Browse PHF8 in pancancer
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02373 JmjC domain PF00628 PHD-finger |
||||||||||
| Function |
Histone lysine demethylase with selectivity for the di- and monomethyl states that plays a key role cell cycle progression, rDNA transcription and brain development. Demethylates mono- and dimethylated histone H3 'Lys-9' residue (H3K9Me1 and H3K9Me2), dimethylated H3 'Lys-27' (H3K27Me2) and monomethylated histone H4 'Lys-20' residue (H4K20Me1). Acts as a transcription activator as H3K9Me1, H3K9Me2, H3K27Me2 and H4K20Me1 are epigenetic repressive marks. Involved in cell cycle progression by being required to control G1-S transition. Acts as a coactivator of rDNA transcription, by activating polymerase I (pol I) mediated transcription of rRNA genes. Required for brain development, probably by regulating expression of neuron-specific genes. Only has activity toward H4K20Me1 when nucleosome is used as a substrate and when not histone octamer is used as substrate. May also have weak activity toward dimethylated H3 'Lys-36' (H3K36Me2), however, the relevance of this result remains unsure in vivo. Specifically binds trimethylated 'Lys-4' of histone H3 (H3K4me3), affecting histone demethylase specificity: has weak activity toward H3K9Me2 in absence of H3K4me3, while it has high activity toward H3K9me2 when binding H3K4me3. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000082 G1/S transition of mitotic cell cycle GO:0000183 chromatin silencing at rDNA GO:0006342 chromatin silencing GO:0006356 regulation of transcription from RNA polymerase I promoter GO:0006360 transcription from RNA polymerase I promoter GO:0006482 protein demethylation GO:0008214 protein dealkylation GO:0016458 gene silencing GO:0016570 histone modification GO:0016577 histone demethylation GO:0031935 regulation of chromatin silencing GO:0031936 negative regulation of chromatin silencing GO:0033169 histone H3-K9 demethylation GO:0035574 histone H4-K20 demethylation GO:0040029 regulation of gene expression, epigenetic GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044843 cell cycle G1/S phase transition GO:0045814 negative regulation of gene expression, epigenetic GO:0045815 positive regulation of gene expression, epigenetic GO:0045943 positive regulation of transcription from RNA polymerase I promoter GO:0060968 regulation of gene silencing GO:0060969 negative regulation of gene silencing GO:0061187 regulation of chromatin silencing at rDNA GO:0061188 negative regulation of chromatin silencing at rDNA GO:0070076 histone lysine demethylation GO:0070544 histone H3-K36 demethylation GO:0070988 demethylation GO:0071557 histone H3-K27 demethylation GO:1902275 regulation of chromatin organization GO:1905268 negative regulation of chromatin organization |
| Molecular Function |
GO:0003682 chromatin binding GO:0005506 iron ion binding GO:0016705 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen GO:0016706 oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors GO:0032451 demethylase activity GO:0032452 histone demethylase activity GO:0032454 histone demethylase activity (H3-K9 specific) GO:0035064 methylated histone binding GO:0035575 histone demethylase activity (H4-K20 specific) GO:0042393 histone binding GO:0051213 dioxygenase activity GO:0051864 histone demethylase activity (H3-K36 specific) GO:0071558 histone demethylase activity (H3-K27 specific) |
| Cellular Component |
GO:0005635 nuclear envelope GO:0031965 nuclear membrane |
| KEGG | - |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2299718: Condensation of Prophase Chromosomes R-HSA-3214842: HDMs demethylate histones R-HSA-68886: M Phase R-HSA-68875: Mitotic Prophase |
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for PHF8. |
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
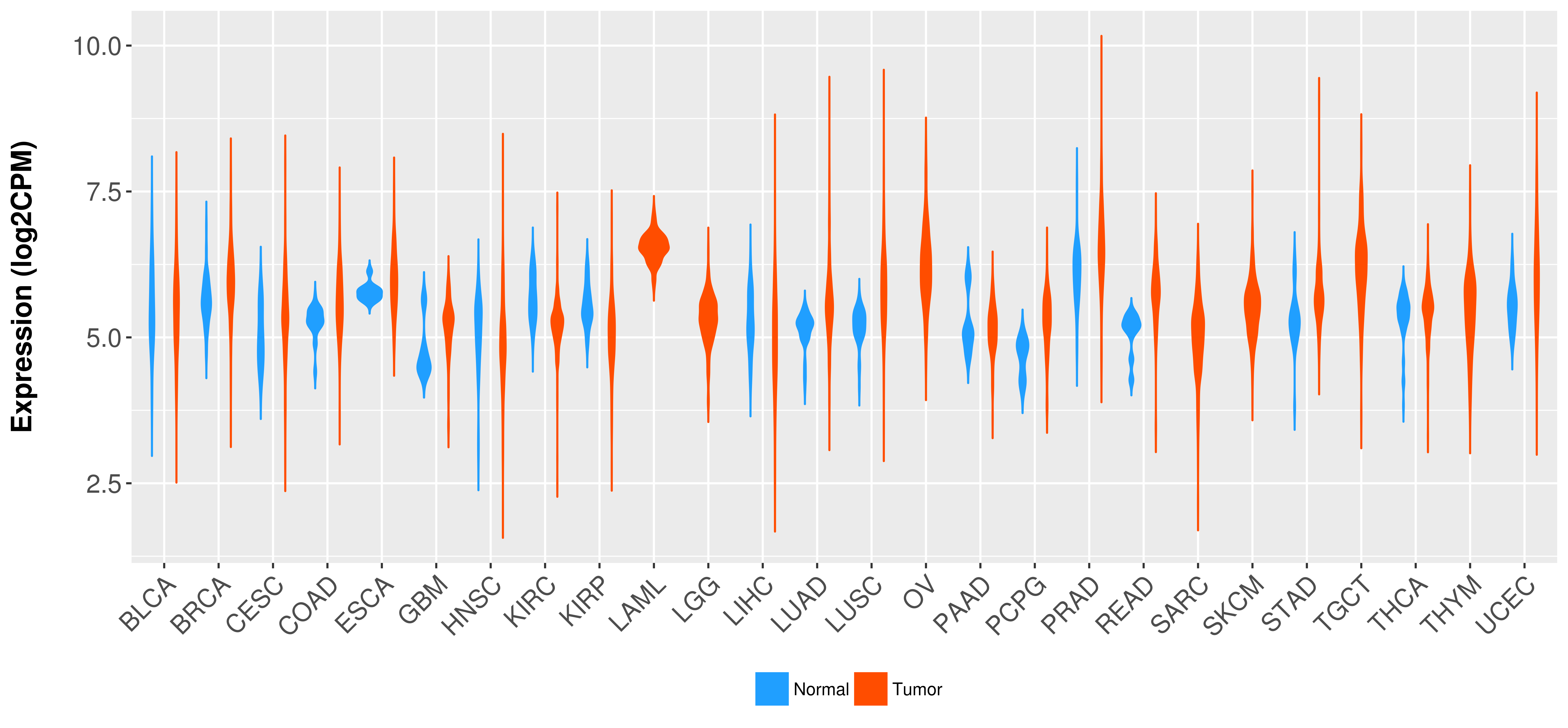
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
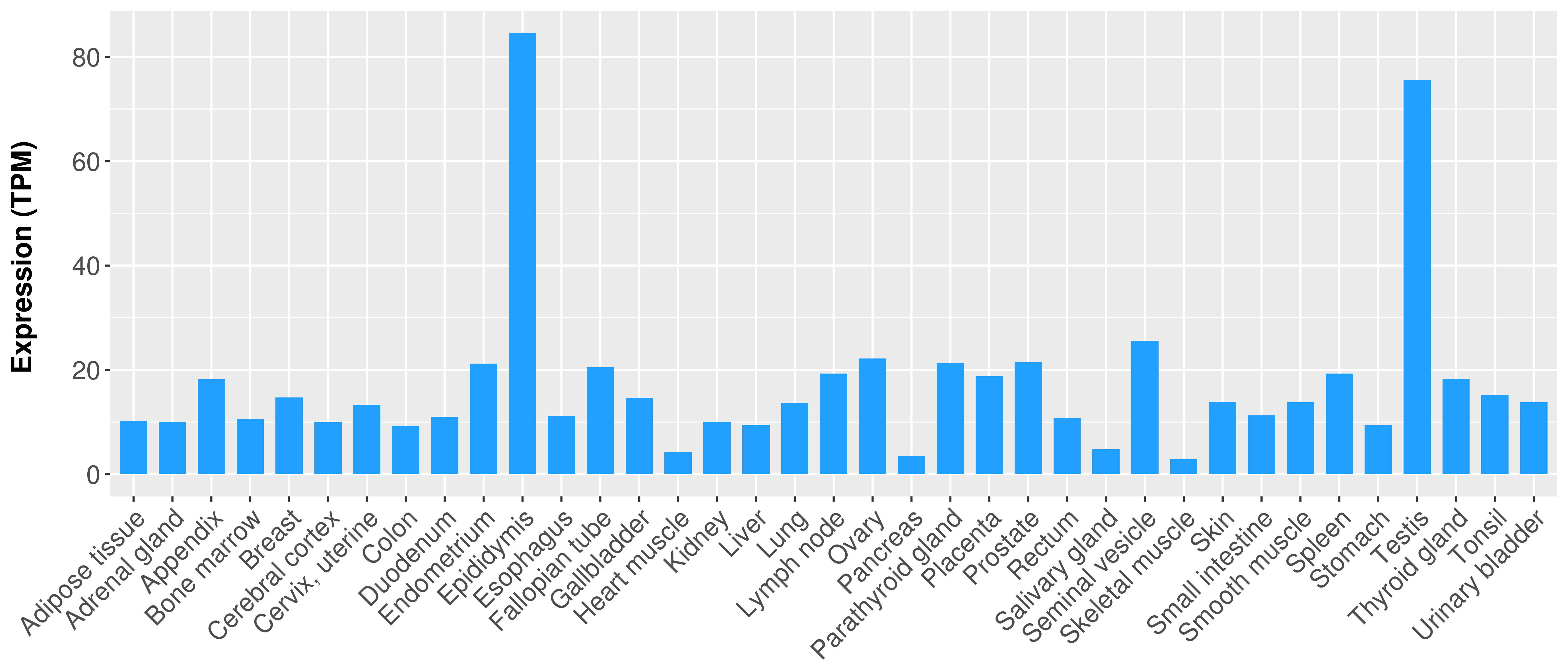
|
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
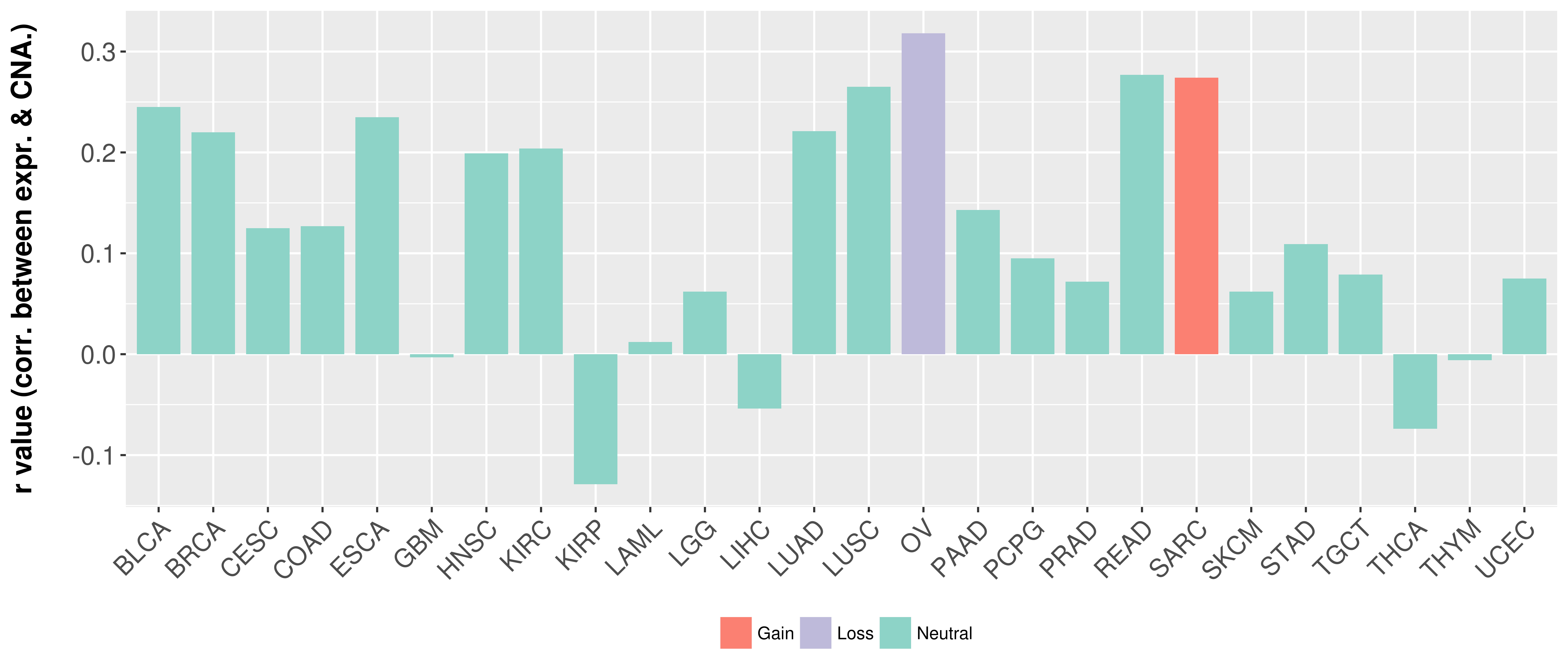
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
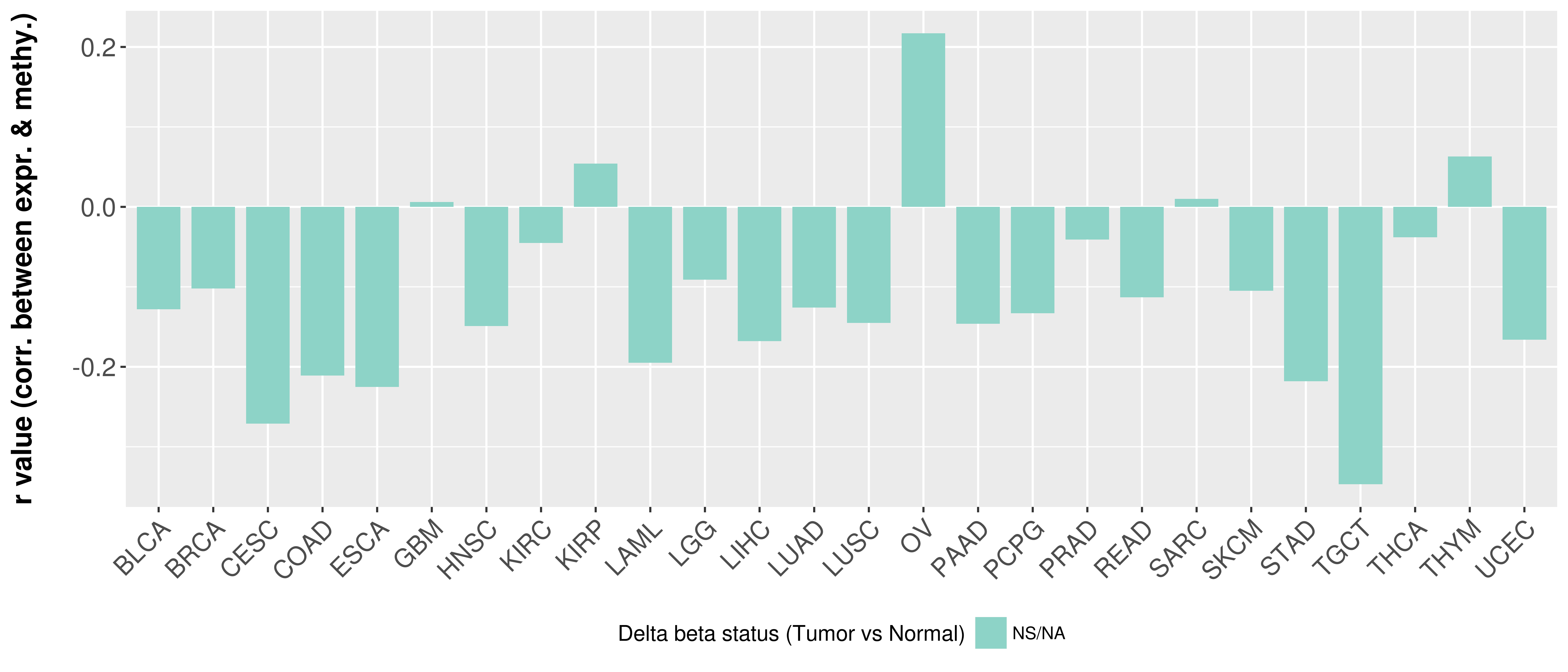
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
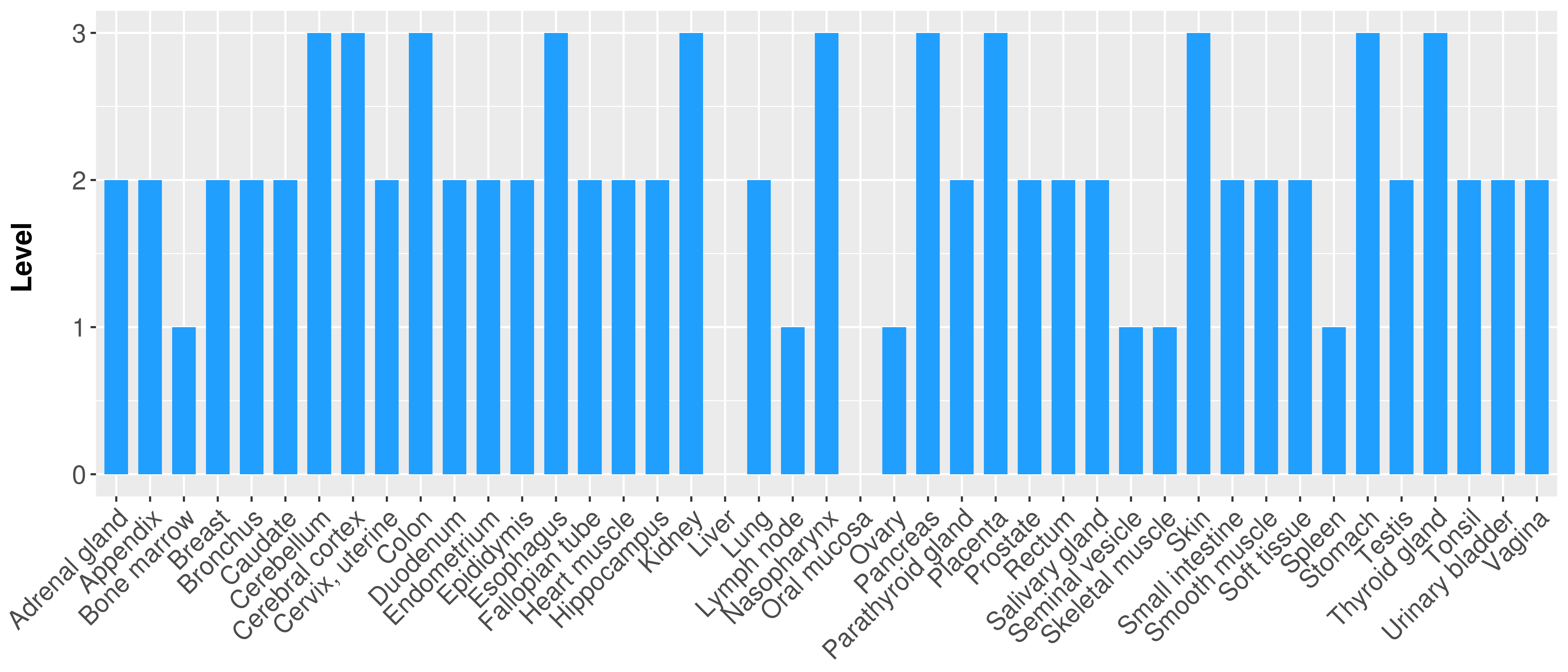
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
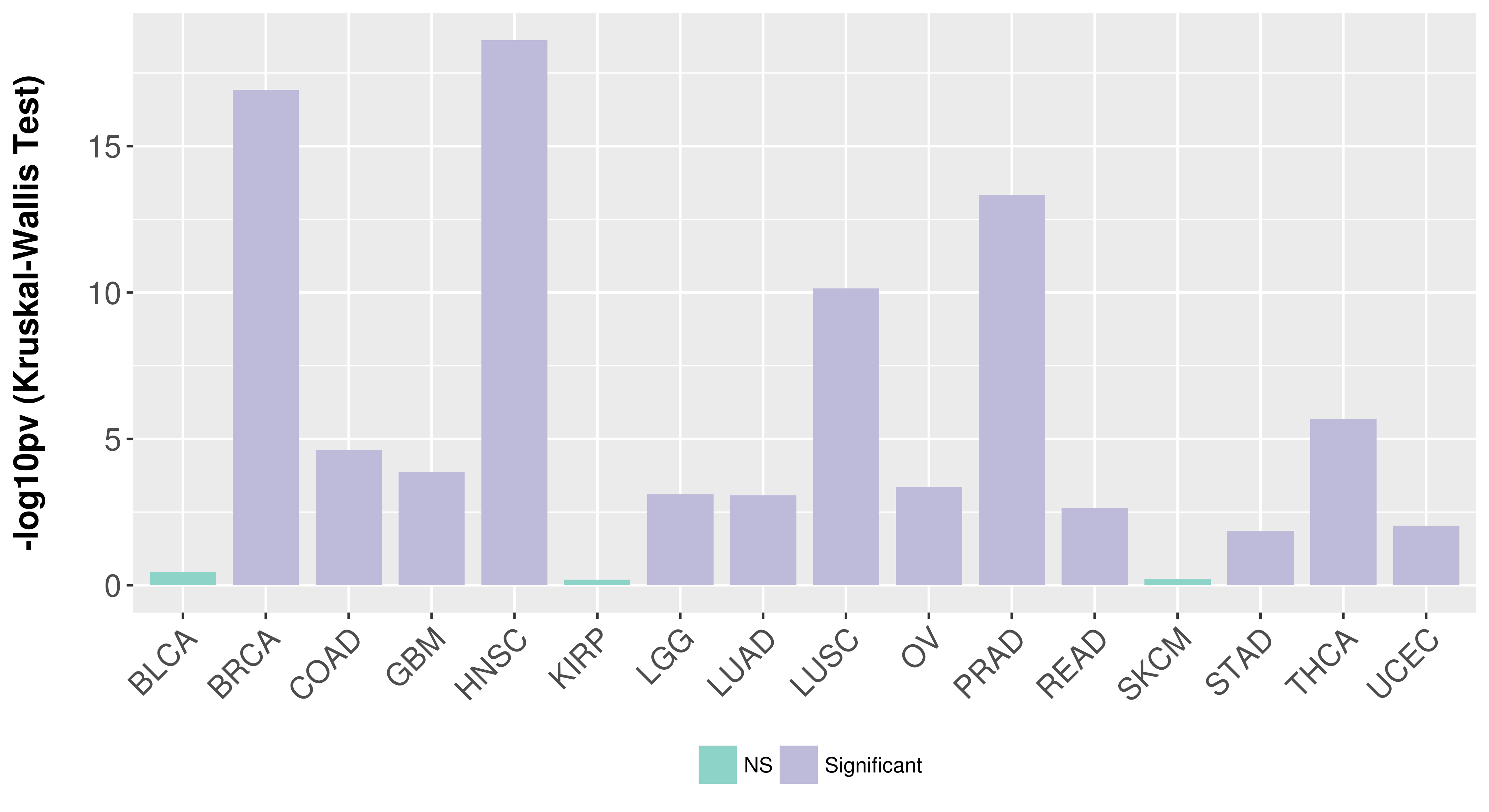
Association between expresson and subtype.
|
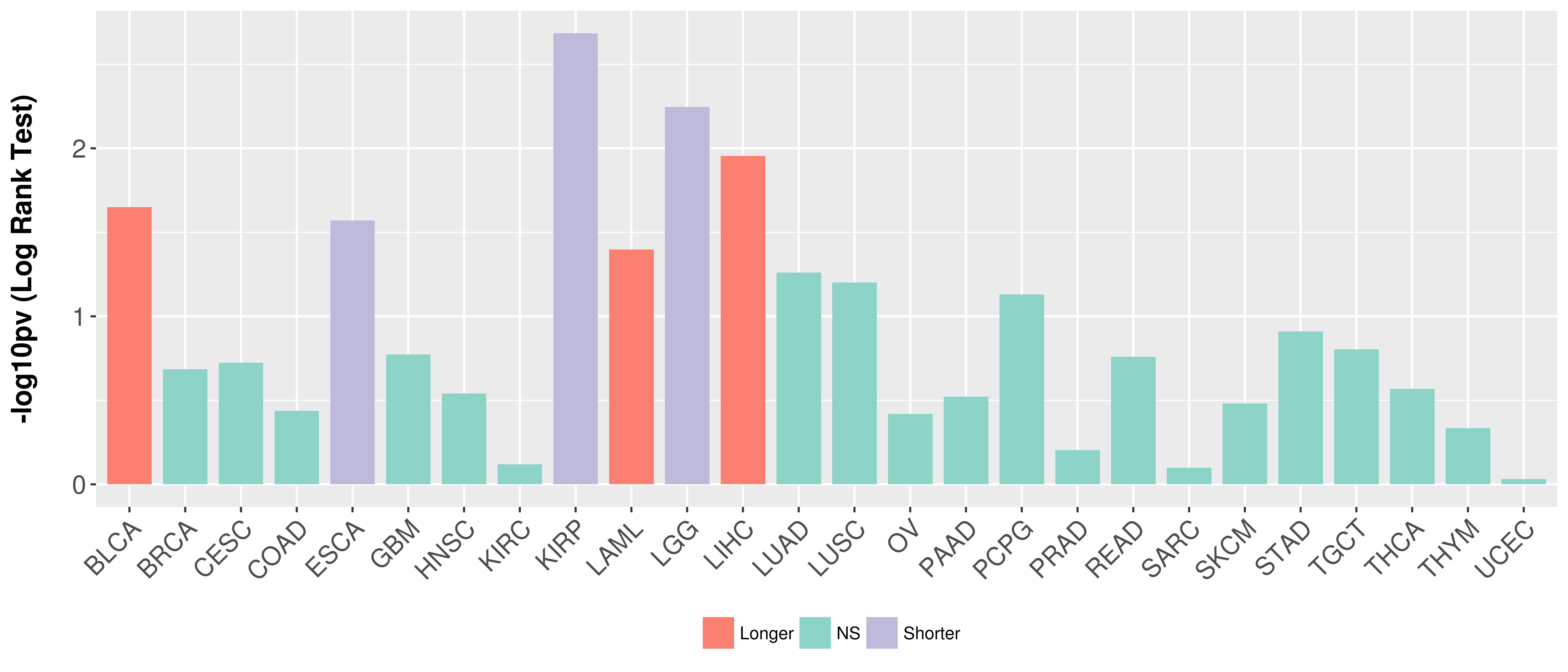
Overall survival analysis based on expression.
|
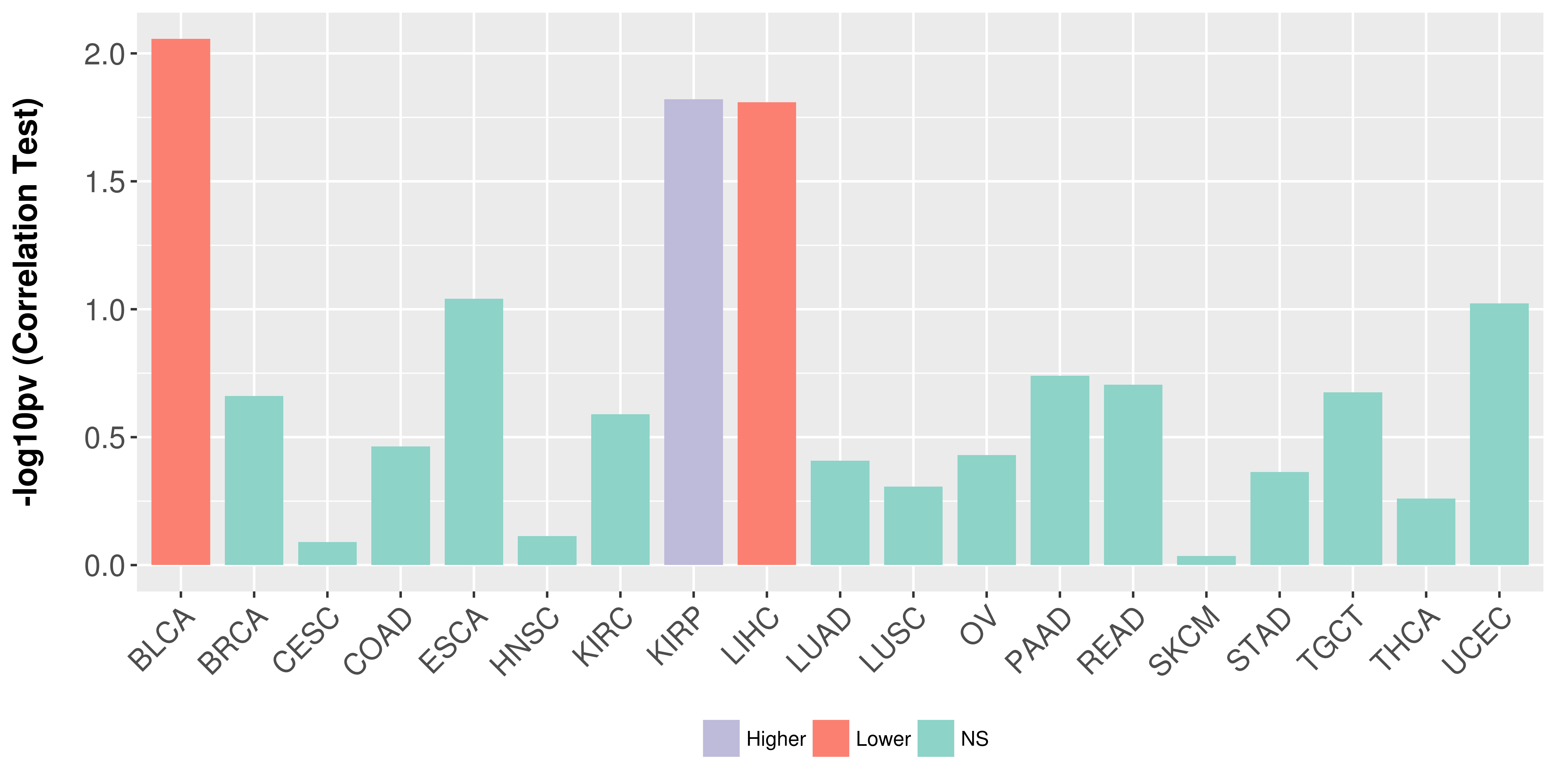
Association between expresson and stage.
|
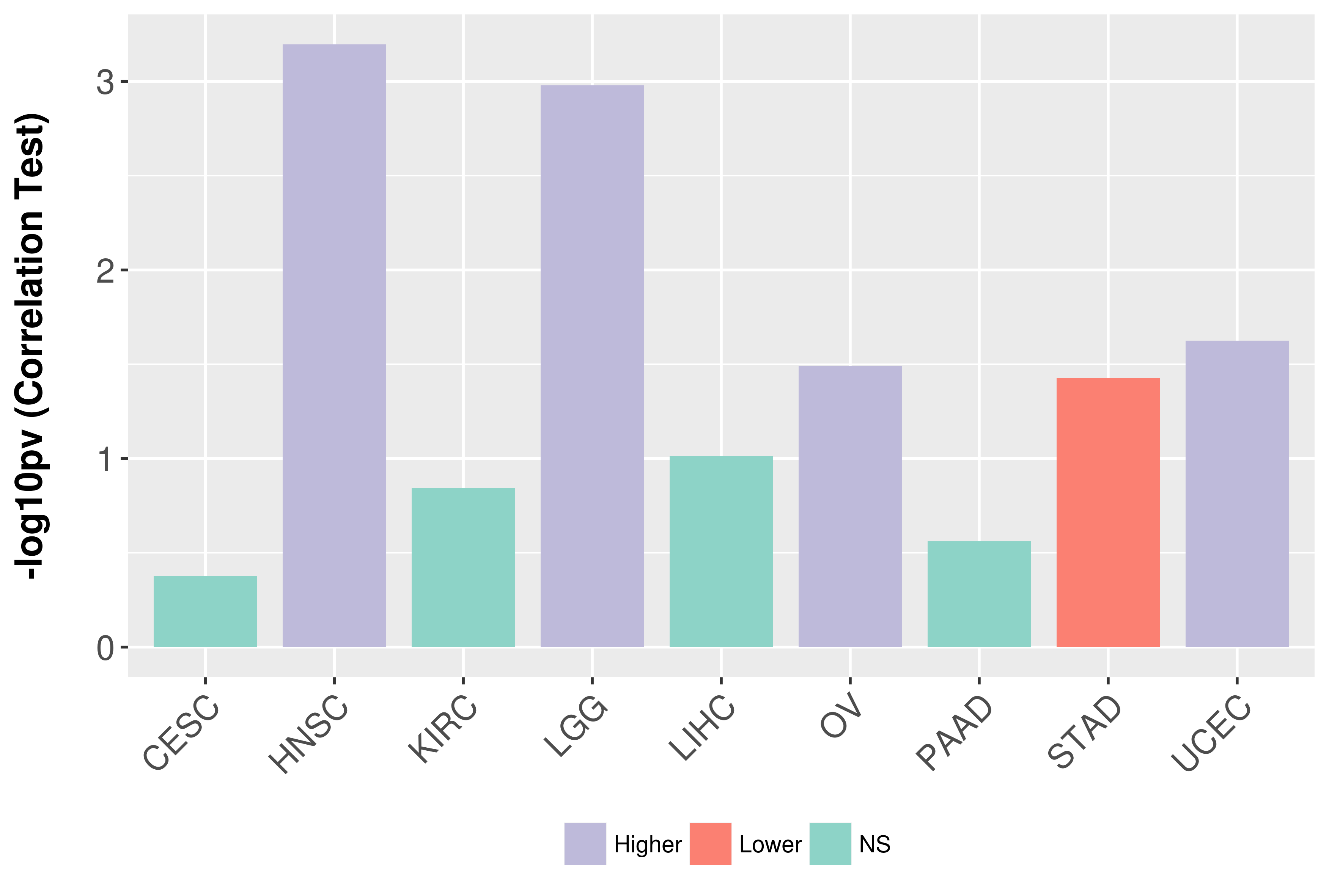
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for PHF8. |
| Summary | |
|---|---|
| Symbol | PHF8 |
| Name | PHD finger protein 8 |
| Aliases | KIAA1111; JHDM1F; jumonji C domain-containing histone demethylase 1F; MRXSSD; Histone lysine demethylase PHF ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|