Browse PRDM1 in pancancer
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00856 SET domain |
||||||||||
| Function |
Transcription factor that mediates a transcriptional program in various innate and adaptive immune tissue-resident lymphocyte T cell types such as tissue-resident memory T (Trm), natural killer (trNK) and natural killer T (NKT) cells and negatively regulates gene expression of proteins that promote the egress of tissue-resident T-cell populations from non-lymphoid organs. Plays a role in the development, retention and long-term establishment of adaptive and innate tissue-resident lymphocyte T cell types in non-lymphoid organs, such as the skin and gut, but also in other nonbarrier tissues like liver and kidney, and therefore may provide immediate immunological protection against reactivating infections or viral reinfection (By similarity). Binds specifically to the PRDI element in the promoter of the beta-interferon gene (PubMed:1851123). Drives the maturation of B-lymphocytes into Ig secreting cells (PubMed:12626569). Associates with the transcriptional repressor ZNF683 to chromatin at gene promoter regions (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001654 eye development GO:0001701 in utero embryonic development GO:0001754 eye photoreceptor cell differentiation GO:0001763 morphogenesis of a branching structure GO:0001779 natural killer cell differentiation GO:0001865 NK T cell differentiation GO:0001890 placenta development GO:0001892 embryonic placenta development GO:0001893 maternal placenta development GO:0002064 epithelial cell development GO:0002065 columnar/cuboidal epithelial cell differentiation GO:0002066 columnar/cuboidal epithelial cell development GO:0002237 response to molecule of bacterial origin GO:0002250 adaptive immune response GO:0002521 leukocyte differentiation GO:0002683 negative regulation of immune system process GO:0002694 regulation of leukocyte activation GO:0002695 negative regulation of leukocyte activation GO:0002696 positive regulation of leukocyte activation GO:0002831 regulation of response to biotic stimulus GO:0002832 negative regulation of response to biotic stimulus GO:0003170 heart valve development GO:0003205 cardiac chamber development GO:0003231 cardiac ventricle development GO:0003279 cardiac septum development GO:0003281 ventricular septum development GO:0007159 leukocyte cell-cell adhesion GO:0007281 germ cell development GO:0007423 sensory organ development GO:0007507 heart development GO:0007565 female pregnancy GO:0009791 post-embryonic development GO:0022407 regulation of cell-cell adhesion GO:0022412 cellular process involved in reproduction in multicellular organism GO:0030098 lymphocyte differentiation GO:0030101 natural killer cell activation GO:0030183 B cell differentiation GO:0030217 T cell differentiation GO:0030888 regulation of B cell proliferation GO:0030889 negative regulation of B cell proliferation GO:0031663 lipopolysaccharide-mediated signaling pathway GO:0031664 regulation of lipopolysaccharide-mediated signaling pathway GO:0031665 negative regulation of lipopolysaccharide-mediated signaling pathway GO:0032102 negative regulation of response to external stimulus GO:0032259 methylation GO:0032496 response to lipopolysaccharide GO:0032814 regulation of natural killer cell activation GO:0032823 regulation of natural killer cell differentiation GO:0032943 mononuclear cell proliferation GO:0032944 regulation of mononuclear cell proliferation GO:0032945 negative regulation of mononuclear cell proliferation GO:0033078 extrathymic T cell differentiation GO:0033082 regulation of extrathymic T cell differentiation GO:0035904 aorta development GO:0042100 B cell proliferation GO:0042110 T cell activation GO:0042113 B cell activation GO:0042461 photoreceptor cell development GO:0042462 eye photoreceptor cell development GO:0043900 regulation of multi-organism process GO:0043901 negative regulation of multi-organism process GO:0044706 multi-multicellular organism process GO:0045165 cell fate commitment GO:0045577 regulation of B cell differentiation GO:0045579 positive regulation of B cell differentiation GO:0045580 regulation of T cell differentiation GO:0045619 regulation of lymphocyte differentiation GO:0045621 positive regulation of lymphocyte differentiation GO:0046530 photoreceptor cell differentiation GO:0046631 alpha-beta T cell activation GO:0046632 alpha-beta T cell differentiation GO:0046634 regulation of alpha-beta T cell activation GO:0046637 regulation of alpha-beta T cell differentiation GO:0046651 lymphocyte proliferation GO:0048514 blood vessel morphogenesis GO:0048565 digestive tract development GO:0048568 embryonic organ development GO:0048592 eye morphogenesis GO:0048608 reproductive structure development GO:0048844 artery morphogenesis GO:0050670 regulation of lymphocyte proliferation GO:0050672 negative regulation of lymphocyte proliferation GO:0050863 regulation of T cell activation GO:0050864 regulation of B cell activation GO:0050865 regulation of cell activation GO:0050866 negative regulation of cell activation GO:0050867 positive regulation of cell activation GO:0050869 negative regulation of B cell activation GO:0050871 positive regulation of B cell activation GO:0051136 regulation of NK T cell differentiation GO:0051249 regulation of lymphocyte activation GO:0051250 negative regulation of lymphocyte activation GO:0051251 positive regulation of lymphocyte activation GO:0055123 digestive system development GO:0060135 maternal process involved in female pregnancy GO:0060575 intestinal epithelial cell differentiation GO:0060576 intestinal epithelial cell development GO:0060706 cell differentiation involved in embryonic placenta development GO:0060707 trophoblast giant cell differentiation GO:0060840 artery development GO:0060976 coronary vasculature development GO:0061458 reproductive system development GO:0070486 leukocyte aggregation GO:0070489 T cell aggregation GO:0070661 leukocyte proliferation GO:0070663 regulation of leukocyte proliferation GO:0070664 negative regulation of leukocyte proliferation GO:0071216 cellular response to biotic stimulus GO:0071219 cellular response to molecule of bacterial origin GO:0071222 cellular response to lipopolysaccharide GO:0071396 cellular response to lipid GO:0071593 lymphocyte aggregation GO:0090596 sensory organ morphogenesis GO:1902105 regulation of leukocyte differentiation GO:1902107 positive regulation of leukocyte differentiation GO:1903037 regulation of leukocyte cell-cell adhesion GO:1903706 regulation of hemopoiesis GO:1903708 positive regulation of hemopoiesis GO:1990654 sebum secreting cell proliferation |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000982 transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001078 transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0001159 core promoter proximal region DNA binding GO:0001227 transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding GO:0003682 chromatin binding GO:0008168 methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0042826 histone deacetylase binding GO:1990841 promoter-specific chromatin binding |
| Cellular Component | - |
| KEGG | - |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804754: Regulation of TP53 Expression R-HSA-6806003: Regulation of TP53 Expression and Degradation R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|

|
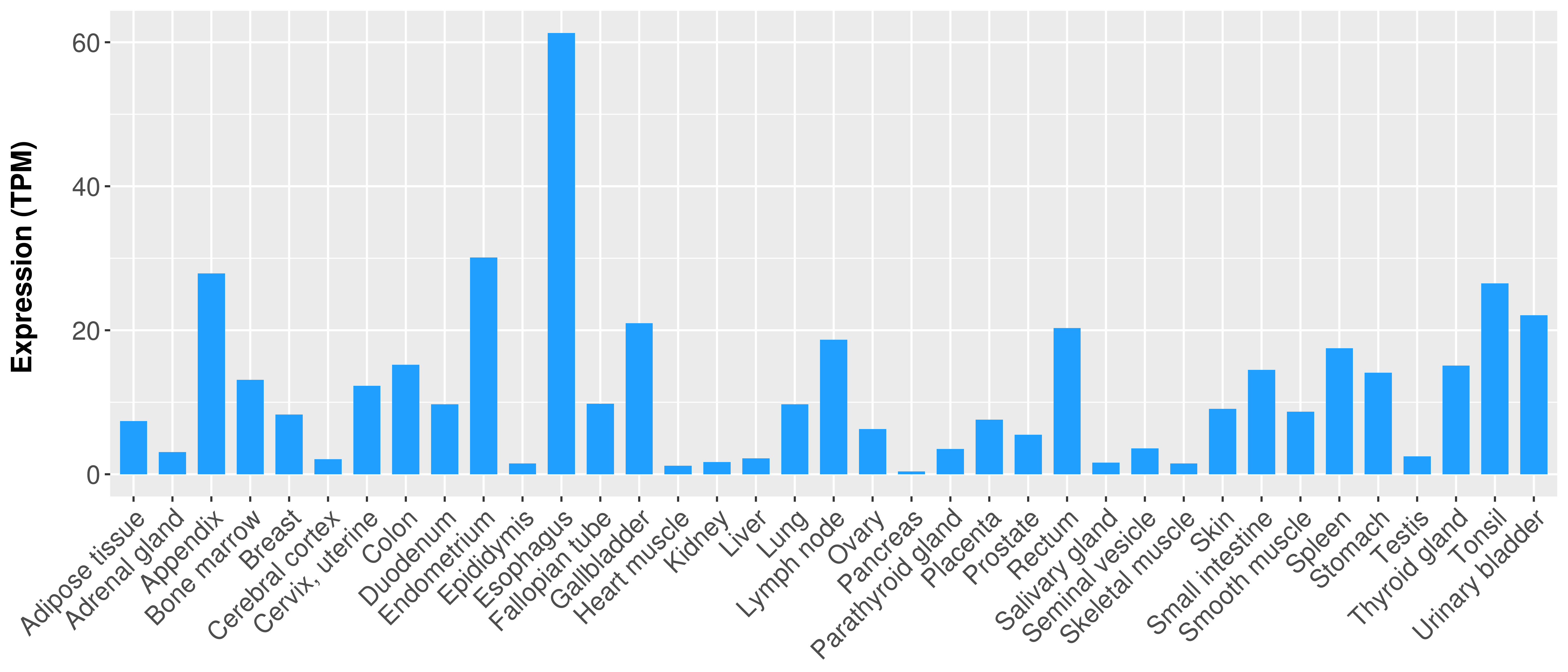
|
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
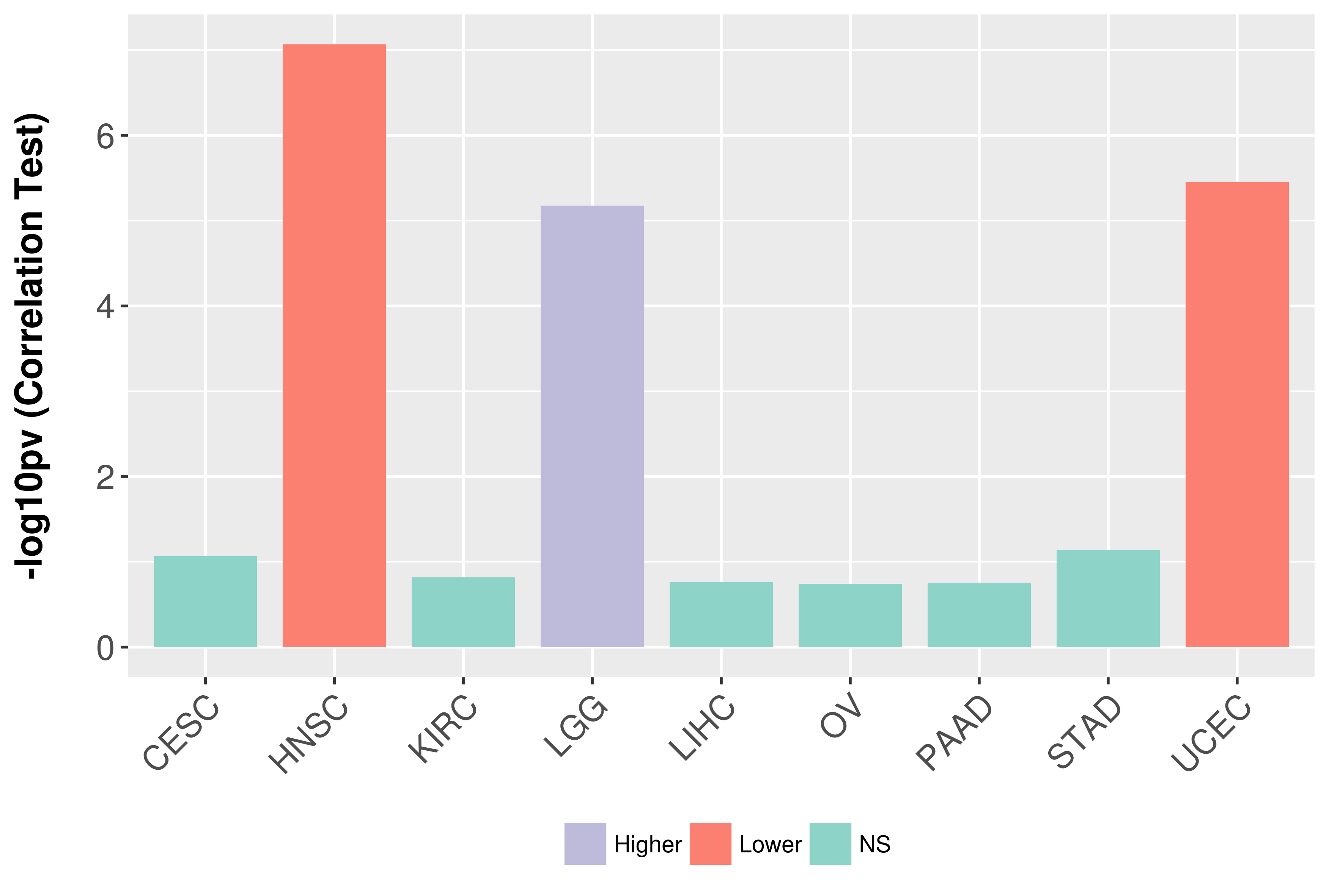
| Content |
> Somatic copy number alteration in primary tomur tissue |
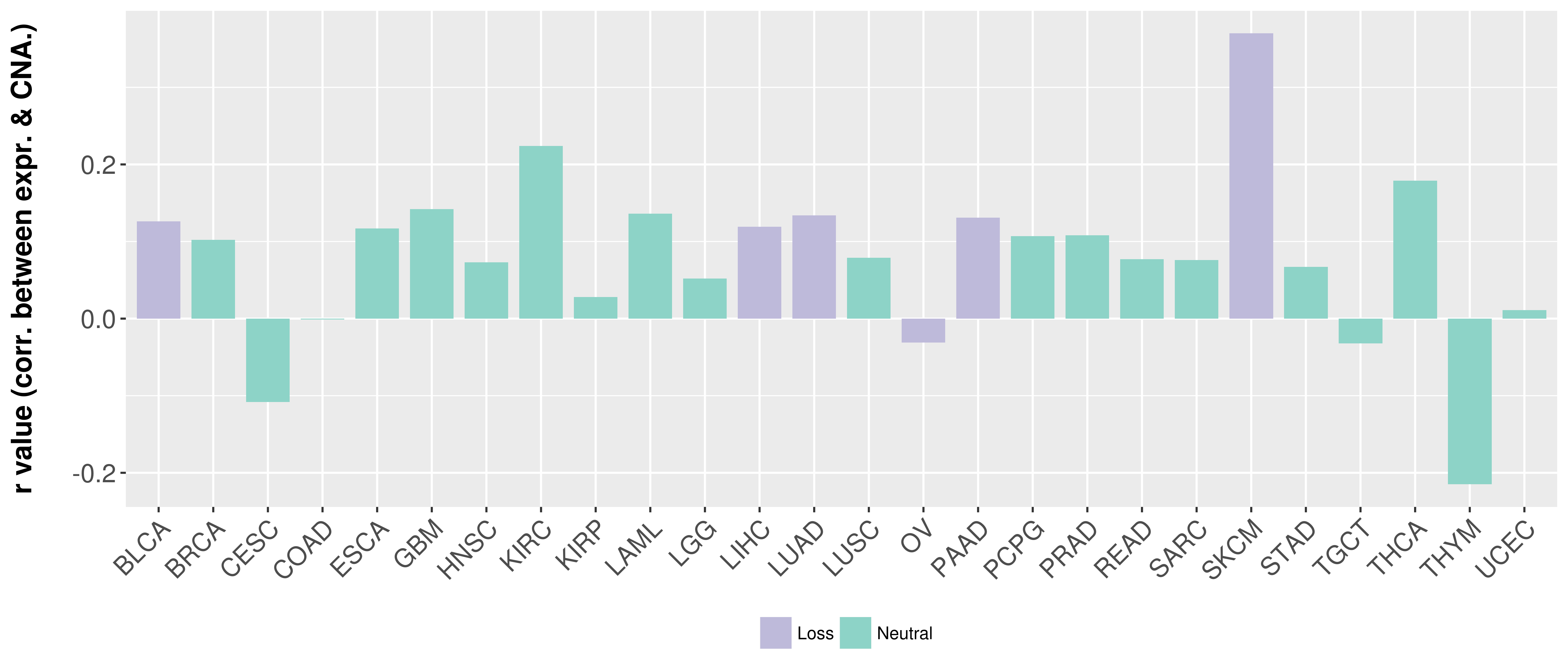
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
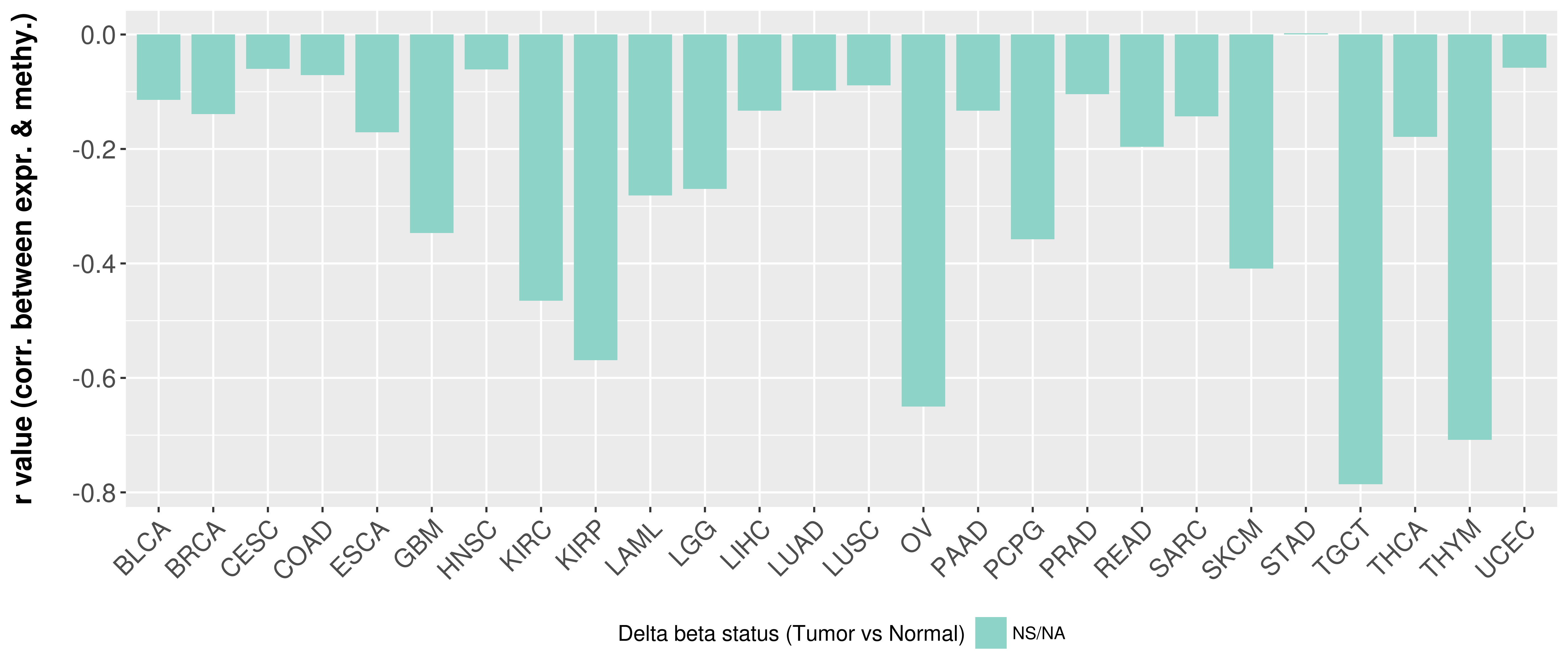
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
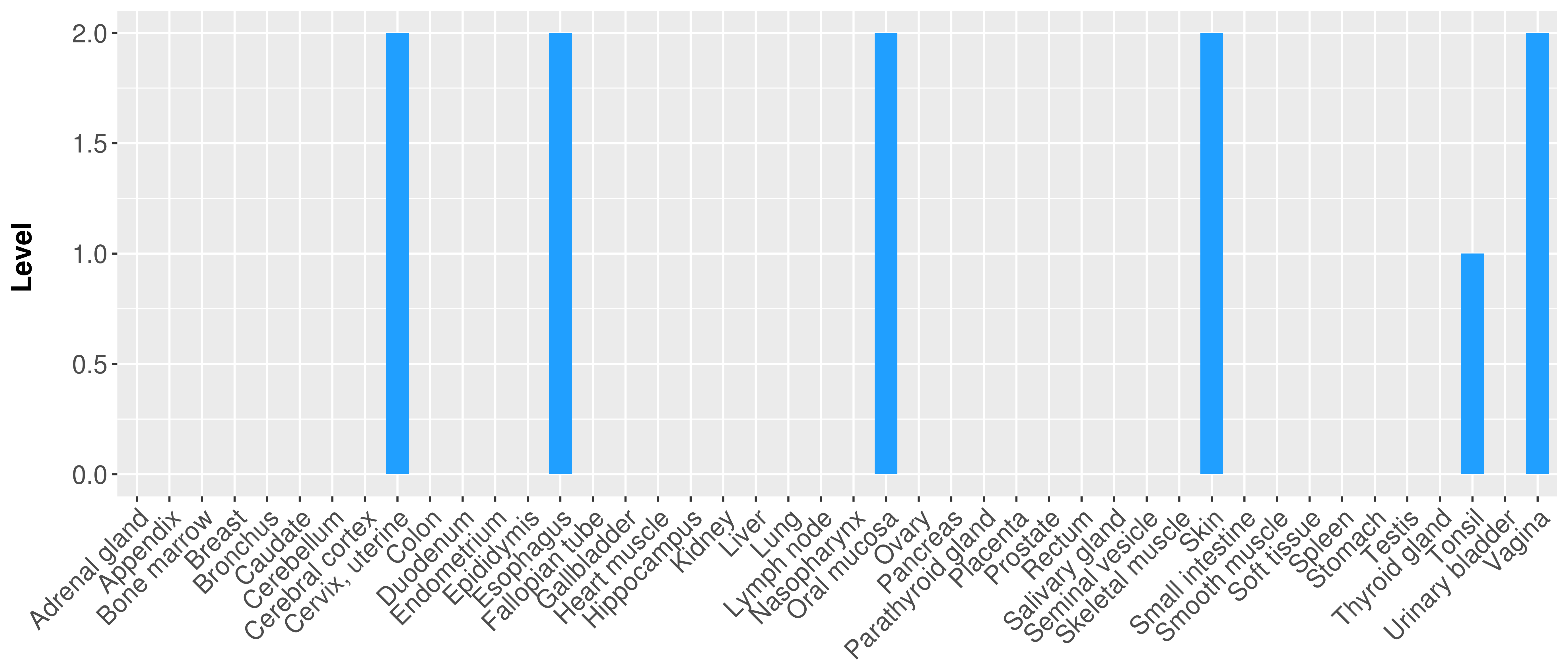
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
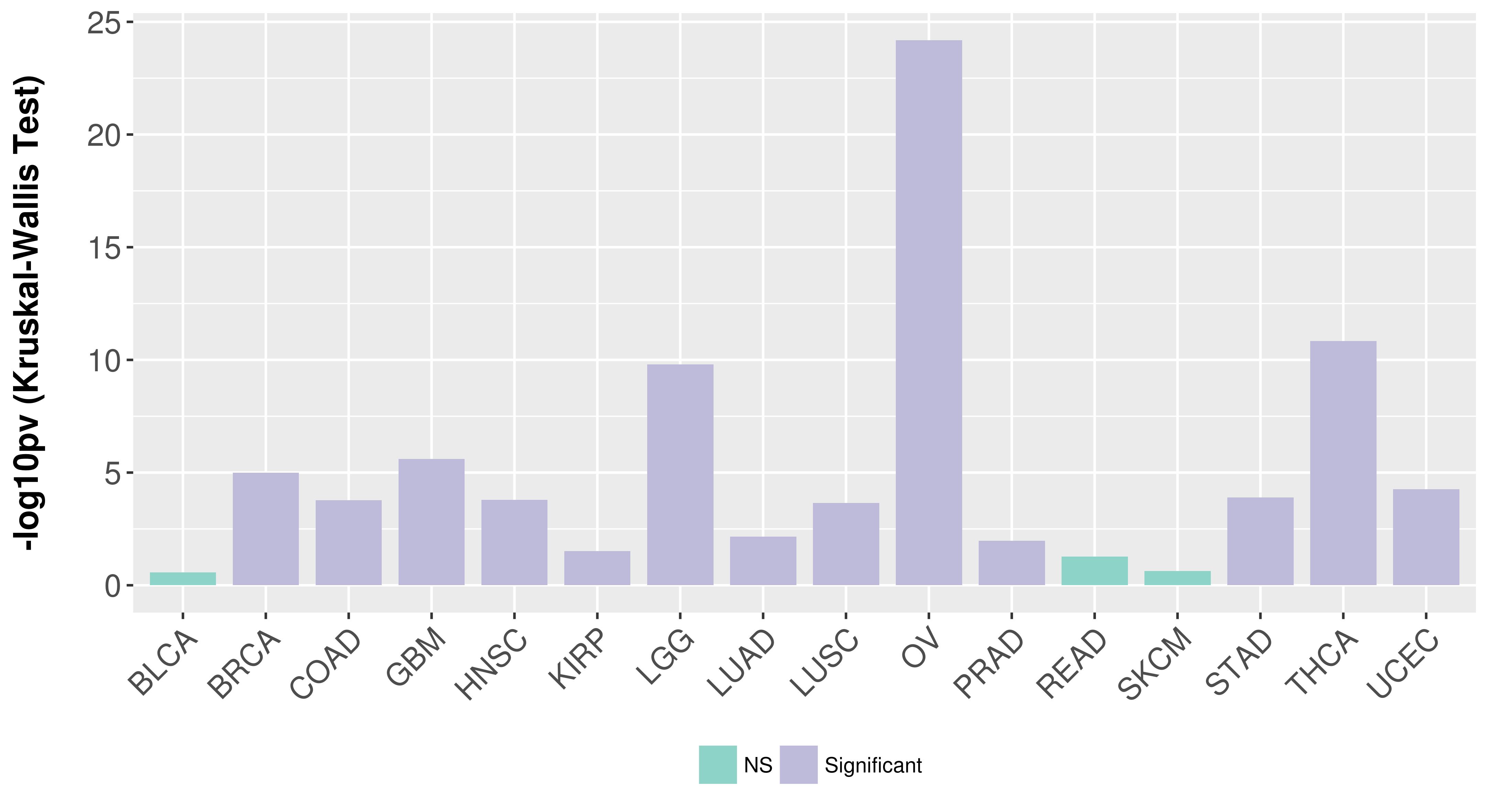
Association between expresson and subtype.
|
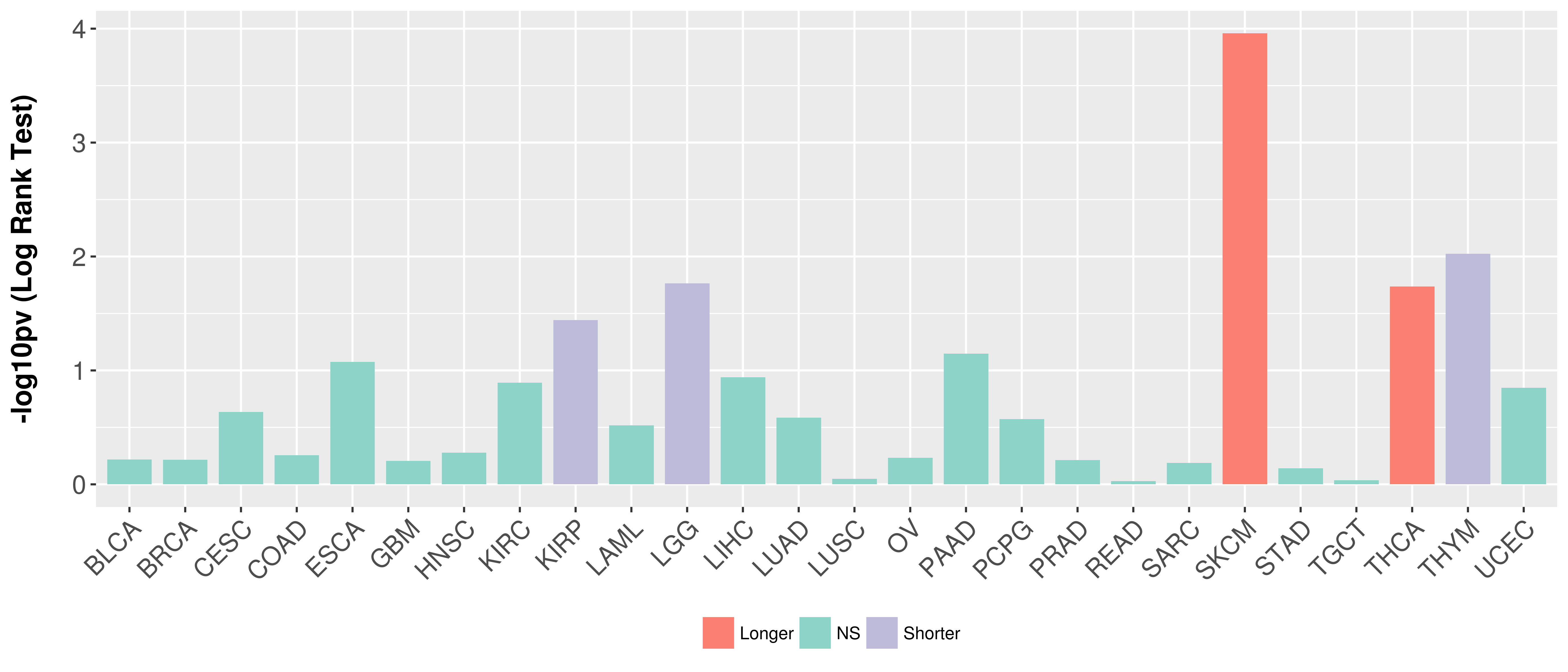
Overall survival analysis based on expression.
|

Association between expresson and stage.
|
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for PRDM1. |
| Summary | |
|---|---|
| Symbol | PRDM1 |
| Name | PR/SET domain 1 |
| Aliases | PRDI-BF1; BLIMP1; B-lymphocyte-induced maturation protein 1; BLIMP-1; PRDI-binding factor-1; beta-interferon ...... |
| Location | 6q21 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|