Browse ORC1 in pancancer
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00004 ATPase family associated with various cellular activities (AAA) PF01426 BAH domain PF09079 CDC6 |
||||||||||
| Function |
Component of the origin recognition complex (ORC) that binds origins of replication. DNA-binding is ATP-dependent. The DNA sequences that define origins of replication have not been identified yet. ORC is required to assemble the pre-replication complex necessary to initiate DNA replication. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000082 G1/S transition of mitotic cell cycle GO:0000083 regulation of transcription involved in G1/S transition of mitotic cell cycle GO:0006260 DNA replication GO:0006261 DNA-dependent DNA replication GO:0006270 DNA replication initiation GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044843 cell cycle G1/S phase transition |
| Molecular Function |
GO:0003682 chromatin binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0000808 origin recognition complex GO:0005664 nuclear origin of replication recognition complex GO:0044454 nuclear chromosome part GO:0098687 chromosomal region |
| KEGG |
hsa04110 Cell cycle |
| Reactome |
R-HSA-176187: Activation of ATR in response to replication stress R-HSA-68962: Activation of the pre-replicative complex R-HSA-68616: Assembly of the ORC complex at the origin of replication R-HSA-68867: Assembly of the pre-replicative complex R-HSA-69298: Association of licensing factors with the pre-replicative complex R-HSA-68689: CDC6 association with the ORC R-HSA-68827: CDT1 association with the CDC6 R-HSA-1640170: Cell Cycle R-HSA-69620: Cell Cycle Checkpoints R-HSA-69278: Cell Cycle, Mitotic R-HSA-69306: DNA Replication R-HSA-69002: DNA Replication Pre-Initiation R-HSA-113510: E2F mediated regulation of DNA replication R-HSA-113507: E2F-enabled inhibition of pre-replication complex formation R-HSA-69206: G1/S Transition R-HSA-69205: G1/S-Specific Transcription R-HSA-69481: G2/M Checkpoints R-HSA-68874: M/G1 Transition R-HSA-453279: Mitotic G1-G1/S phases R-HSA-68949: Orc1 removal from chromatin R-HSA-69304: Regulation of DNA replication R-HSA-69300: Removal of licensing factors from origins R-HSA-69242: S Phase R-HSA-69052: Switching of origins to a post-replicative state R-HSA-69239: Synthesis of DNA |
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for ORC1. |
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
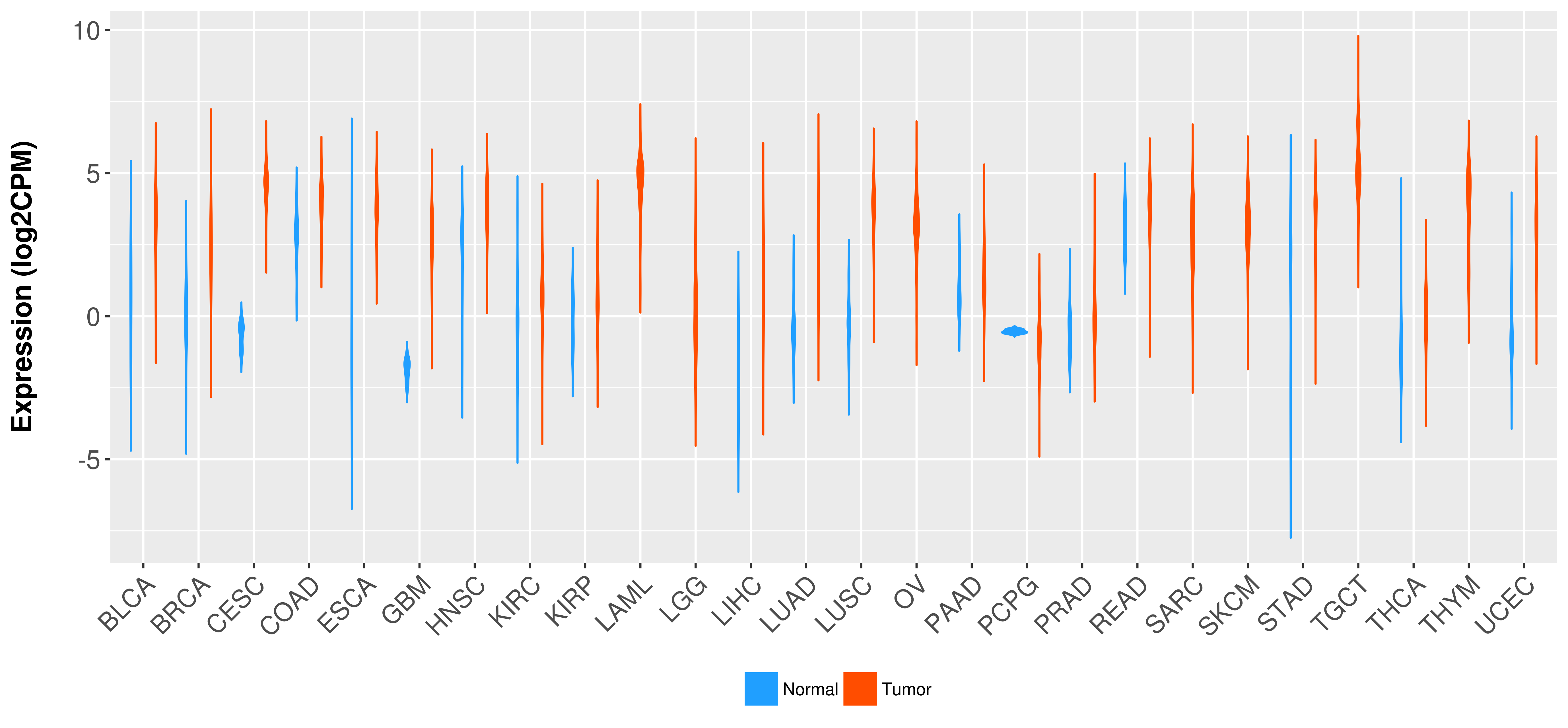
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
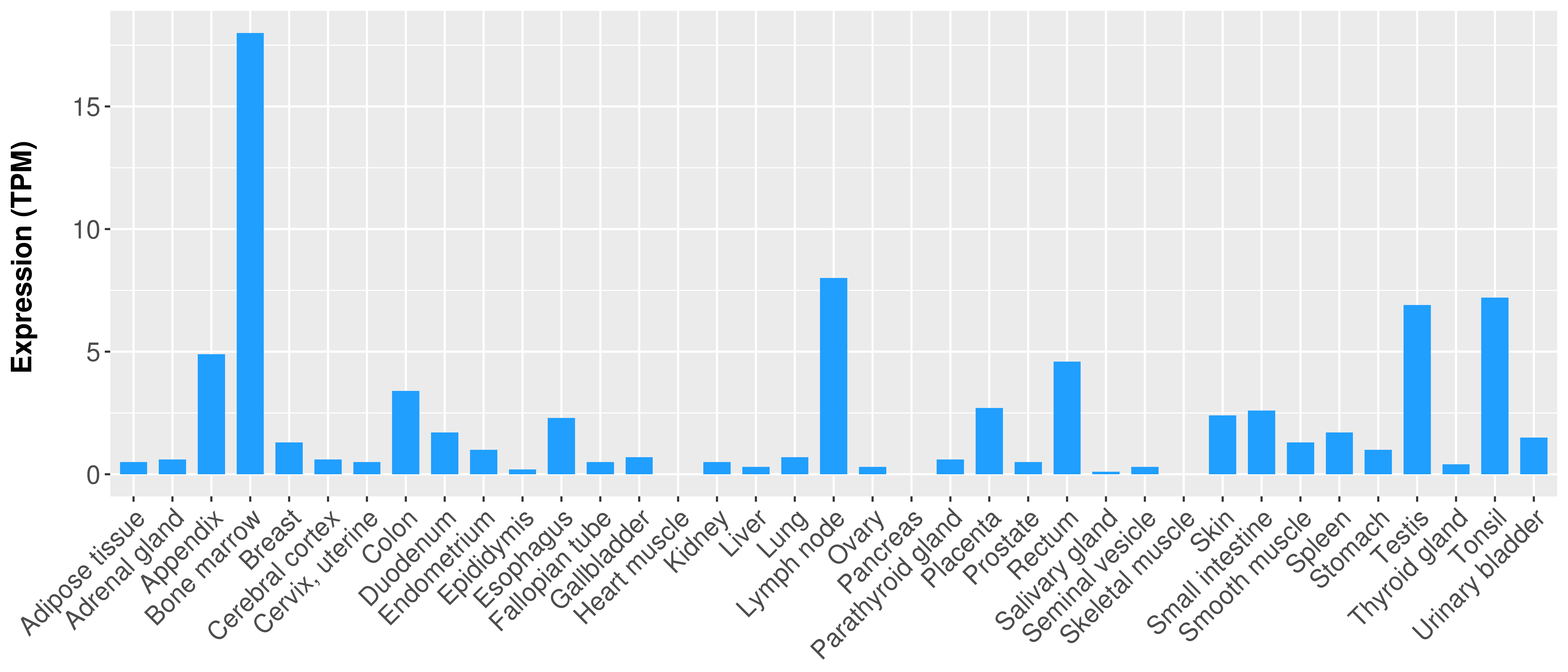
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
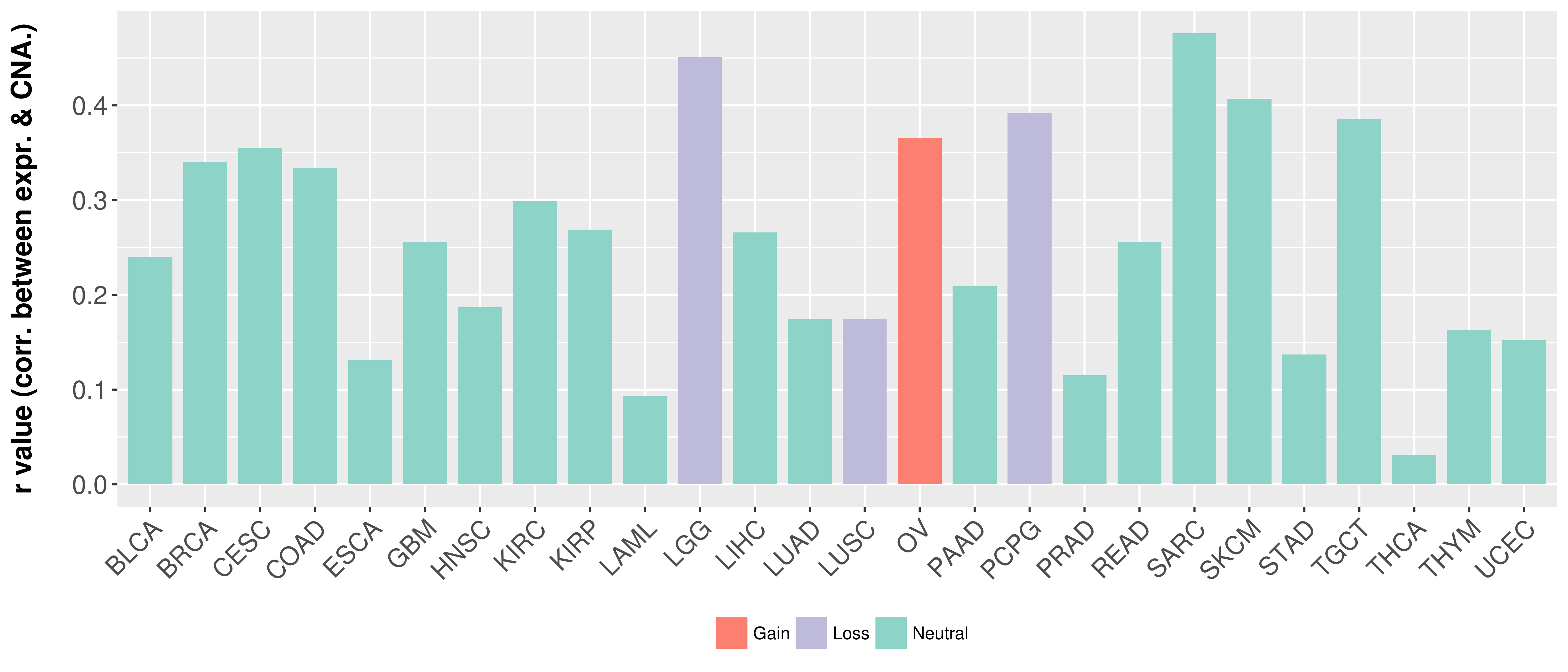
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
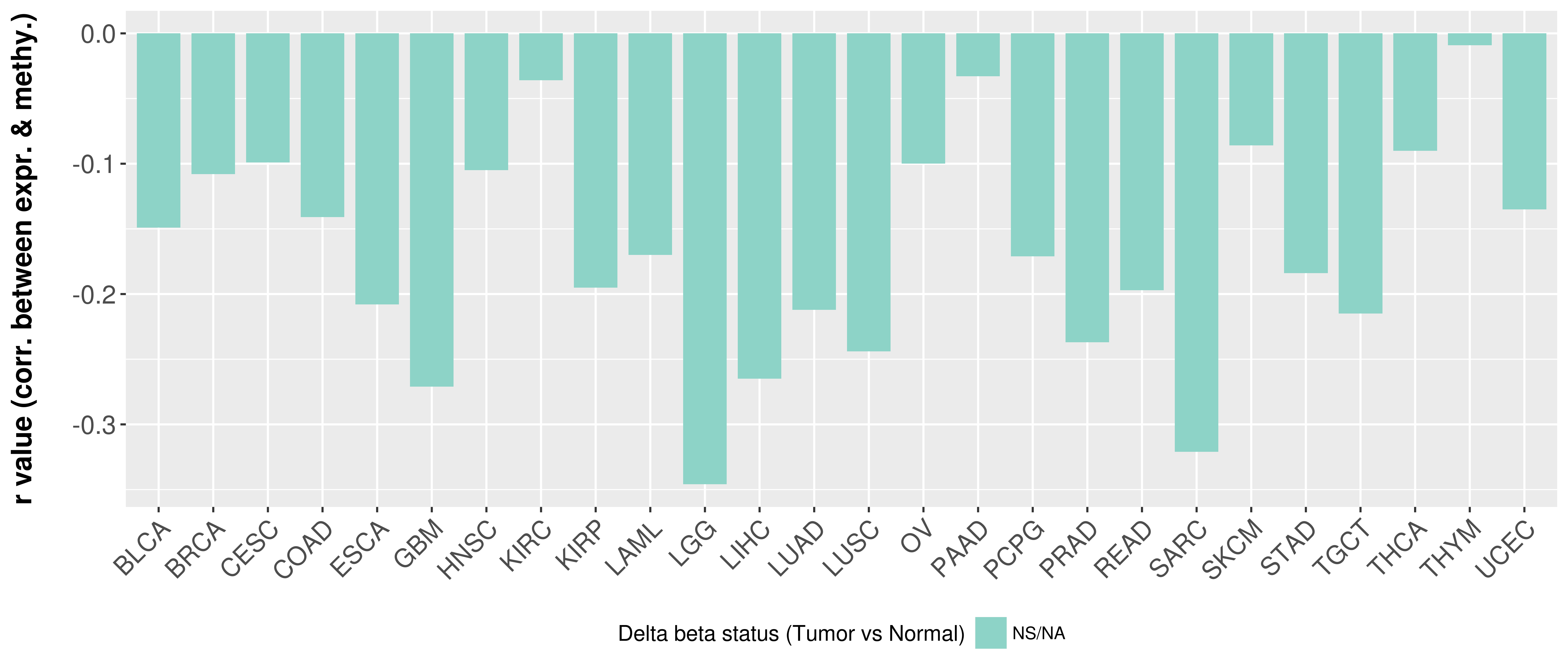
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
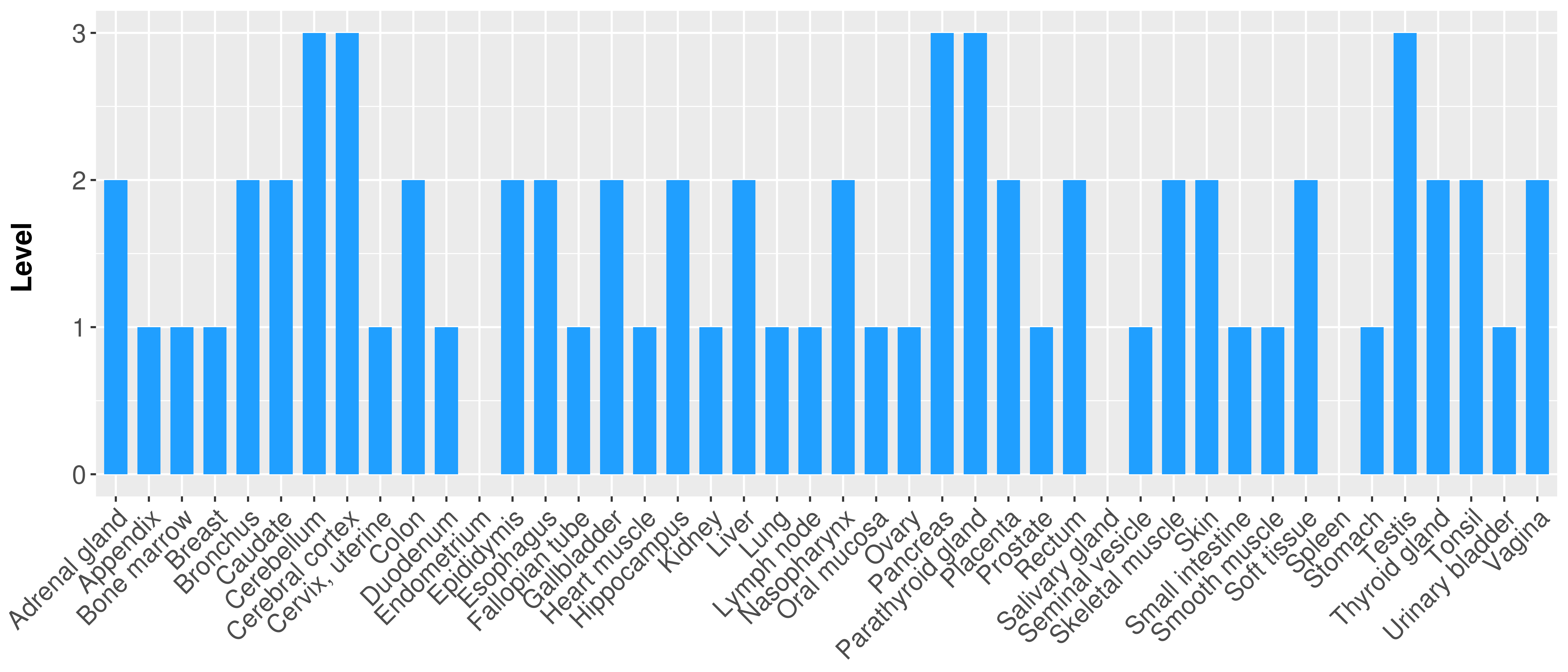
|
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
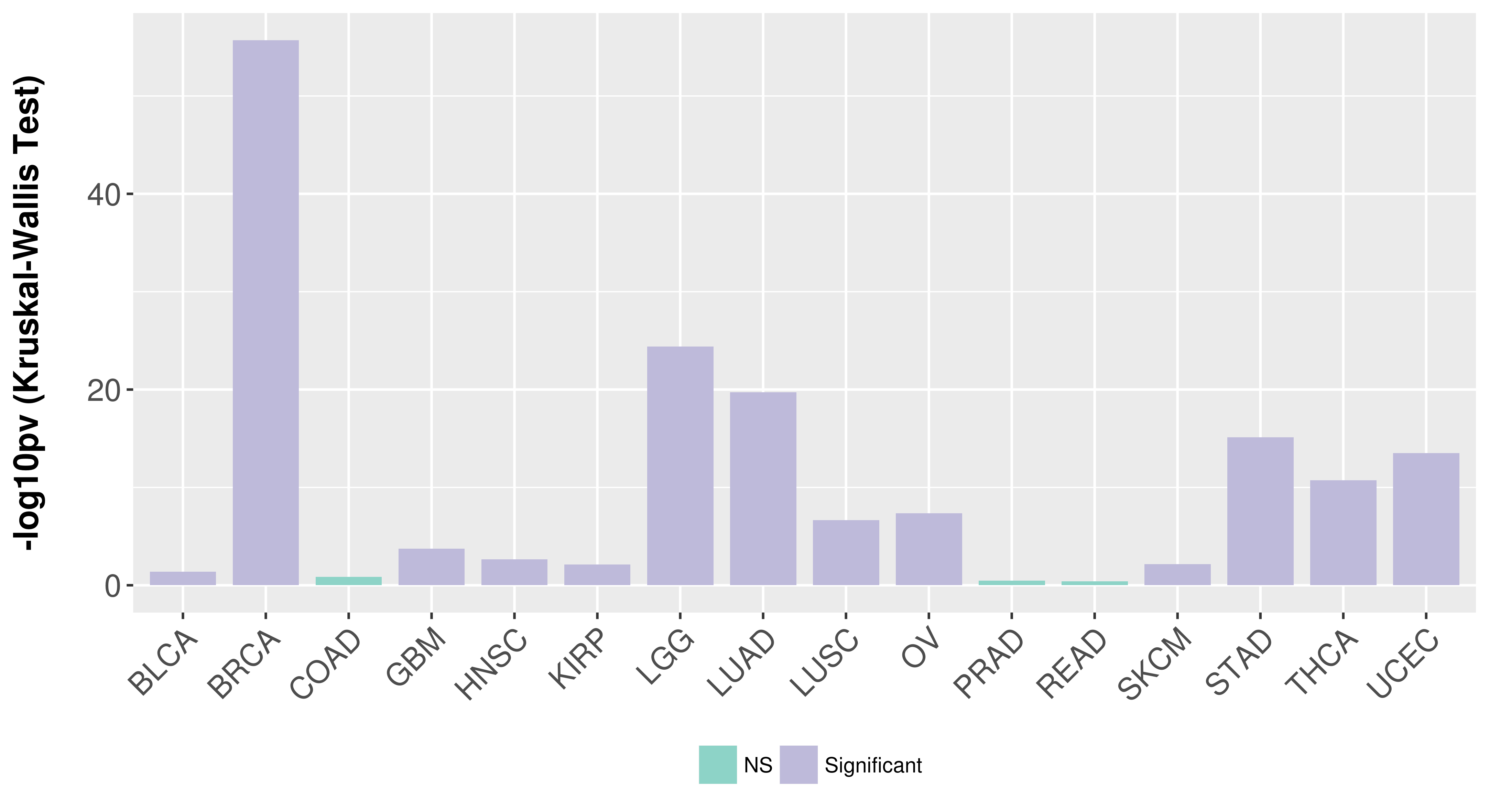
Association between expresson and subtype.
|
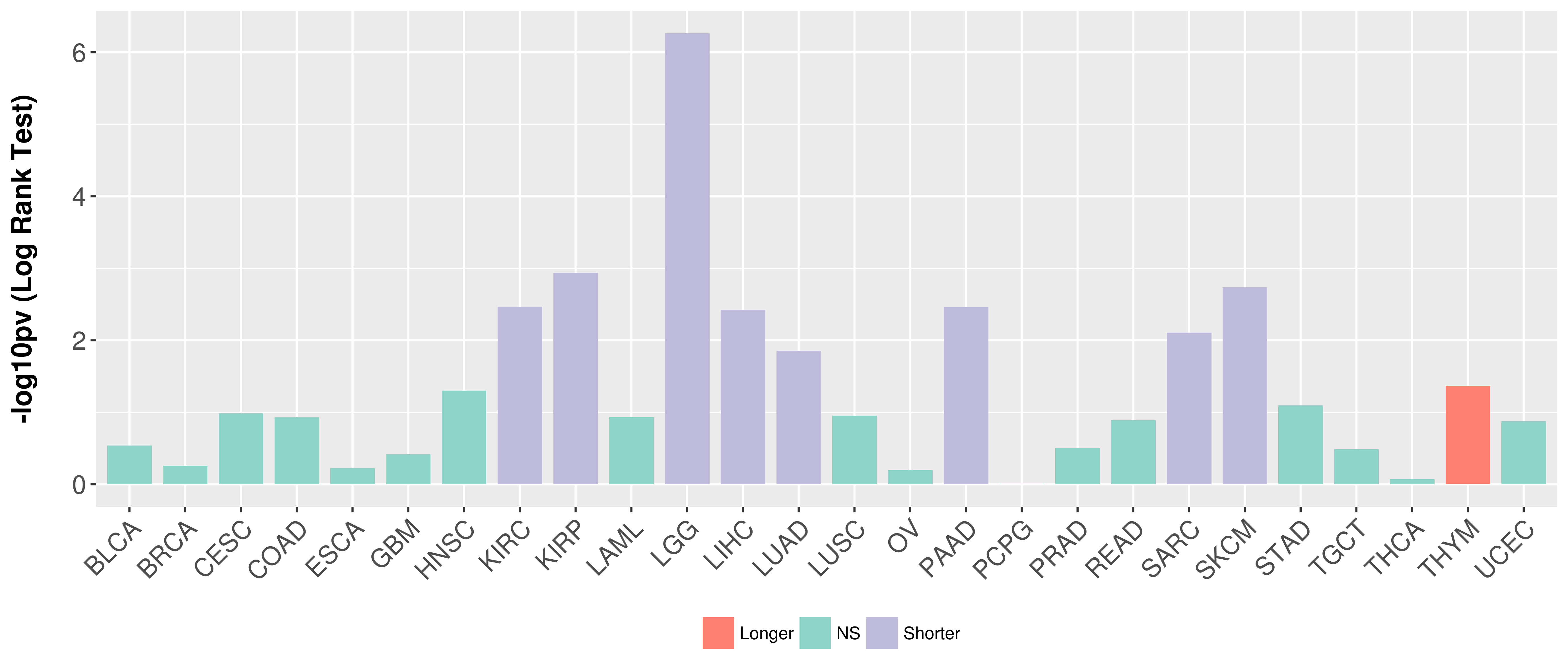
Overall survival analysis based on expression.
|
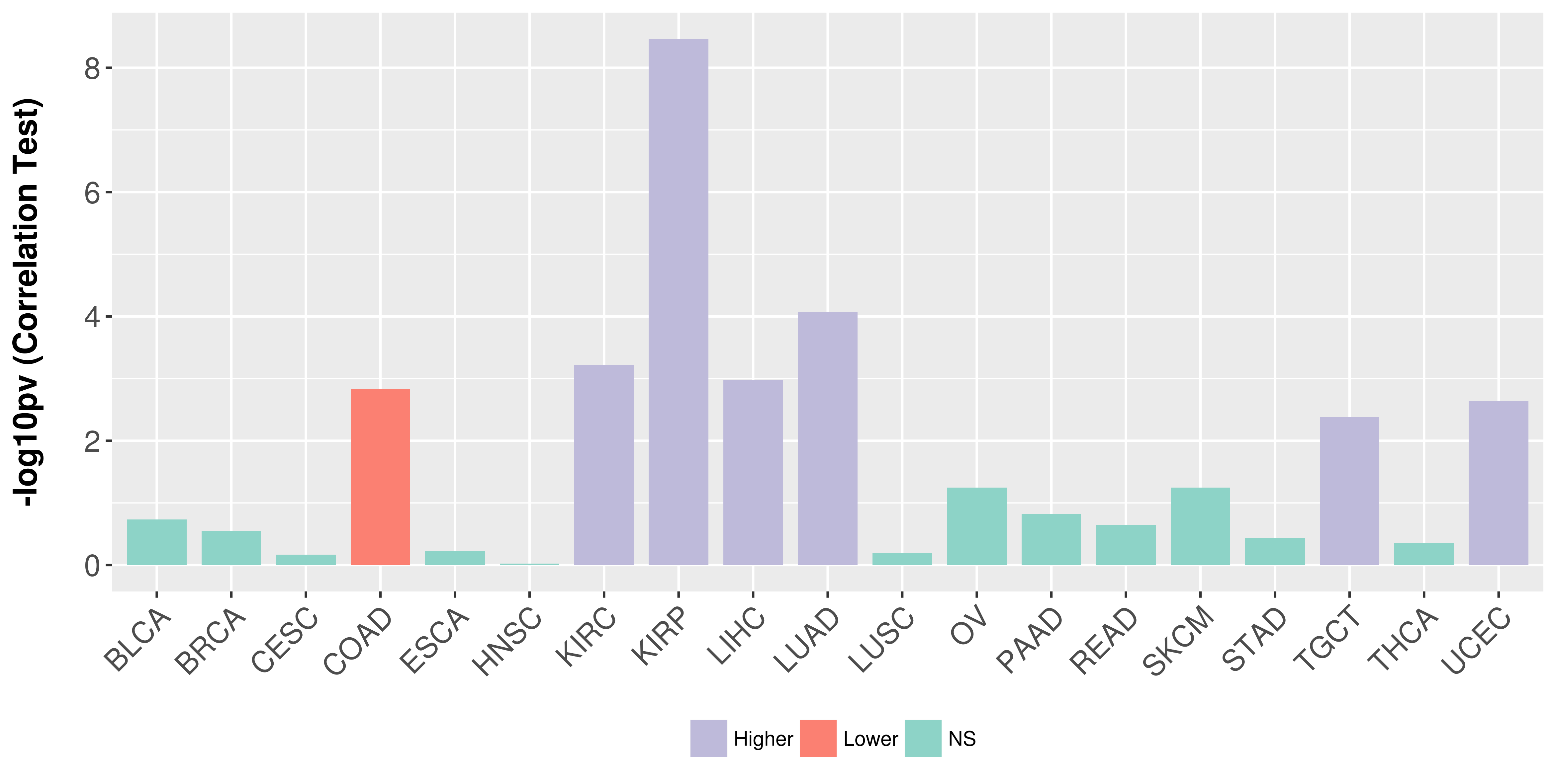
Association between expresson and stage.
|
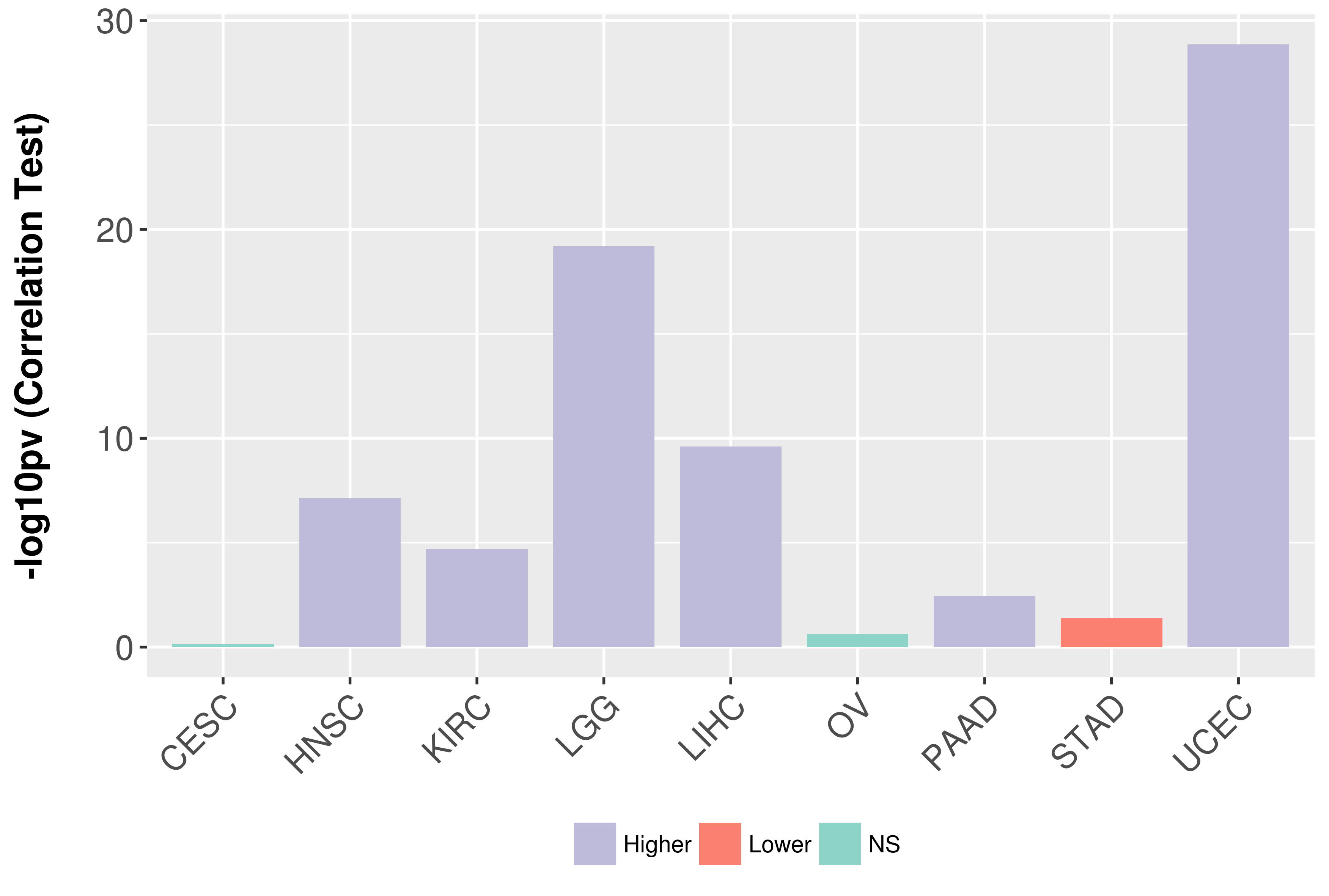
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ORC1. |
| Summary | |
|---|---|
| Symbol | ORC1 |
| Name | origin recognition complex subunit 1 |
| Aliases | HSORC1; PARC1; origin recognition complex, subunit 1, S. cerevisiae, homolog-like; origin recognition comple ...... |
| Location | 1p32.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for ORC1. |
| There is no record for ORC1. |