Browse PRMT1 in pancancer
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain | - | ||||||||||
| Function |
Arginine methyltransferase that methylates (mono and asymmetric dimethylation) the guanidino nitrogens of arginyl residues present in proteins such as ESR1, histone H2, H3 and H4, PIAS1, HNRNPA1, HNRNPD, NFATC2IP, SUPT5H, TAF15 and EWS. Constitutes the main enzyme that mediates monomethylation and asymmetric dimethylation of histone H4 'Arg-4' (H4R3me1 and H4R3me2a, respectively), a specific tag for epigenetic transcriptional activation. Together with dimethylated PIAS1, represses STAT1 transcriptional activity, in the late phase of interferon gamma (IFN-gamma) signaling. May be involved in the regulation of TAF15 transcriptional activity, act as an activator of estrogen receptor (ER)-mediated transactivation, play a key role in neurite outgrowth and act as a negative regulator of megakaryocytic differentiation, by modulating p38 MAPK pathway. Methylates FOXO1 and retains it in the nucleus increasing its transcriptional activity. Methylates CHTOP and this methylation is critical for its 5-hydroxymethylcytosine (5hmC)-binding activity (PubMed:25284789). Methylates H4R3 in genes involved in glioblastomagenesis in a CHTOP- and/or TET1-dependent manner (PubMed:25284789). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000077 DNA damage checkpoint GO:0000082 G1/S transition of mitotic cell cycle GO:0002262 myeloid cell homeostasis GO:0002683 negative regulation of immune system process GO:0006479 protein methylation GO:0006977 DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest GO:0007050 cell cycle arrest GO:0007093 mitotic cell cycle checkpoint GO:0007346 regulation of mitotic cell cycle GO:0008213 protein alkylation GO:0010948 negative regulation of cell cycle process GO:0016570 histone modification GO:0016571 histone methylation GO:0018195 peptidyl-arginine modification GO:0018216 peptidyl-arginine methylation GO:0019919 peptidyl-arginine methylation, to asymmetrical-dimethyl arginine GO:0020027 hemoglobin metabolic process GO:0030099 myeloid cell differentiation GO:0030218 erythrocyte differentiation GO:0030219 megakaryocyte differentiation GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0031098 stress-activated protein kinase signaling cascade GO:0031570 DNA integrity checkpoint GO:0031571 mitotic G1 DNA damage checkpoint GO:0032259 methylation GO:0032844 regulation of homeostatic process GO:0032846 positive regulation of homeostatic process GO:0032872 regulation of stress-activated MAPK cascade GO:0032874 positive regulation of stress-activated MAPK cascade GO:0034101 erythrocyte homeostasis GO:0034969 histone arginine methylation GO:0035246 peptidyl-arginine N-methylation GO:0035247 peptidyl-arginine omega-N-methylation GO:0038066 p38MAPK cascade GO:0042541 hemoglobin biosynthetic process GO:0042770 signal transduction in response to DNA damage GO:0043410 positive regulation of MAPK cascade GO:0043414 macromolecule methylation GO:0043985 histone H4-R3 methylation GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044773 mitotic DNA damage checkpoint GO:0044774 mitotic DNA integrity checkpoint GO:0044783 G1 DNA damage checkpoint GO:0044819 mitotic G1/S transition checkpoint GO:0044843 cell cycle G1/S phase transition GO:0045637 regulation of myeloid cell differentiation GO:0045638 negative regulation of myeloid cell differentiation GO:0045639 positive regulation of myeloid cell differentiation GO:0045646 regulation of erythrocyte differentiation GO:0045648 positive regulation of erythrocyte differentiation GO:0045652 regulation of megakaryocyte differentiation GO:0045653 negative regulation of megakaryocyte differentiation GO:0045786 negative regulation of cell cycle GO:0045787 positive regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0046984 regulation of hemoglobin biosynthetic process GO:0046985 positive regulation of hemoglobin biosynthetic process GO:0048872 homeostasis of number of cells GO:0051403 stress-activated MAPK cascade GO:0070302 regulation of stress-activated protein kinase signaling cascade GO:0070304 positive regulation of stress-activated protein kinase signaling cascade GO:0071156 regulation of cell cycle arrest GO:0071158 positive regulation of cell cycle arrest GO:0072331 signal transduction by p53 class mediator GO:0072395 signal transduction involved in cell cycle checkpoint GO:0072401 signal transduction involved in DNA integrity checkpoint GO:0072413 signal transduction involved in mitotic cell cycle checkpoint GO:0072422 signal transduction involved in DNA damage checkpoint GO:0072431 signal transduction involved in mitotic G1 DNA damage checkpoint GO:0090068 positive regulation of cell cycle process GO:1900744 regulation of p38MAPK cascade GO:1900745 positive regulation of p38MAPK cascade GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901991 negative regulation of mitotic cell cycle phase transition GO:1902400 intracellular signal transduction involved in G1 DNA damage checkpoint GO:1902402 signal transduction involved in mitotic DNA damage checkpoint GO:1902403 signal transduction involved in mitotic DNA integrity checkpoint GO:1902806 regulation of cell cycle G1/S phase transition GO:1902807 negative regulation of cell cycle G1/S phase transition GO:1903706 regulation of hemopoiesis GO:1903707 negative regulation of hemopoiesis GO:1903708 positive regulation of hemopoiesis GO:2000045 regulation of G1/S transition of mitotic cell cycle GO:2000134 negative regulation of G1/S transition of mitotic cell cycle |
| Molecular Function |
GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008327 methyl-CpG binding GO:0008469 histone-arginine N-methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016273 arginine N-methyltransferase activity GO:0016274 protein-arginine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0035242 protein-arginine omega-N asymmetric methyltransferase activity GO:0042054 histone methyltransferase activity GO:0044020 histone methyltransferase activity (H4-R3 specific) GO:0048273 mitogen-activated protein kinase p38 binding GO:0051019 mitogen-activated protein kinase binding |
| Cellular Component |
GO:0034708 methyltransferase complex GO:0034709 methylosome |
| KEGG |
hsa04068 FoxO signaling pathway hsa04922 Glucagon signaling pathway |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214858: RMTs methylate histone arginines R-HSA-6791312: TP53 Regulates Transcription of Cell Cycle Genes R-HSA-6804114: TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for PRMT1. |
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
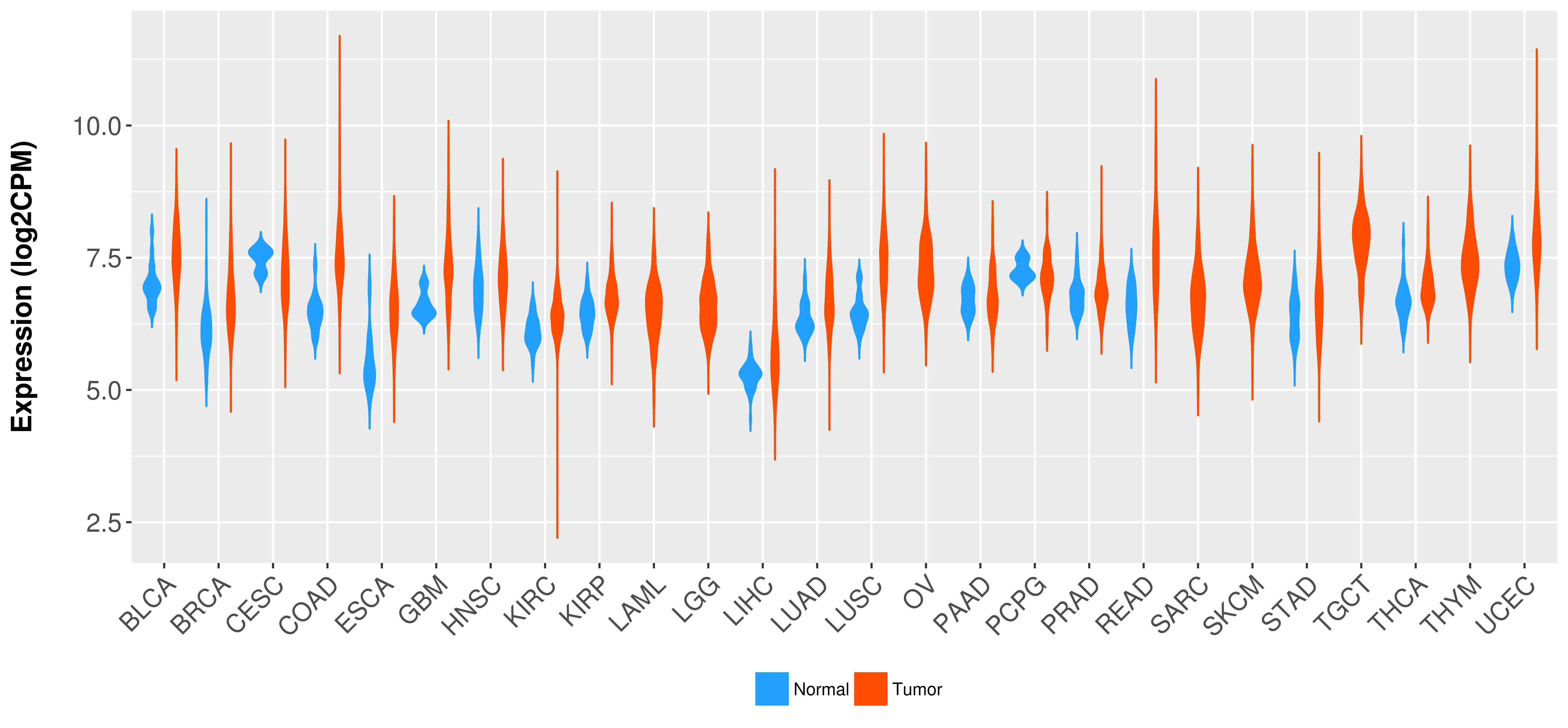
Differential expression analysis for cancers with more than 10 normal samples
|
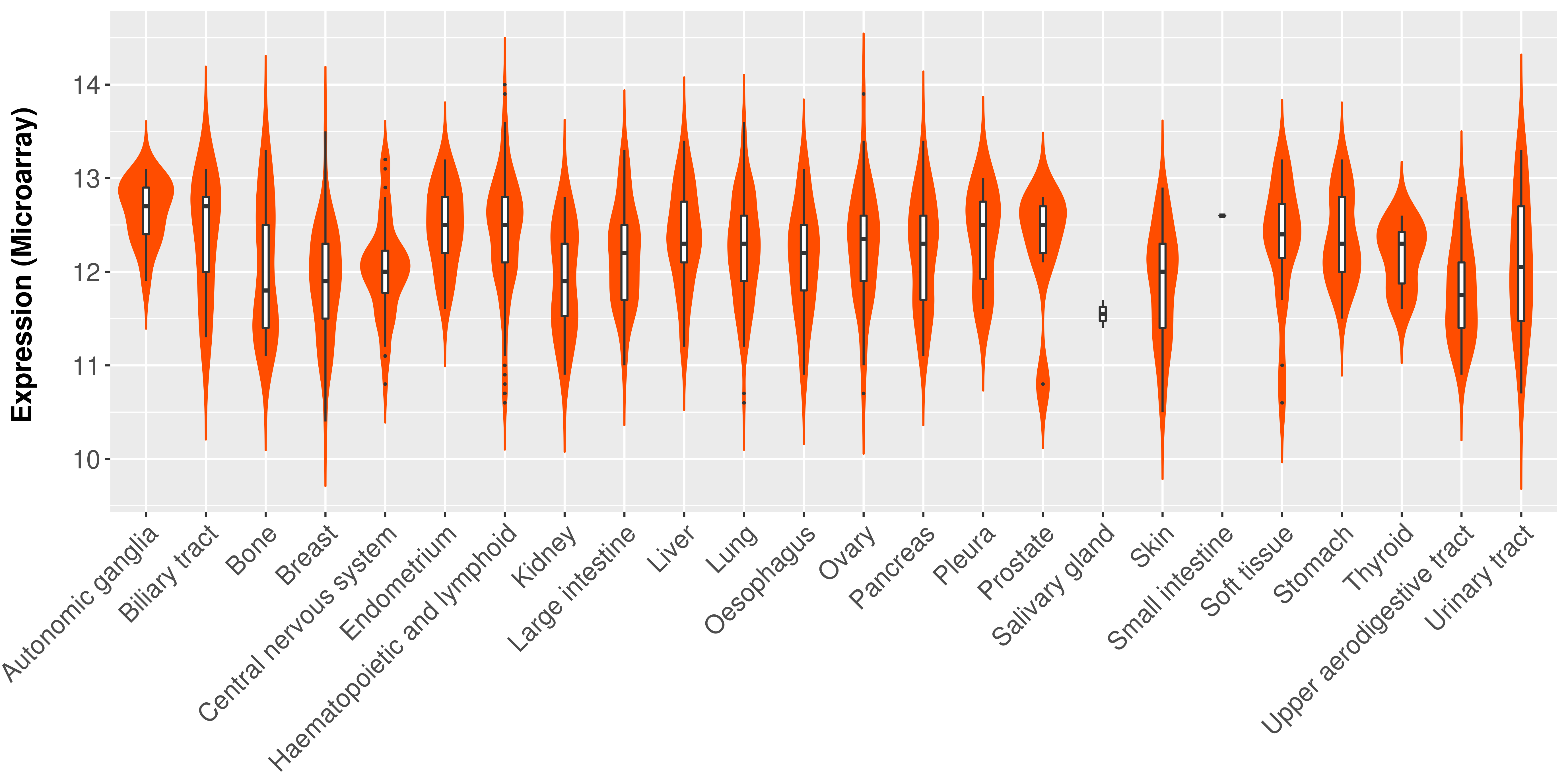
|

|
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
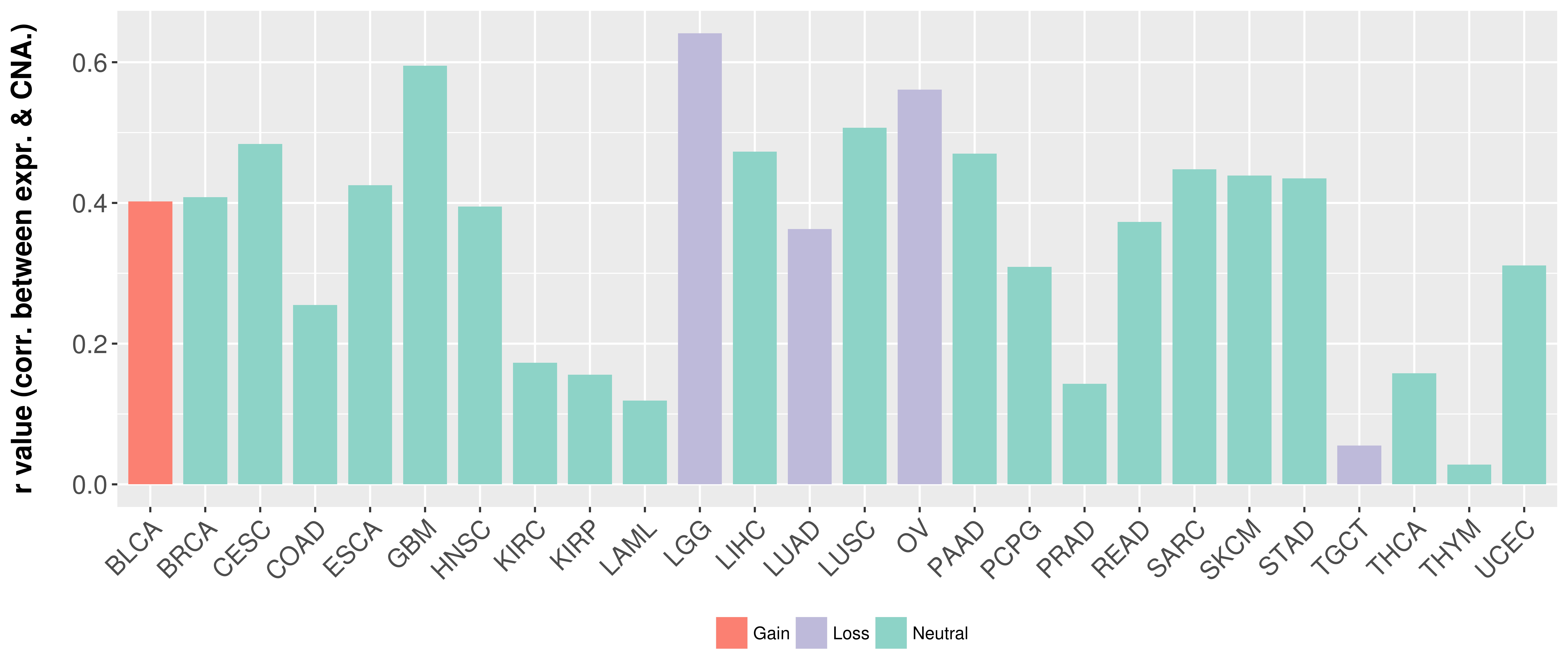
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
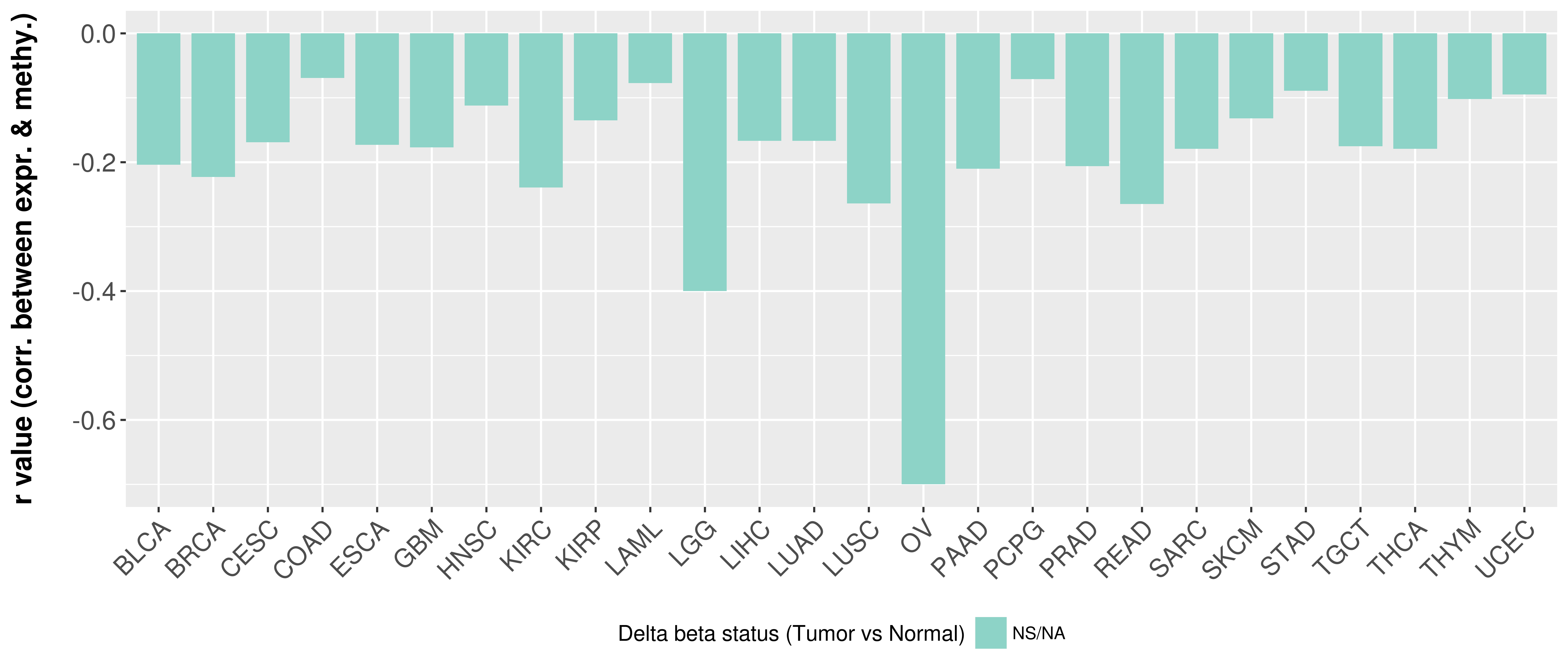
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
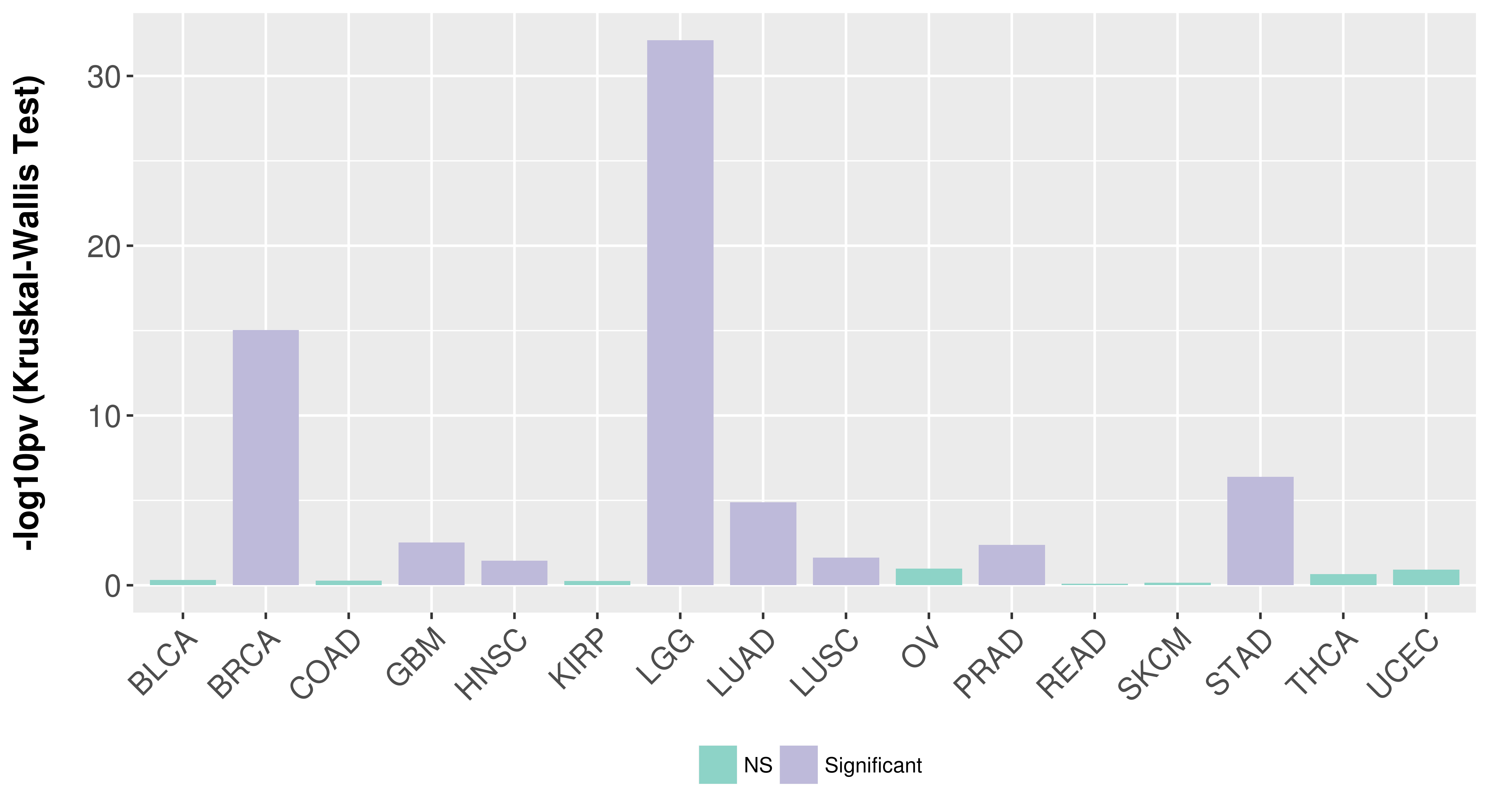
Association between expresson and subtype.
|
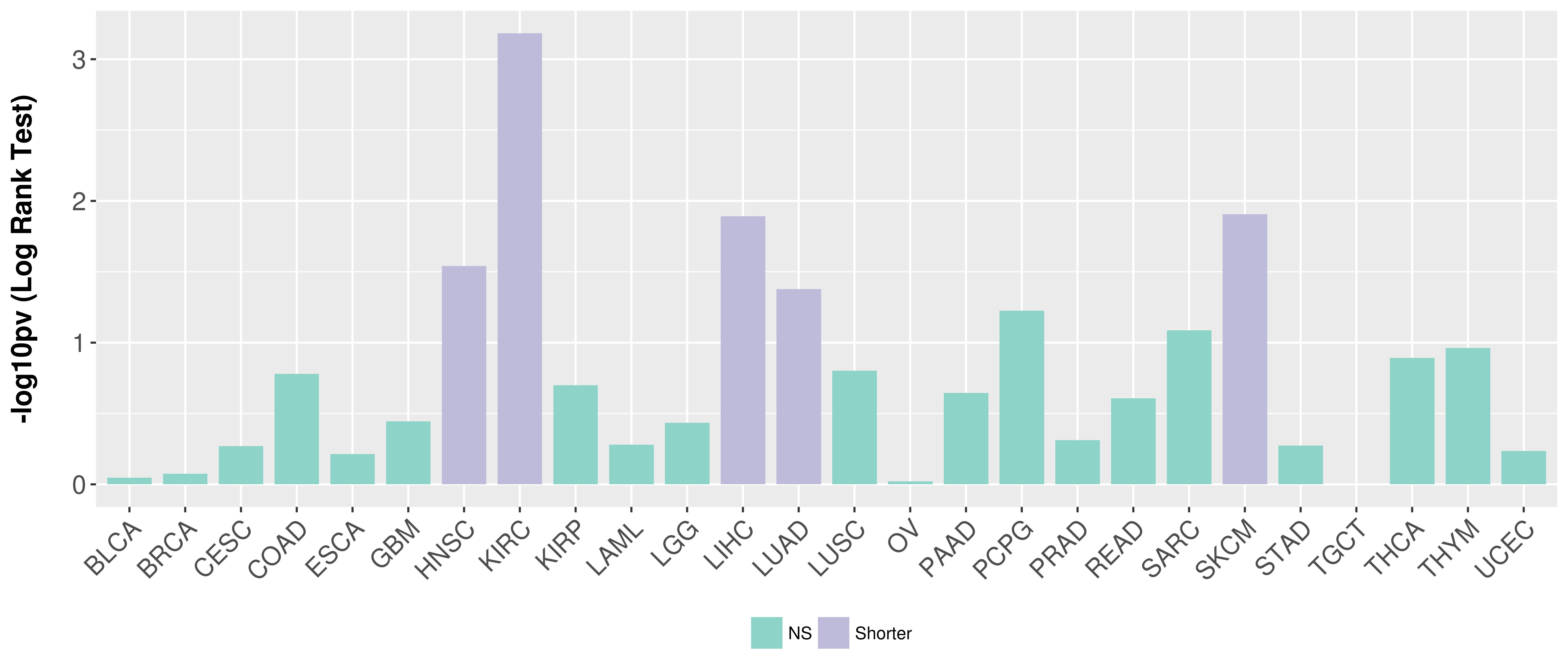
Overall survival analysis based on expression.
|
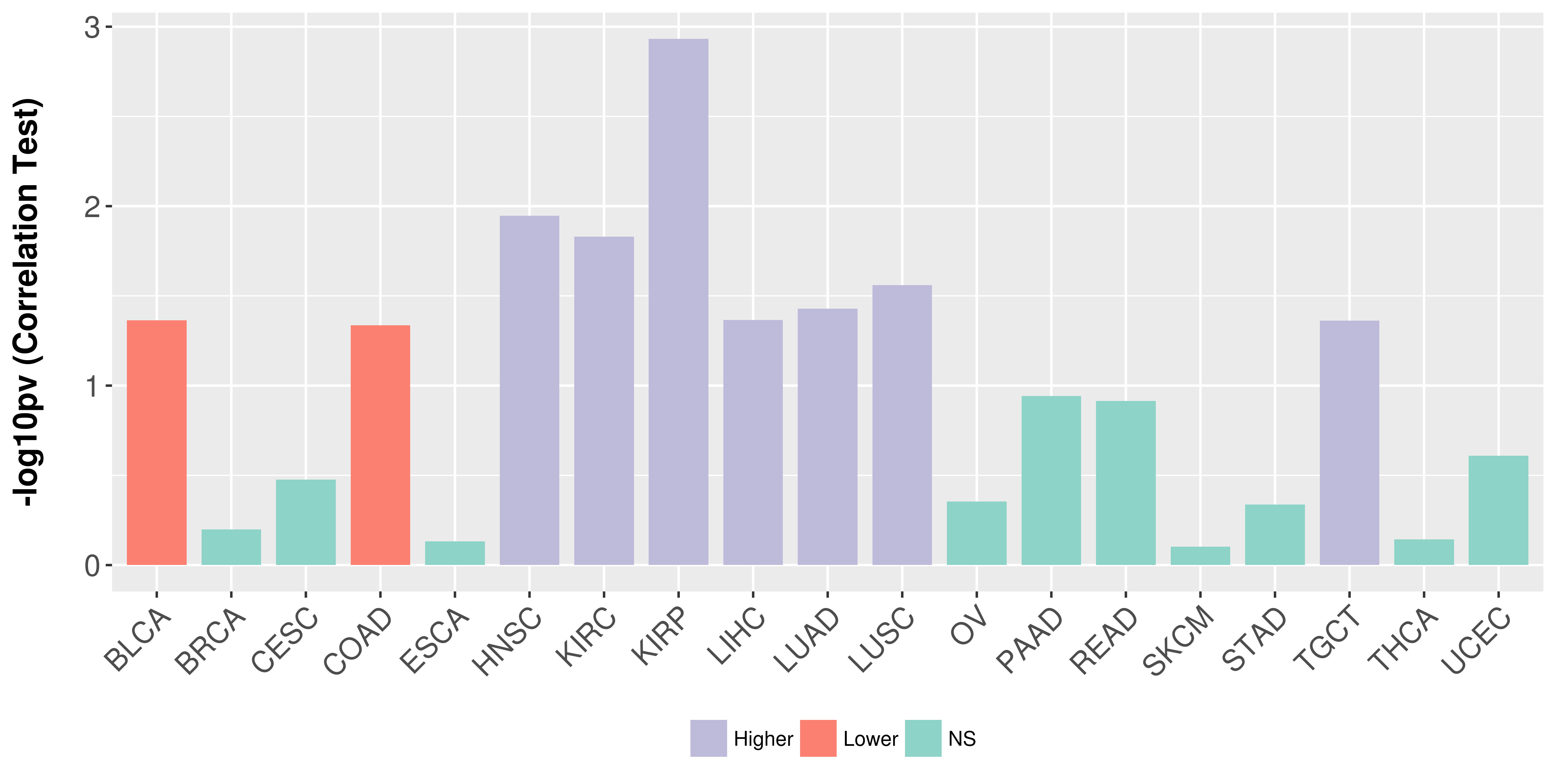
Association between expresson and stage.
|
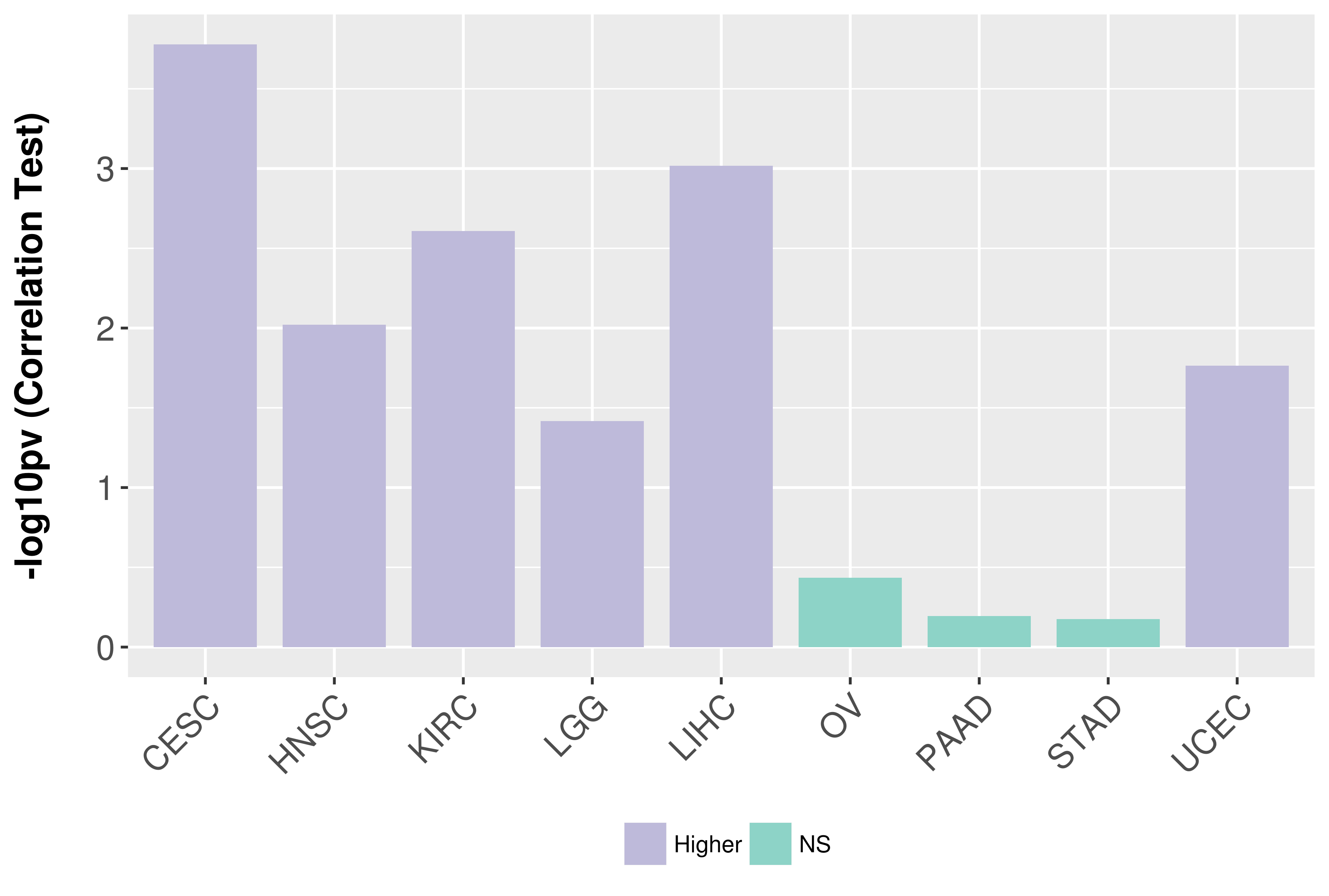
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
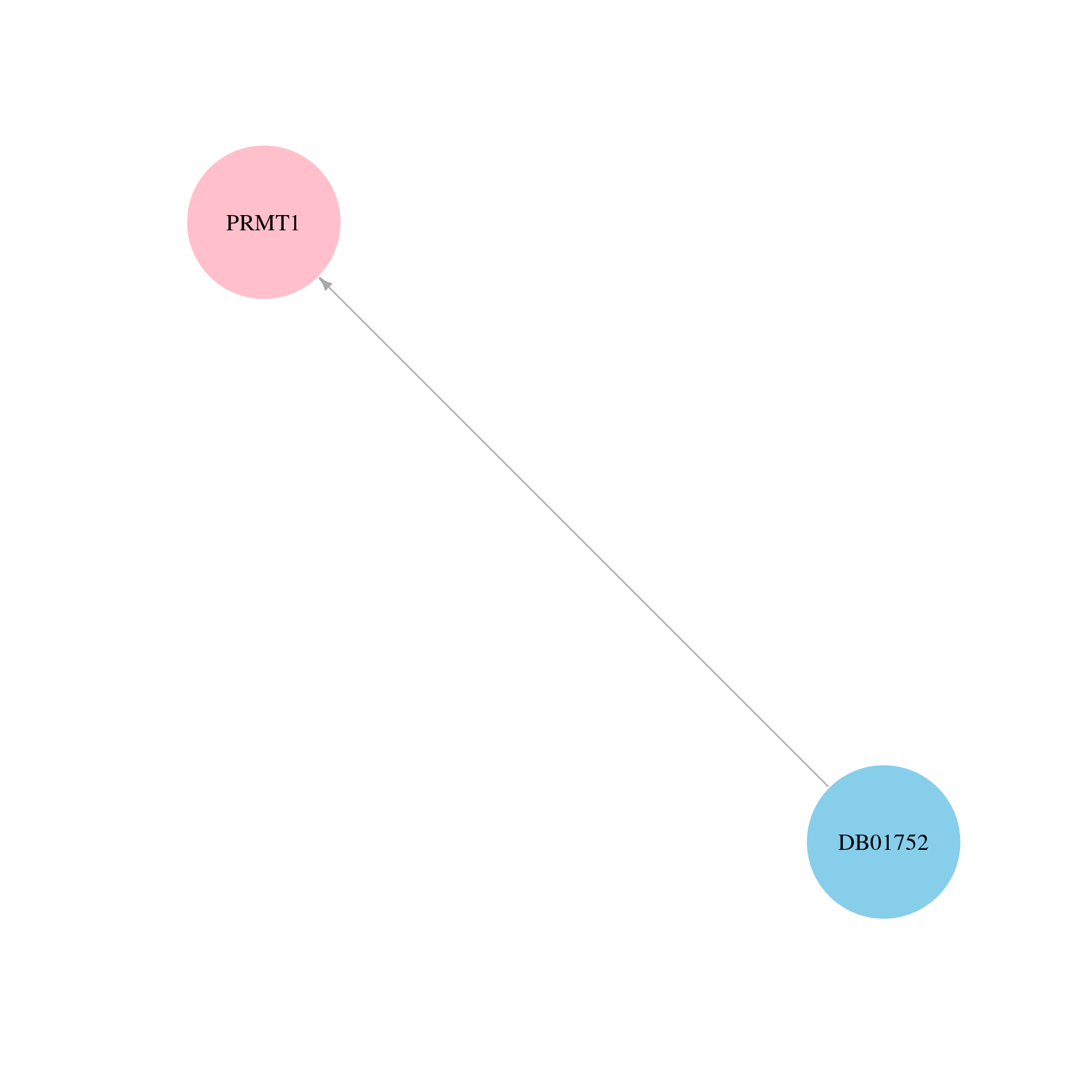
|
| Summary | |
|---|---|
| Symbol | PRMT1 |
| Name | protein arginine methyltransferase 1 |
| Aliases | ANM1; HRMT1L2; HMT1 (hnRNP methyltransferase, S. cerevisiae)-like 2; HMT1 hnRNP methyltransferase-like 2 (S. ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|