Browse PCNA in pancancer
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02747 Proliferating cell nuclear antigen PF00705 Proliferating cell nuclear antigen |
||||||||||
| Function |
Auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand. Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways (PubMed:24939902). Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000077 DNA damage checkpoint GO:0000082 G1/S transition of mitotic cell cycle GO:0000083 regulation of transcription involved in G1/S transition of mitotic cell cycle GO:0000302 response to reactive oxygen species GO:0000722 telomere maintenance via recombination GO:0000723 telomere maintenance GO:0000731 DNA synthesis involved in DNA repair GO:0001889 liver development GO:0006260 DNA replication GO:0006261 DNA-dependent DNA replication GO:0006271 DNA strand elongation involved in DNA replication GO:0006272 leading strand elongation GO:0006275 regulation of DNA replication GO:0006282 regulation of DNA repair GO:0006283 transcription-coupled nucleotide-excision repair GO:0006289 nucleotide-excision repair GO:0006296 nucleotide-excision repair, DNA incision, 5'-to lesion GO:0006297 nucleotide-excision repair, DNA gap filling GO:0006298 mismatch repair GO:0006301 postreplication repair GO:0006310 DNA recombination GO:0006312 mitotic recombination GO:0006977 DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest GO:0006979 response to oxidative stress GO:0007050 cell cycle arrest GO:0007093 mitotic cell cycle checkpoint GO:0007346 regulation of mitotic cell cycle GO:0007507 heart development GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0010035 response to inorganic substance GO:0010038 response to metal ion GO:0010948 negative regulation of cell cycle process GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0019985 translesion synthesis GO:0022616 DNA strand elongation GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0031099 regeneration GO:0031100 animal organ regeneration GO:0031297 replication fork processing GO:0031570 DNA integrity checkpoint GO:0031571 mitotic G1 DNA damage checkpoint GO:0031960 response to corticosteroid GO:0032069 regulation of nuclease activity GO:0032070 regulation of deoxyribonuclease activity GO:0032075 positive regulation of nuclease activity GO:0032077 positive regulation of deoxyribonuclease activity GO:0032200 telomere organization GO:0032201 telomere maintenance via semi-conservative replication GO:0032355 response to estradiol GO:0033260 nuclear DNA replication GO:0033683 nucleotide-excision repair, DNA incision GO:0034599 cellular response to oxidative stress GO:0034614 cellular response to reactive oxygen species GO:0034644 cellular response to UV GO:0042276 error-prone translesion synthesis GO:0042542 response to hydrogen peroxide GO:0042698 ovulation cycle GO:0042769 DNA damage response, detection of DNA damage GO:0042770 signal transduction in response to DNA damage GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044773 mitotic DNA damage checkpoint GO:0044774 mitotic DNA integrity checkpoint GO:0044783 G1 DNA damage checkpoint GO:0044786 cell cycle DNA replication GO:0044819 mitotic G1/S transition checkpoint GO:0044843 cell cycle G1/S phase transition GO:0044849 estrous cycle GO:0045005 DNA-dependent DNA replication maintenance of fidelity GO:0045739 positive regulation of DNA repair GO:0045740 positive regulation of DNA replication GO:0045786 negative regulation of cell cycle GO:0045787 positive regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0046686 response to cadmium ion GO:0048511 rhythmic process GO:0048545 response to steroid hormone GO:0048732 gland development GO:0051052 regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051384 response to glucocorticoid GO:0060249 anatomical structure homeostasis GO:0061008 hepaticobiliary system development GO:0070301 cellular response to hydrogen peroxide GO:0070987 error-free translesion synthesis GO:0071156 regulation of cell cycle arrest GO:0071158 positive regulation of cell cycle arrest GO:0071214 cellular response to abiotic stimulus GO:0071478 cellular response to radiation GO:0071482 cellular response to light stimulus GO:0071548 response to dexamethasone GO:0071897 DNA biosynthetic process GO:0072331 signal transduction by p53 class mediator GO:0072395 signal transduction involved in cell cycle checkpoint GO:0072401 signal transduction involved in DNA integrity checkpoint GO:0072413 signal transduction involved in mitotic cell cycle checkpoint GO:0072422 signal transduction involved in DNA damage checkpoint GO:0072431 signal transduction involved in mitotic G1 DNA damage checkpoint GO:0090068 positive regulation of cell cycle process GO:0090305 nucleic acid phosphodiester bond hydrolysis GO:0097421 liver regeneration GO:1901654 response to ketone GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901991 negative regulation of mitotic cell cycle phase transition GO:1902065 response to L-glutamate GO:1902400 intracellular signal transduction involved in G1 DNA damage checkpoint GO:1902402 signal transduction involved in mitotic DNA damage checkpoint GO:1902403 signal transduction involved in mitotic DNA integrity checkpoint GO:1902806 regulation of cell cycle G1/S phase transition GO:1902807 negative regulation of cell cycle G1/S phase transition GO:1902990 mitotic telomere maintenance via semi-conservative replication GO:2000045 regulation of G1/S transition of mitotic cell cycle GO:2000134 negative regulation of G1/S transition of mitotic cell cycle GO:2001020 regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus |
| Molecular Function |
GO:0000700 mismatch base pair DNA N-glycosylase activity GO:0000701 purine-specific mismatch base pair DNA N-glycosylase activity GO:0003682 chromatin binding GO:0003684 damaged DNA binding GO:0016798 hydrolase activity, acting on glycosyl bonds GO:0016799 hydrolase activity, hydrolyzing N-glycosyl compounds GO:0019104 DNA N-glycosylase activity GO:0030331 estrogen receptor binding GO:0030337 DNA polymerase processivity factor activity GO:0030971 receptor tyrosine kinase binding GO:0030983 mismatched DNA binding GO:0032135 DNA insertion or deletion binding GO:0032139 dinucleotide insertion or deletion binding GO:0032404 mismatch repair complex binding GO:0032405 MutLalpha complex binding GO:0035035 histone acetyltransferase binding GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0051427 hormone receptor binding GO:0070182 DNA polymerase binding GO:1990782 protein tyrosine kinase binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0005657 replication fork GO:0005663 DNA replication factor C complex GO:0005813 centrosome GO:0030894 replisome GO:0032993 protein-DNA complex GO:0043596 nuclear replication fork GO:0043626 PCNA complex GO:0044454 nuclear chromosome part GO:0044796 DNA polymerase processivity factor complex GO:0070557 PCNA-p21 complex GO:0098687 chromosomal region |
| KEGG |
hsa03030 DNA replication hsa03410 Base excision repair hsa03420 Nucleotide excision repair hsa03430 Mismatch repair hsa04110 Cell cycle hsa04530 Tight junction |
| Reactome |
R-HSA-73884: Base Excision Repair R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-73886: Chromosome Maintenance R-HSA-73893: DNA Damage Bypass R-HSA-5693532: DNA Double-Strand Break Repair R-HSA-73894: DNA Repair R-HSA-69306: DNA Replication R-HSA-69190: DNA strand elongation R-HSA-5696400: Dual Incision in GG-NER R-HSA-6782135: Dual incision in TC-NER R-HSA-113510: E2F mediated regulation of DNA replication R-HSA-180786: Extension of Telomeres R-HSA-1538133: G0 and Early G1 R-HSA-69206: G1/S Transition R-HSA-69205: G1/S-Specific Transcription R-HSA-5696397: Gap-filling DNA repair synthesis and ligation in GG-NER R-HSA-6782210: Gap-filling DNA repair synthesis and ligation in TC-NER R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-5696399: Global Genome Nucleotide Excision Repair (GG-NER) R-HSA-5693567: HDR through Homologous Recombination (HR) or Single Strand Annealing (SSA) R-HSA-5685942: HDR through Homologous Recombination (HRR) R-HSA-5693538: Homology Directed Repair R-HSA-69186: Lagging Strand Synthesis R-HSA-69109: Leading Strand Synthesis R-HSA-392499: Metabolism of proteins R-HSA-5358508: Mismatch Repair R-HSA-5358606: Mismatch repair (MMR) directed by MSH2 R-HSA-5358565: Mismatch repair (MMR) directed by MSH2 R-HSA-453279: Mitotic G1-G1/S phases R-HSA-5696398: Nucleotide Excision Repair R-HSA-5651801: PCNA-Dependent Long Patch Base Excision Repair R-HSA-69091: Polymerase switching R-HSA-174411: Polymerase switching on the C-strand of the telomere R-HSA-597592: Post-translational protein modification R-HSA-174414: Processive synthesis on the C-strand of the telomere R-HSA-69183: Processive synthesis on the lagging strand R-HSA-110314: Recognition of DNA damage by PCNA-containing replication complex R-HSA-69166: Removal of the Flap Intermediate R-HSA-174437: Removal of the Flap Intermediate from the C-strand R-HSA-110373: Resolution of AP sites via the multiple-nucleotide patch replacement pathway R-HSA-73933: Resolution of Abasic Sites (AP sites) R-HSA-69242: S Phase R-HSA-3108232: SUMO E3 ligases SUMOylate target proteins R-HSA-2990846: SUMOylation R-HSA-4615885: SUMOylation of DNA replication proteins R-HSA-69239: Synthesis of DNA R-HSA-6791312: TP53 Regulates Transcription of Cell Cycle Genes R-HSA-6804114: TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest R-HSA-174417: Telomere C-strand (Lagging Strand) Synthesis R-HSA-157579: Telomere Maintenance R-HSA-5656169: Termination of translesion DNA synthesis R-HSA-6781827: Transcription-Coupled Nucleotide Excision Repair (TC-NER) R-HSA-3700989: Transcriptional Regulation by TP53 R-HSA-110320: Translesion Synthesis by POLH R-HSA-5656121: Translesion synthesis by POLI R-HSA-5655862: Translesion synthesis by POLK R-HSA-110312: Translesion synthesis by REV1 R-HSA-110313: Translesion synthesis by Y family DNA polymerases bypasses lesions on DNA template |
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for PCNA. |
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
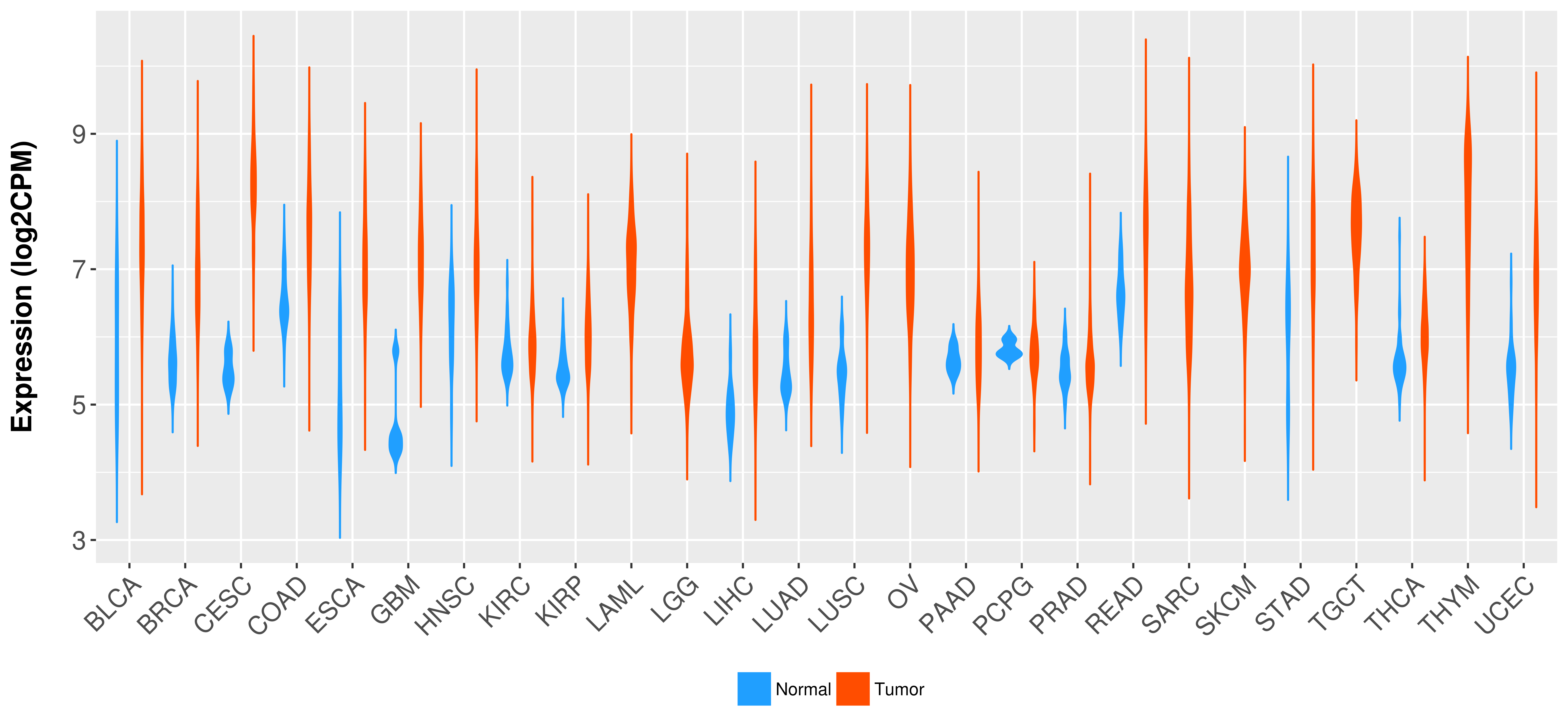
Differential expression analysis for cancers with more than 10 normal samples
|
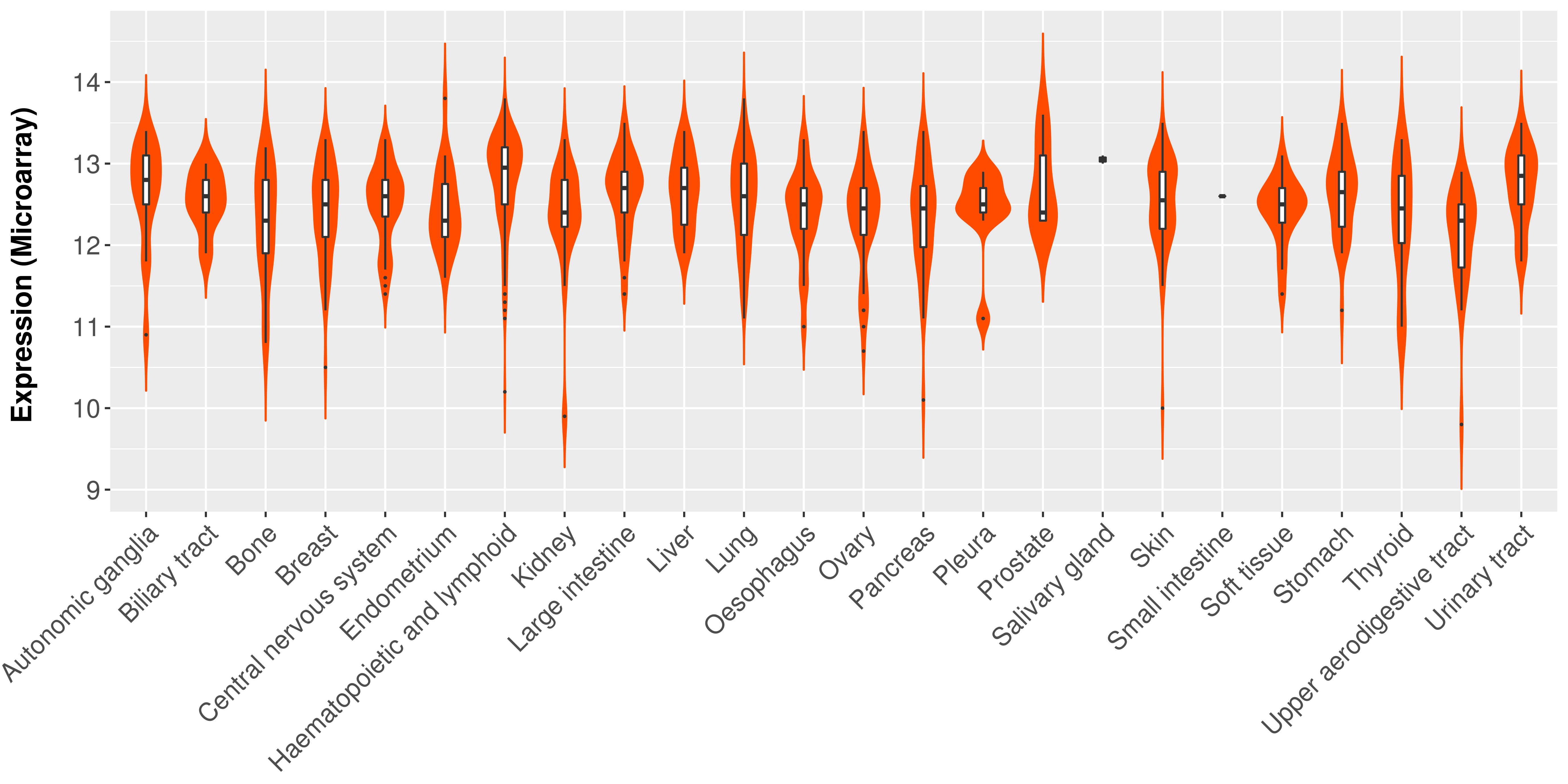
|
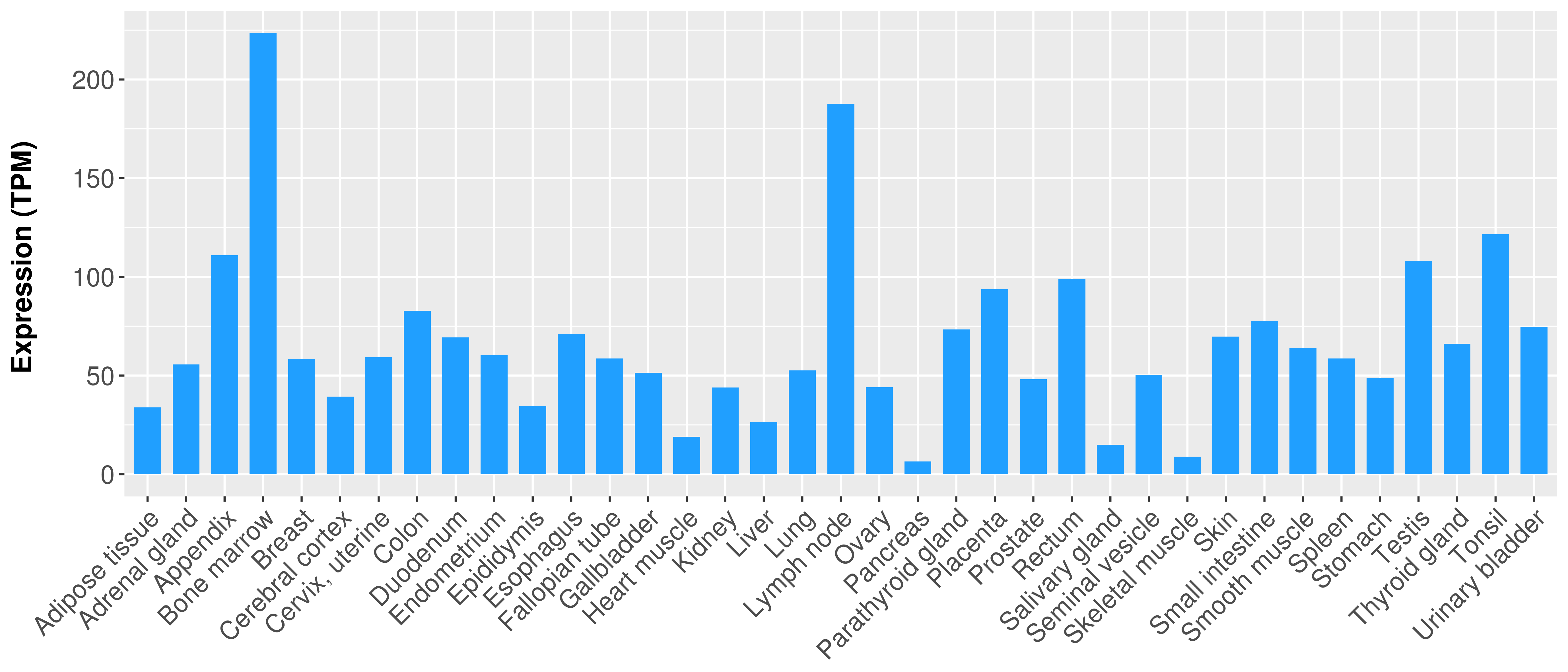
|
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
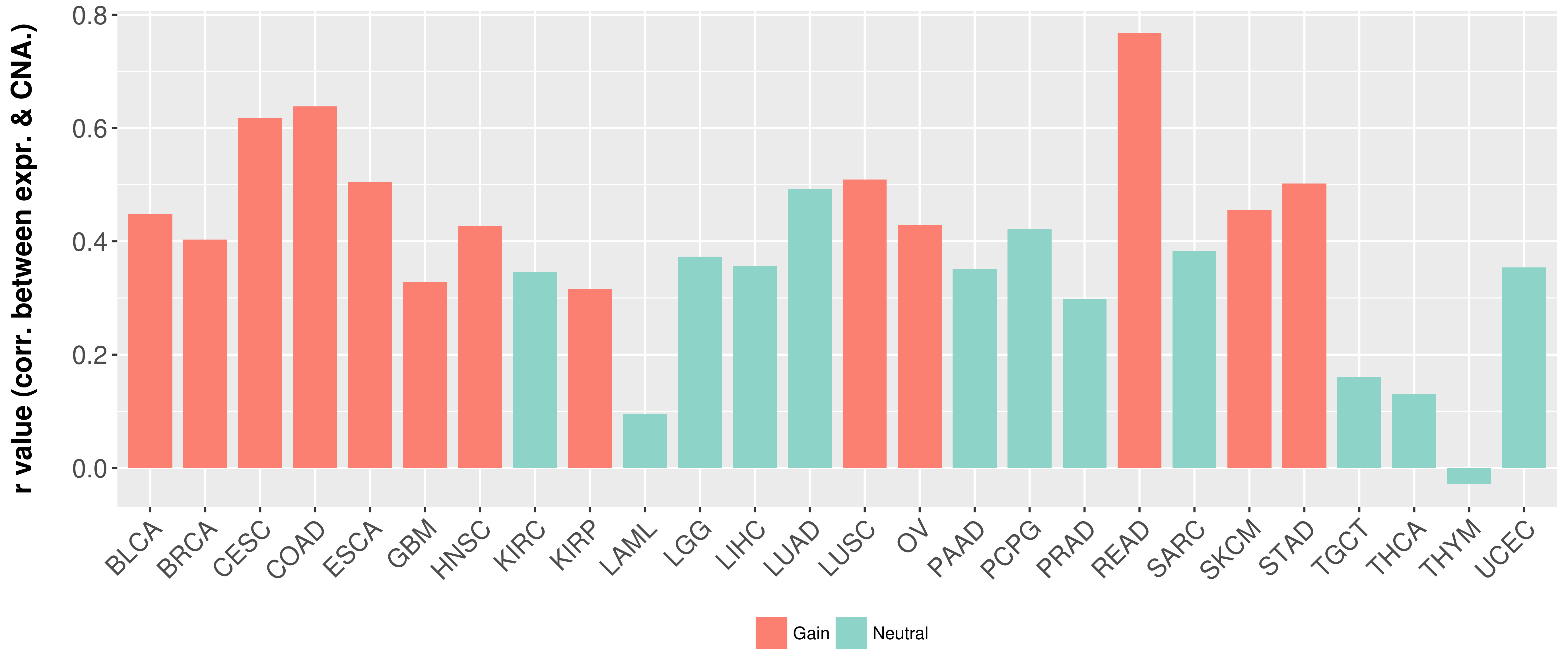
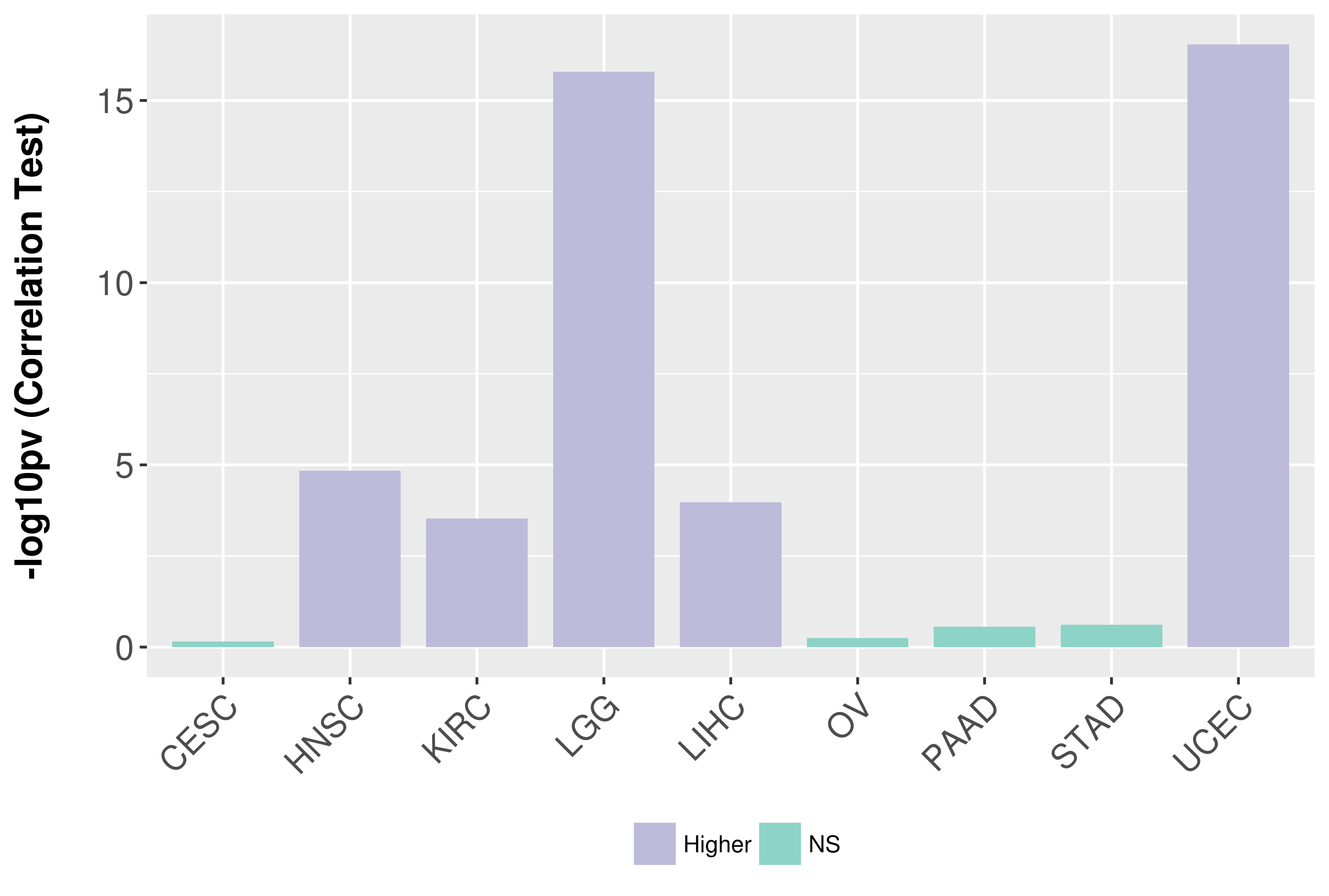
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
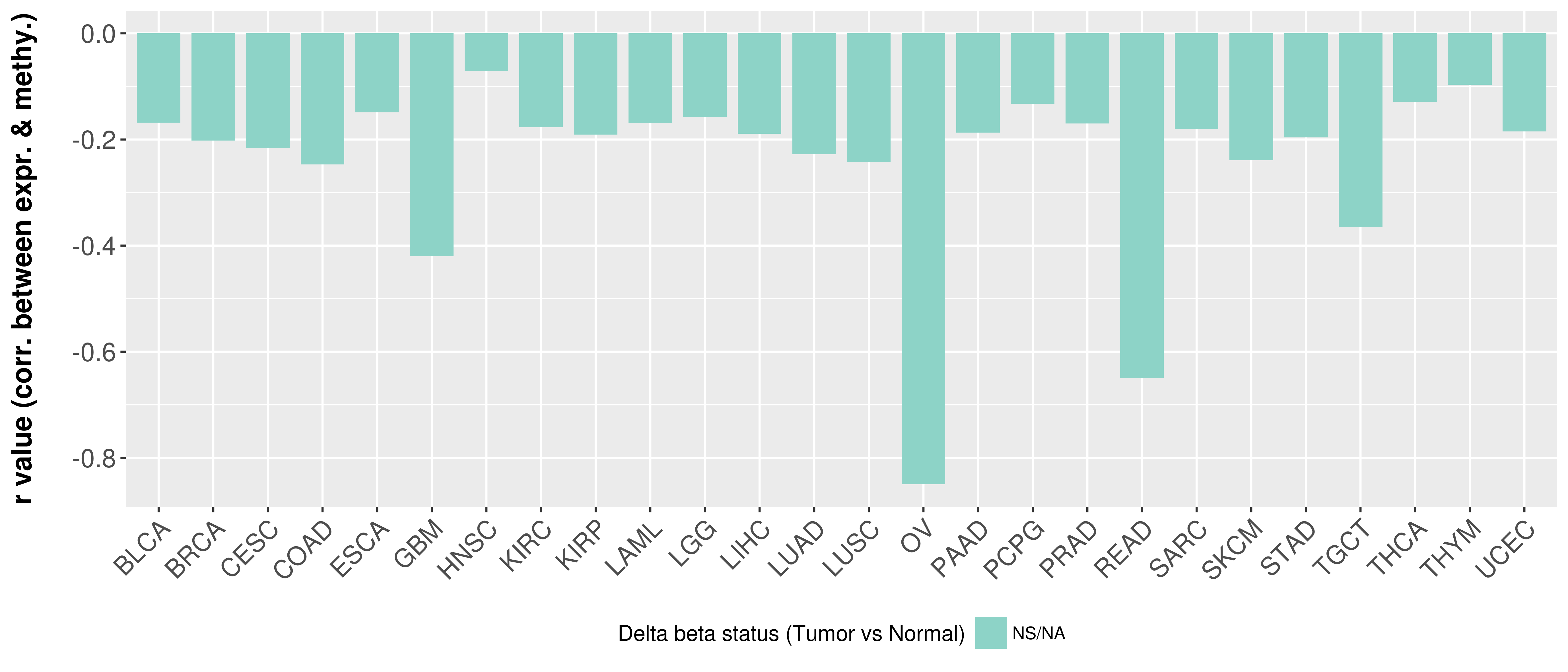
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
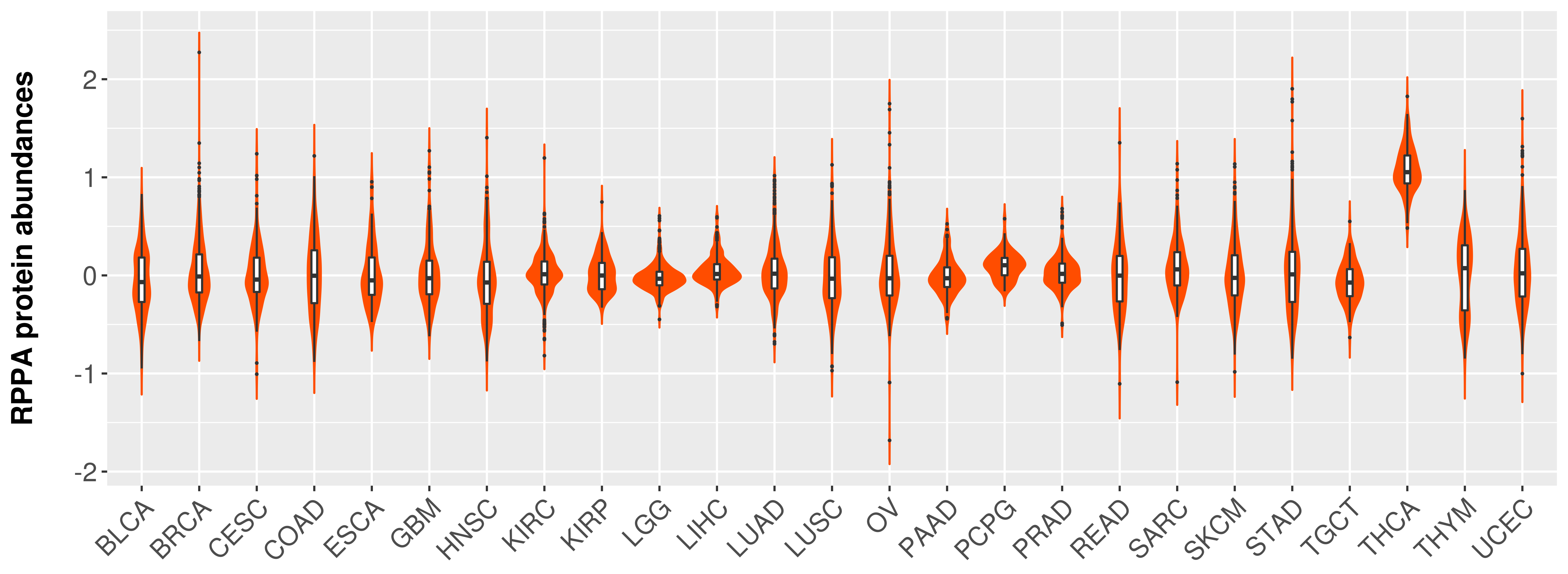
|
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
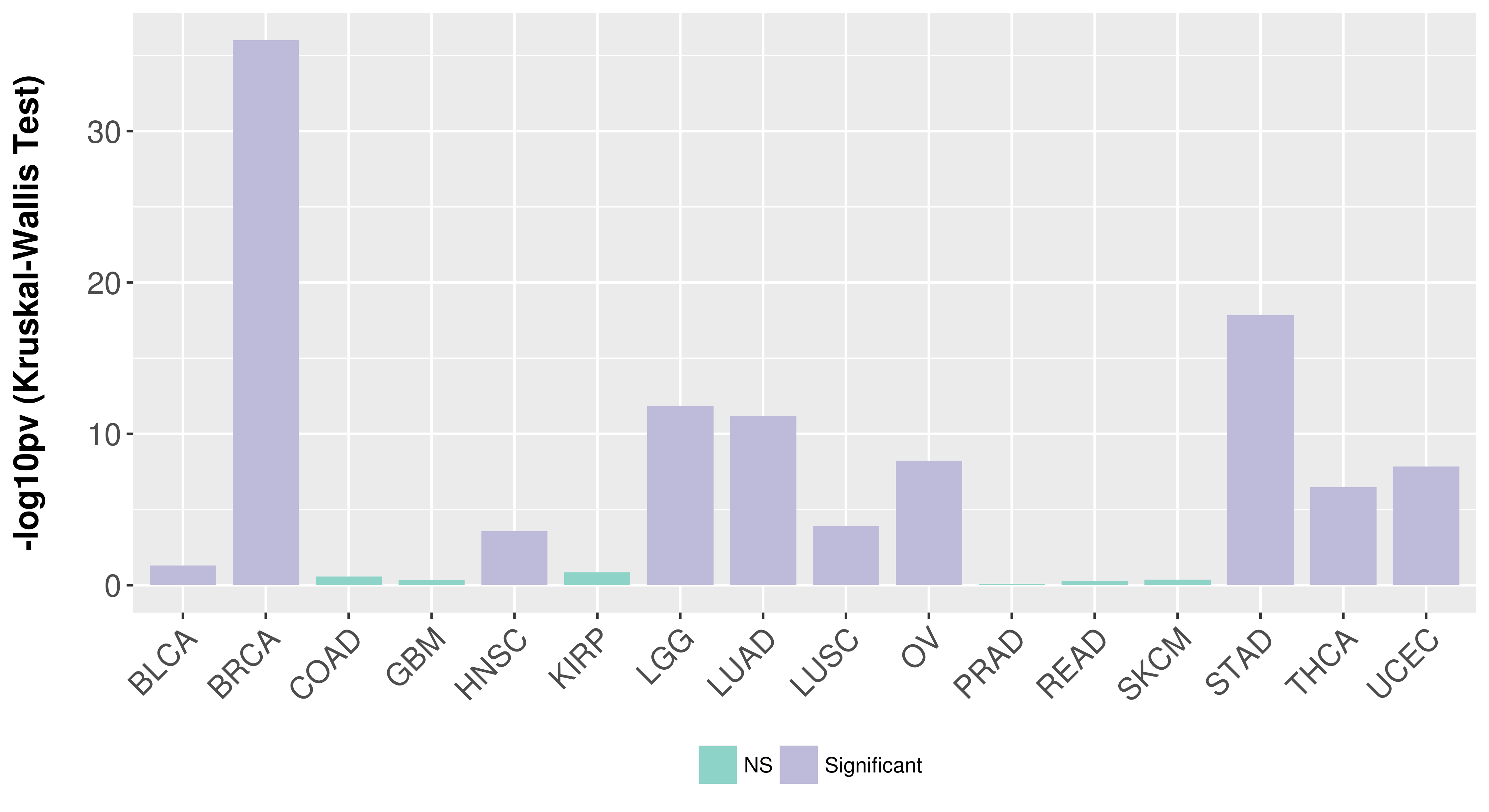
Association between expresson and subtype.
|
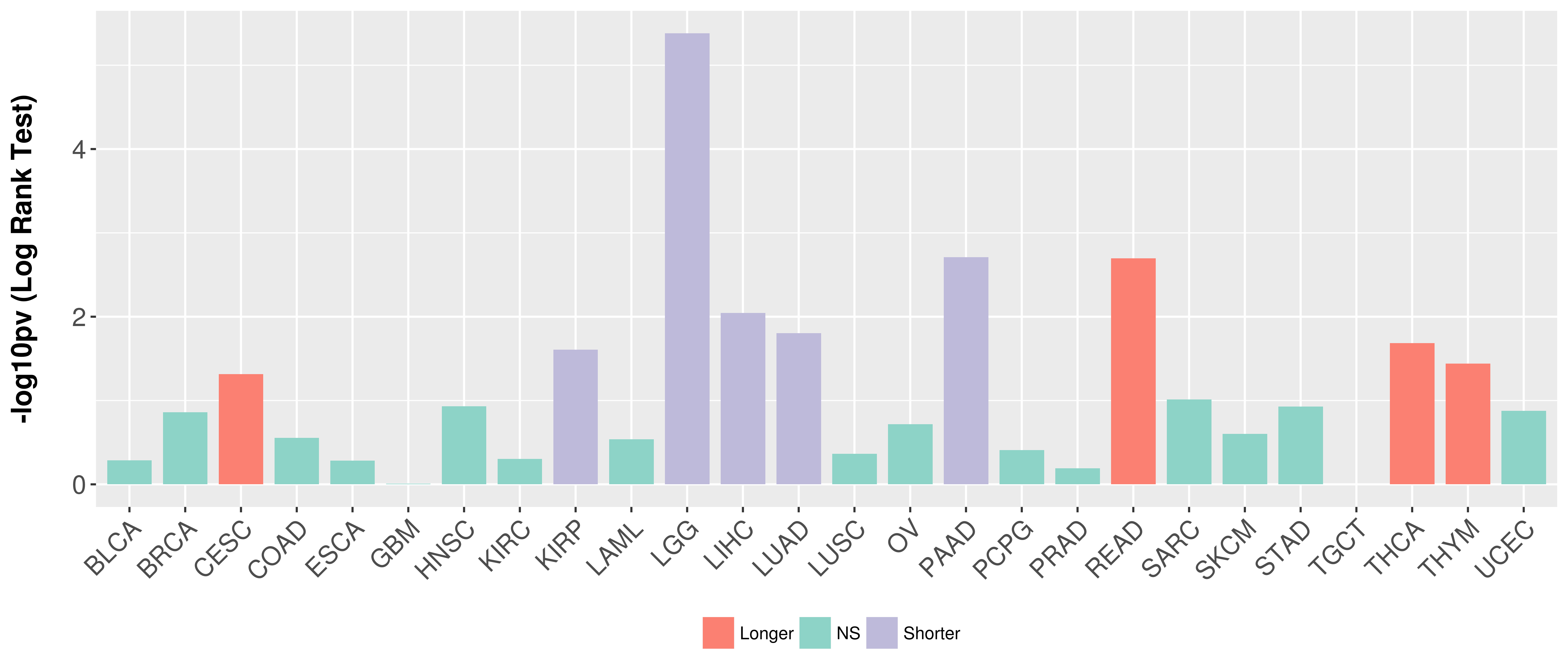
Overall survival analysis based on expression.
|

Association between expresson and stage.
|
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for PCNA. |
| Summary | |
|---|---|
| Symbol | PCNA |
| Name | proliferating cell nuclear antigen |
| Aliases | ATLD2; DNA polymerase delta auxiliary protein; cyclin |
| Location | 20p12.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|