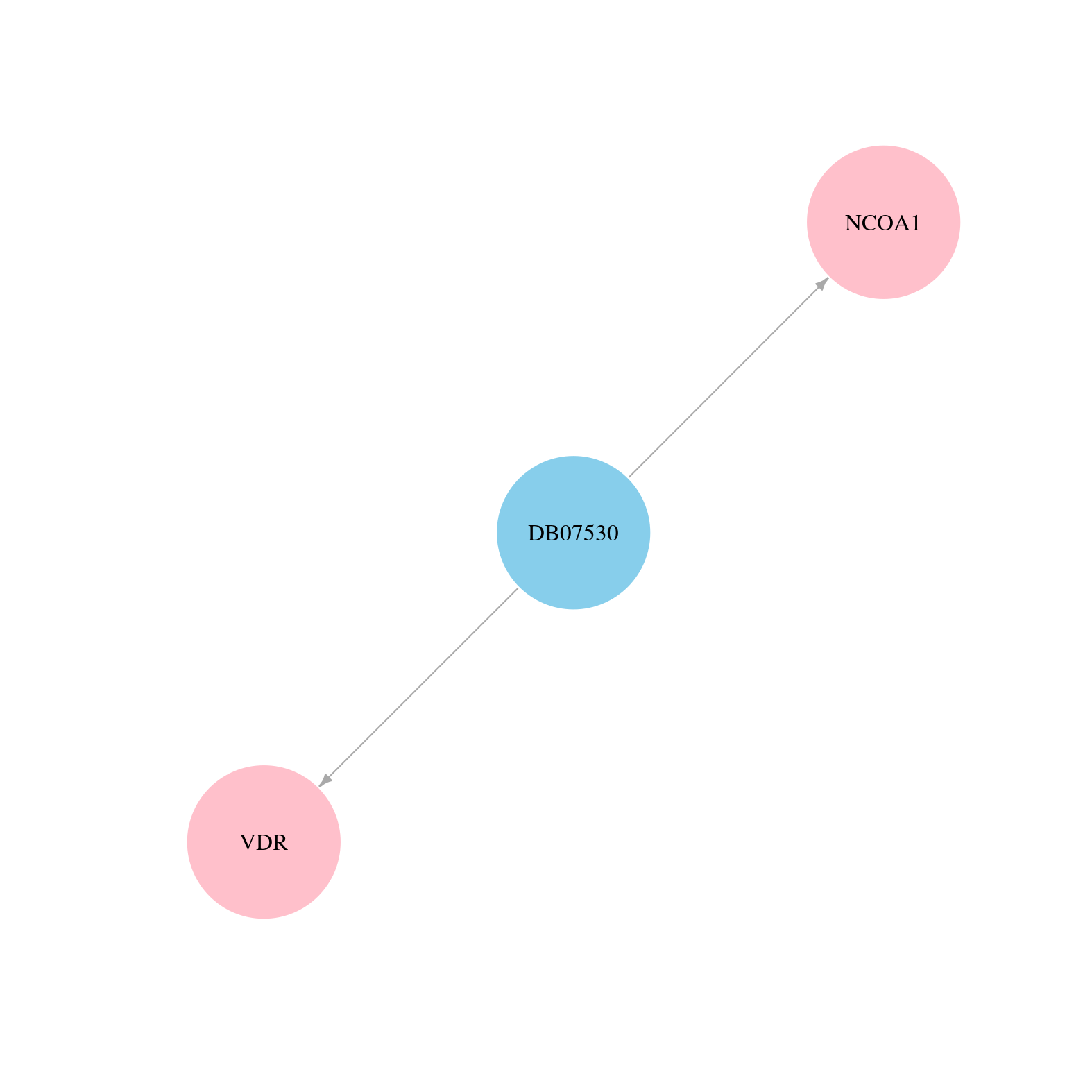
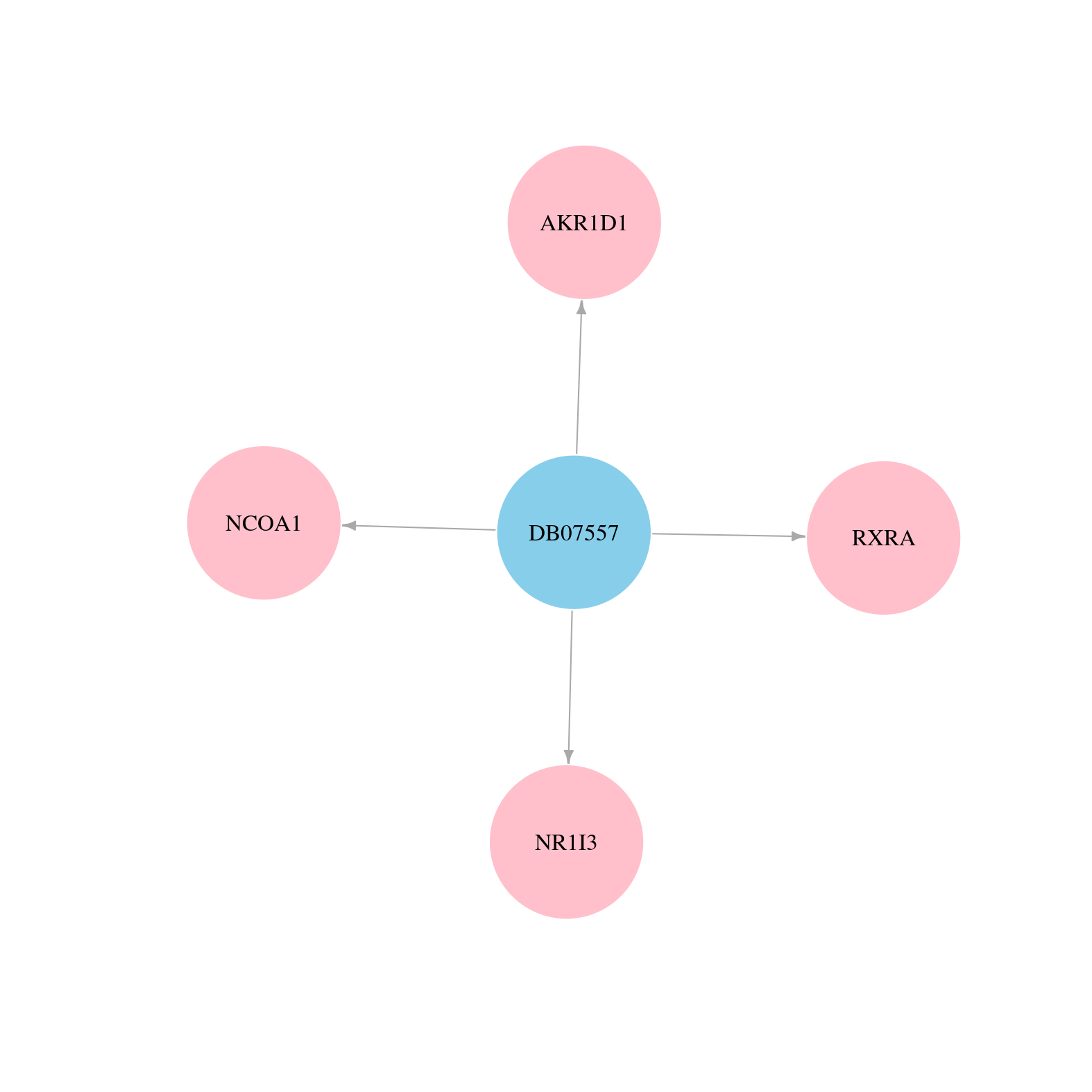
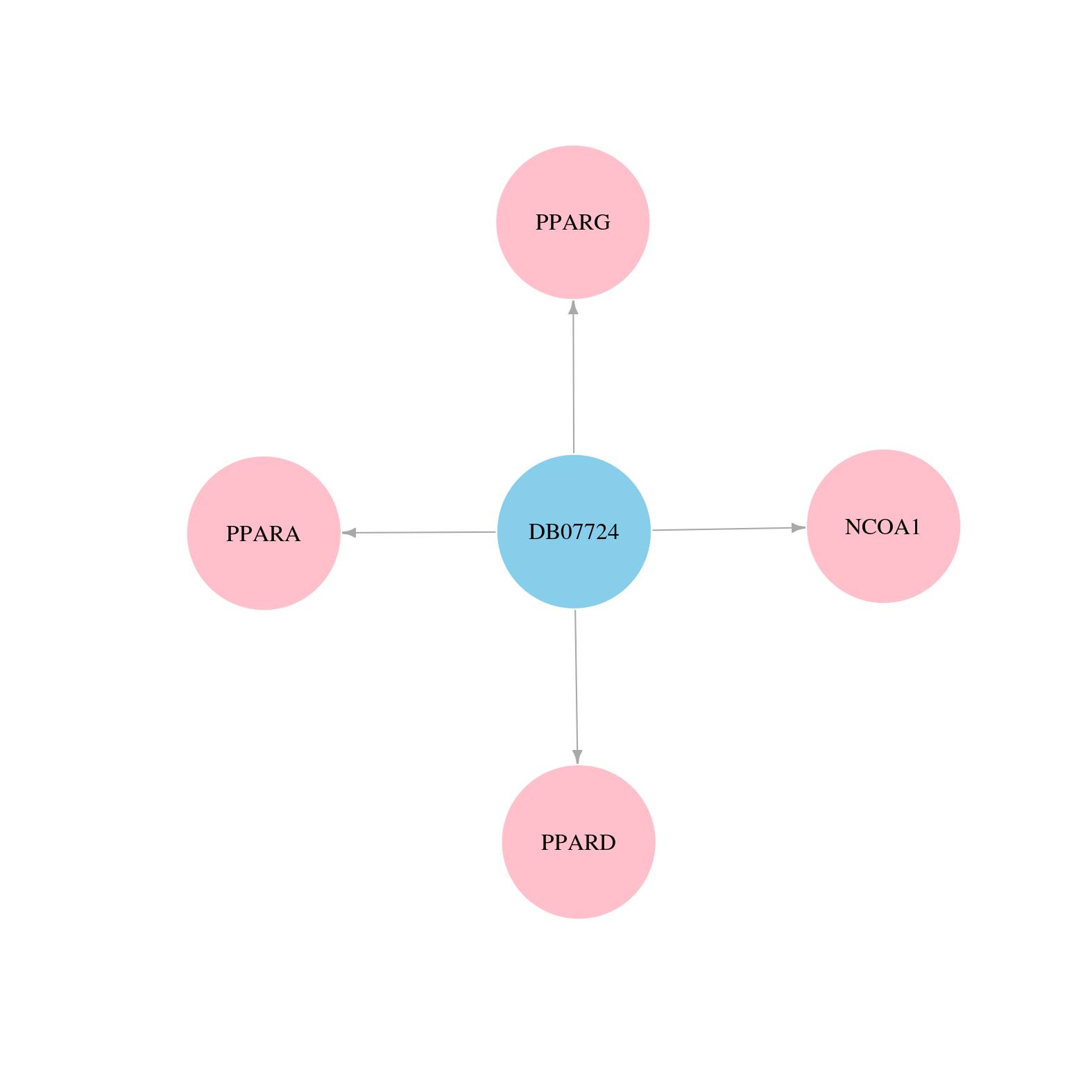
> Drugs from DrugBank database |
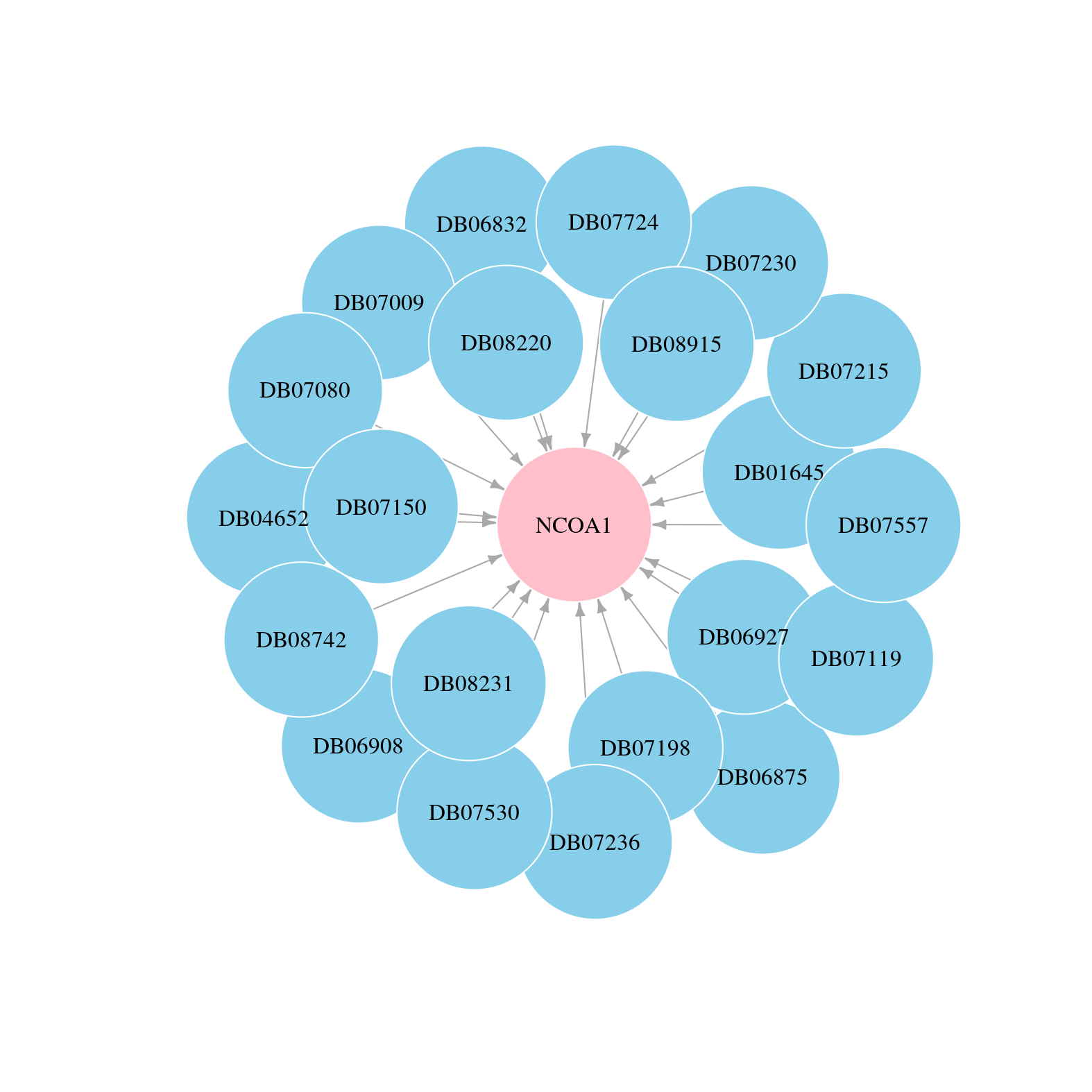
|
| Summary | |
|---|---|
| Symbol | NCOA1 |
| Name | nuclear receptor coactivator 1 |
| Aliases | F-SRC-1; NCoA-1; KAT13A; RIP160; bHLHe74; Hin-2 protein; PAX3/NCOA1 fusion protein; class E basic helix-loop ...... |
| Location | 2p23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|