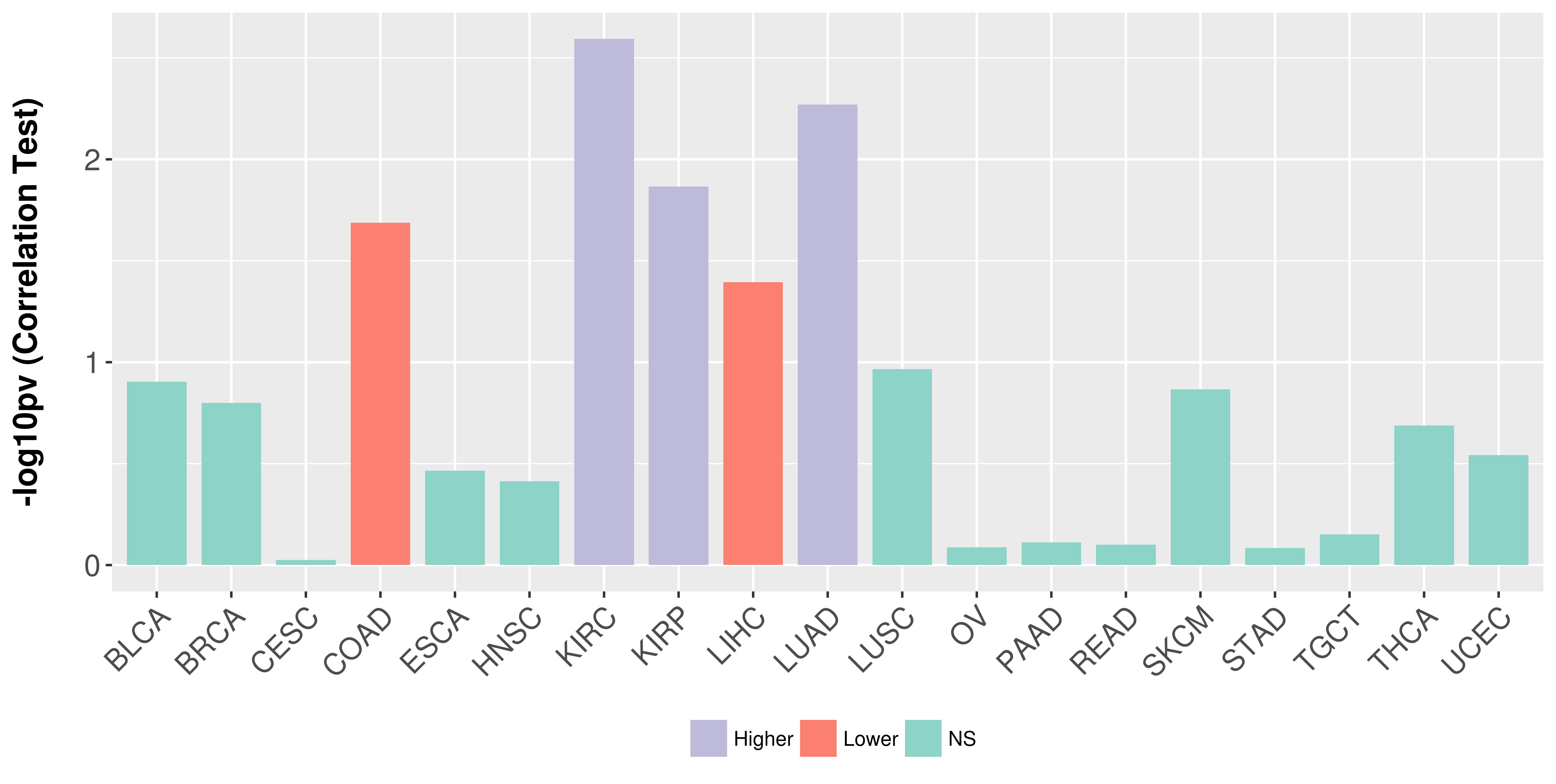
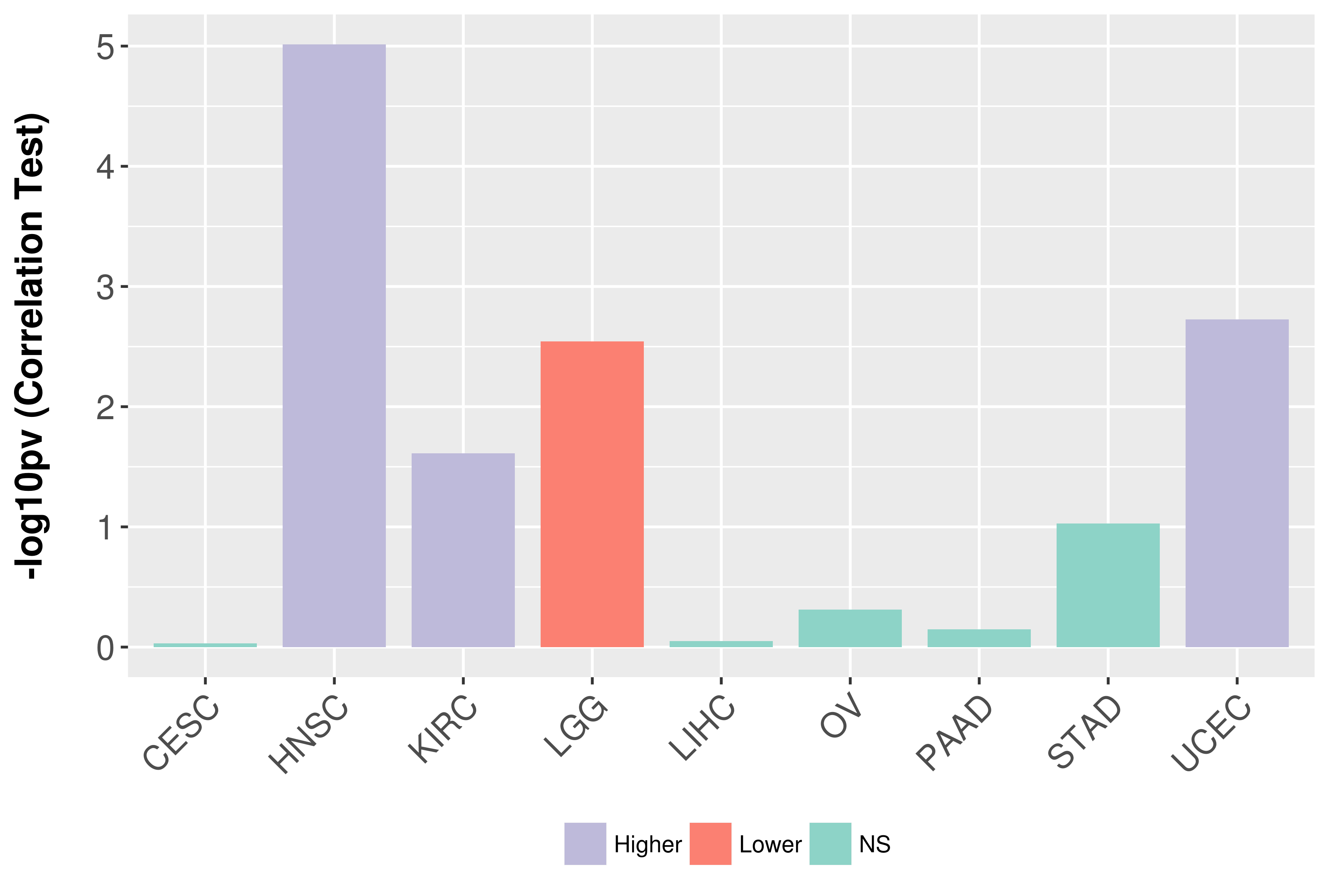
| COSM5657023 | c.86T>C | p.V29A | Substitution - Missense | Soft_tissue
|
| COSM1504169 | c.270G>A | p.L90L | Substitution - coding silent | Large_intestine
|
| COSM2059751 | c.388C>G | p.P130A | Substitution - Missense | Large_intestine
|
| COSM5698149 | c.103A>C | p.T35P | Substitution - Missense | Soft_tissue
|
| COSM2059758 | c.297_299delGGA | p.E99delE | Deletion - In frame | Breast
|
| COSM5698148 | c.103A>C | p.T35P | Substitution - Missense | Soft_tissue
|
| COSM914323 | c.261_263delAGA | p.E89delE | Deletion - In frame | Endometrium
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4787506 | c.763G>A | p.A255T | Substitution - Missense | Liver
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4787505 | c.988G>A | p.A330T | Substitution - Missense | Liver
|
| COSM683490 | c.4G>C | p.E2Q | Substitution - Missense | Lung
|
| COSM2059735 | c.558-3C>T | p.? | Unknown | Skin
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM1296894 | c.141T>G | p.P47P | Substitution - coding silent | Urinary_tract
|
| COSM1675527 | c.565C>T | p.R189* | Substitution - Nonsense | Large_intestine
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM2059748 | c.459C>T | p.T153T | Substitution - coding silent | Large_intestine
|
| COSM4912083 | c.770T>C | p.L257P | Substitution - Missense | Liver
|
| COSM3414650 | c.715G>C | p.E239Q | Substitution - Missense | Large_intestine
|
| COSM1675527 | c.565C>T | p.R189* | Substitution - Nonsense | Large_intestine
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM5894926 | c.178C>T | p.R60W | Substitution - Missense | Skin
|
| COSM5901671 | c.656G>A | p.R219K | Substitution - Missense | Skin
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM5692395 | c.274G>A | p.E92K | Substitution - Missense | Soft_tissue
|
| COSM3716374 | c.721T>C | p.Y241H | Substitution - Missense | Prostate
|
| COSM4573327 | c.379G>A | p.E127K | Substitution - Missense | Bone
|
| COSM4787506 | c.763G>A | p.A255T | Substitution - Missense | Liver
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM3716373 | c.946T>C | p.Y316H | Substitution - Missense | Prostate
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4712698 | c.300C>A | p.D100E | Substitution - Missense | Large_intestine
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM1289304 | c.96C>A | p.P32P | Substitution - coding silent | Haematopoietic_and_lymphoid_tissue
|
| COSM3786312 | c.909G>T | p.Q303H | Substitution - Missense | Pancreas
|
| COSM23379 | c.877A>G | p.R293G | Substitution - Missense | Kidney
|
| COSM4433520 | c.945delG | p.Q315fs*5 | Deletion - Frameshift | Oesophagus
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM401104 | c.167G>T | p.C56F | Substitution - Missense | Lung
|
| COSM5631241 | c.414A>G | p.K138K | Substitution - coding silent | Oesophagus
|
| COSM465212 | c.77C>A | p.P26Q | Substitution - Missense | Kidney
|
| COSM4011355 | c.381G>C | p.E127D | Substitution - Missense | Stomach
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM5657024 | c.86T>C | p.V29A | Substitution - Missense | Soft_tissue
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM5831383 | c.68_69delTG | p.L23fs*19 | Deletion - Frameshift | Breast
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM368494 | c.912A>G | p.V304V | Substitution - coding silent | Lung
|
| COSM465211 | c.77C>A | p.P26Q | Substitution - Missense | Kidney
|
| COSM683492 | c.822G>C | p.K274N | Substitution - Missense | Lung
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM1474325 | c.967C>G | p.R323G | Substitution - Missense | Breast
|
| COSM4849890 | c.574C>G | p.Q192E | Substitution - Missense | Cervix
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4712697 | c.614-3_614-2insT | p.? | Unknown | Large_intestine
|
| COSM5901670 | c.881G>A | p.R294K | Substitution - Missense | Skin
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM5445985 | c.69_70insCCGCCT | p.P28_V29insPP | Insertion - In frame | Thyroid
|
| COSM5940064 | c.772-8C>T | p.? | Unknown | Skin
|
| COSM2059759 | c.297_299delGGA | p.E99delE | Deletion - In frame | Breast
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM3731481 | c.666T>C | p.P222P | Substitution - coding silent | Stomach
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM3414649 | c.940G>C | p.E314Q | Substitution - Missense | Large_intestine
|
| COSM1296893 | c.141T>G | p.P47P | Substitution - coding silent | Urinary_tract
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM23379 | c.877A>G | p.R293G | Substitution - Missense | Kidney
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM914322 | c.261_263delAGA | p.E89delE | Deletion - In frame | Endometrium
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM2059734 | c.783-3C>T | p.? | Unknown | Skin
|
| COSM1261519 | c.814T>C | p.L272L | Substitution - coding silent | Stomach
|
| COSM5692396 | c.274G>A | p.E92K | Substitution - Missense | Soft_tissue
|
| COSM3978051 | c.640G>C | p.E214Q | Substitution - Missense | Lung
|
| COSM5691119 | c.84delT | p.V29fs*73 | Deletion - Frameshift | Soft_tissue
|
| COSM1638428 | c.614-3delT | p.? | Unknown | Stomach
|
| COSM1675527 | c.565C>T | p.R189* | Substitution - Nonsense | Large_intestine
|
| COSM4626616 | c.943C>T | p.Q315* | Substitution - Nonsense | Large_intestine
|
| COSM2154707 | c.553A>G | p.I185V | Substitution - Missense | Central_nervous_system
|
| COSM2154706 | c.553A>G | p.I185V | Substitution - Missense | Central_nervous_system
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4457525 | c.1049C>G | p.T350S | Substitution - Missense | Skin
|
| COSM4626617 | c.718C>T | p.Q240* | Substitution - Nonsense | Large_intestine
|
| COSM1289303 | c.96C>A | p.P32P | Substitution - coding silent | Haematopoietic_and_lymphoid_tissue
|
| COSM5691118 | c.84delT | p.V29fs*73 | Deletion - Frameshift | Soft_tissue
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM2059749 | c.459C>T | p.T153T | Substitution - coding silent | Large_intestine
|
| COSM5691118 | c.84delT | p.V29fs*73 | Deletion - Frameshift | Soft_tissue
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM3786313 | c.684G>T | p.Q228H | Substitution - Missense | Pancreas
|
| COSM2059750 | c.388C>G | p.P130A | Substitution - Missense | Large_intestine
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM683493 | c.597G>C | p.K199N | Substitution - Missense | Lung
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM1504168 | c.270G>A | p.L90L | Substitution - coding silent | Large_intestine
|
| COSM236191 | c.605T>C | p.L202P | Substitution - Missense | Autonomic_ganglia
|
| COSM4457526 | c.824C>G | p.T275S | Substitution - Missense | Skin
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM1675527 | c.565C>T | p.R189* | Substitution - Nonsense | Large_intestine
|
| COSM914319 | c.361C>T | p.R121C | Substitution - Missense | Endometrium
|
| COSM368495 | c.687A>G | p.V229V | Substitution - coding silent | Lung
|
| COSM5445986 | c.69_70insCCGCCT | p.P28_V29insPP | Insertion - In frame | Thyroid
|
| COSM294683 | c.281A>C | p.E94A | Substitution - Missense | Large_intestine
|
| COSM401105 | c.167G>T | p.C56F | Substitution - Missense | Lung
|
| COSM5657024 | c.86T>C | p.V29A | Substitution - Missense | Skin
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Central_nervous_system
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM343389 | c.761C>T | p.S254F | Substitution - Missense | Skin
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM914318 | c.361C>T | p.R121C | Substitution - Missense | Endometrium
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4787505 | c.988G>A | p.A330T | Substitution - Missense | Liver
|
| COSM5691119 | c.84delT | p.V29fs*73 | Deletion - Frameshift | Soft_tissue
|
| COSM2154707 | c.553A>G | p.I185V | Substitution - Missense | Central_nervous_system
|
| COSM5657023 | c.86T>C | p.V29A | Substitution - Missense | Soft_tissue
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Central_nervous_system
|
| COSM5177983 | c.782+6T>C | p.? | Unknown | Large_intestine
|
| COSM343389 | c.761C>T | p.S254F | Substitution - Missense | Lung
|
| COSM343390 | c.639C>G | p.F213L | Substitution - Missense | Lung
|
| COSM5894925 | c.178C>T | p.R60W | Substitution - Missense | Skin
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4712699 | c.300C>A | p.D100E | Substitution - Missense | Large_intestine
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM5940063 | c.997-8C>T | p.? | Unknown | Skin
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM3978050 | c.865G>C | p.E289Q | Substitution - Missense | Lung
|
| COSM1474326 | c.742C>G | p.R248G | Substitution - Missense | Breast
|
| COSM683491 | c.4G>C | p.E2Q | Substitution - Missense | Lung
|
| COSM3382654 | c.478G>A | p.V160I | Substitution - Missense | Pancreas
|
| COSM5631242 | c.414A>G | p.K138K | Substitution - coding silent | Oesophagus
|
| COSM914320 | c.273G>T | p.E91D | Substitution - Missense | Endometrium
|
| COSM2059758 | c.297_299delGGA | p.E99delE | Deletion - In frame | Large_intestine
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM2059759 | c.297_299delGGA | p.E99delE | Deletion - In frame | Large_intestine
|
| COSM3382655 | c.478G>A | p.V160I | Substitution - Missense | Pancreas
|
| COSM23505 | c.652A>G | p.R218G | Substitution - Missense | Kidney
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM2154706 | c.553A>G | p.I185V | Substitution - Missense | Central_nervous_system
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4433521 | c.720delG | p.Q240fs*5 | Deletion - Frameshift | Oesophagus
|
| COSM4011356 | c.381G>C | p.E127D | Substitution - Missense | Stomach
|
| COSM1560994 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM4573326 | c.379G>A | p.E127K | Substitution - Missense | Bone
|
| COSM3433842 | c.736C>T | p.R246C | Substitution - Missense | Skin
|
| COSM294684 | c.281A>C | p.E94A | Substitution - Missense | Large_intestine
|
| COSM5894925 | c.178C>T | p.R60W | Substitution - Missense | Skin
|
| COSM4603246 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM4603245 | c.84_85insGCCTCC | p.P28_V29insAS | Insertion - In frame | Upper_aerodigestive_tract
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM5657024 | c.86T>C | p.V29A | Substitution - Missense | Soft_tissue
|
| COSM5657023 | c.86T>C | p.V29A | Substitution - Missense | Skin
|
| COSM5831382 | c.68_69delTG | p.L23fs*19 | Deletion - Frameshift | Breast
|
| COSM2059737 | c.674C>T | p.A225V | Substitution - Missense | Bone
|
| COSM1560993 | c.83_84insGCCTCC | p.P28_V29insPP | Insertion - In frame | Large_intestine
|
| COSM914321 | c.273G>T | p.E91D | Substitution - Missense | Endometrium
|
| COSM1675527 | c.565C>T | p.R189* | Substitution - Nonsense | Large_intestine
|
| COSM5894926 | c.178C>T | p.R60W | Substitution - Missense | Skin
|
| COSM1261520 | c.589T>C | p.L197L | Substitution - coding silent | Stomach
|