Browse PRMT5 in pancancer
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05185 PRMT5 arginine-N-methyltransferase |
||||||||||
| Function |
Arginine methyltransferase that can both catalyze the formation of omega-N monomethylarginine (MMA) and symmetrical dimethylarginine (sDMA), with a preference for the formation of MMA (PubMed:10531356, PubMed:11152681, PubMed:11747828, PubMed:12411503, PubMed:15737618, PubMed:17709427, PubMed:20159986, PubMed:20810653, PubMed:21258366, PubMed:21917714, PubMed:22269951). Specifically mediates the symmetrical dimethylation of arginine residues in the small nuclear ribonucleoproteins Sm D1 (SNRPD1) and Sm D3 (SNRPD3); such methylation being required for the assembly and biogenesis of snRNP core particles (PubMed:12411503, PubMed:11747828, PubMed:17709427). Methylates SUPT5H and may regulate its transcriptional elongation properties (PubMed:12718890). Mono- and dimethylates arginine residues of myelin basic protein (MBP) in vitro. May play a role in cytokine-activated transduction pathways. Negatively regulates cyclin E1 promoter activity and cellular proliferation. Methylates histone H2A and H4 'Arg-3' during germ cell development. Methylates histone H3 'Arg-8', which may repress transcription. Methylates the Piwi proteins (PIWIL1, PIWIL2 and PIWIL4), methylation of Piwi proteins being required for the interaction with Tudor domain-containing proteins and subsequent localization to the meiotic nuage (By similarity). Methylates RPS10. Attenuates EGF signaling through the MAPK1/MAPK3 pathway acting at 2 levels. First, monomethylates EGFR; this enhances EGFR 'Tyr-1197' phosphorylation and PTPN6 recruitment, eventually leading to reduced SOS1 phosphorylation (PubMed:21917714, PubMed:21258366). Second, methylates RAF1 and probably BRAF, hence destabilizing these 2 signaling proteins and reducing their catalytic activity (PubMed:21917714). Required for induction of E-selectin and VCAM-1, on the endothelial cells surface at sites of inflammation. Methylates HOXA9 (PubMed:22269951). Methylates and regulates SRGAP2 which is involved in cell migration and differentiation (PubMed:20810653). Acts as a transcriptional corepressor in CRY1-mediated repression of the core circadian component PER1 by regulating the H4R3 dimethylation at the PER1 promoter (By similarity). Methylates GM130/GOLGA2, regulating Golgi ribbon formation (PubMed:20421892). Methylates H4R3 in genes involved in glioblastomagenesis in a CHTOP- and/or TET1-dependent manner (PubMed:25284789). Symmetrically methylates POLR2A, a modification that allows the recruitment to POLR2A of proteins including SMN1/SMN2 and SETX. This is required for resolving RNA-DNA hybrids created by RNA polymerase II, that form R-loop in transcription terminal regions, an important step in proper transcription termination (PubMed:26700805). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000375 RNA splicing, via transesterification reactions GO:0000377 RNA splicing, via transesterification reactions with bulged adenosine as nucleophile GO:0000387 spliceosomal snRNP assembly GO:0000398 mRNA splicing, via spliceosome GO:0001889 liver development GO:0006304 DNA modification GO:0006305 DNA alkylation GO:0006306 DNA methylation GO:0006353 DNA-templated transcription, termination GO:0006397 mRNA processing GO:0006479 protein methylation GO:0007030 Golgi organization GO:0007067 mitotic nuclear division GO:0007088 regulation of mitotic nuclear division GO:0007187 G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger GO:0007188 adenylate cyclase-modulating G-protein coupled receptor signaling pathway GO:0007193 adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway GO:0007195 adenylate cyclase-inhibiting dopamine receptor signaling pathway GO:0007212 dopamine receptor signaling pathway GO:0007346 regulation of mitotic cell cycle GO:0007623 circadian rhythm GO:0008213 protein alkylation GO:0008277 regulation of G-protein coupled receptor protein signaling pathway GO:0008380 RNA splicing GO:0010001 glial cell differentiation GO:0010720 positive regulation of cell development GO:0014013 regulation of gliogenesis GO:0014015 positive regulation of gliogenesis GO:0016570 histone modification GO:0016571 histone methylation GO:0018195 peptidyl-arginine modification GO:0018216 peptidyl-arginine methylation GO:0019918 peptidyl-arginine methylation, to symmetrical-dimethyl arginine GO:0022613 ribonucleoprotein complex biogenesis GO:0022618 ribonucleoprotein complex assembly GO:0031099 regeneration GO:0031100 animal organ regeneration GO:0032259 methylation GO:0032922 circadian regulation of gene expression GO:0034969 histone arginine methylation GO:0035246 peptidyl-arginine N-methylation GO:0035247 peptidyl-arginine omega-N-methylation GO:0042063 gliogenesis GO:0042118 endothelial cell activation GO:0043414 macromolecule methylation GO:0043985 histone H4-R3 methylation GO:0044030 regulation of DNA methylation GO:0044728 DNA methylation or demethylation GO:0045685 regulation of glial cell differentiation GO:0045687 positive regulation of glial cell differentiation GO:0045745 positive regulation of G-protein coupled receptor protein signaling pathway GO:0048511 rhythmic process GO:0048709 oligodendrocyte differentiation GO:0048713 regulation of oligodendrocyte differentiation GO:0048714 positive regulation of oligodendrocyte differentiation GO:0048732 gland development GO:0050769 positive regulation of neurogenesis GO:0051052 regulation of DNA metabolic process GO:0051783 regulation of nuclear division GO:0051962 positive regulation of nervous system development GO:0060159 regulation of dopamine receptor signaling pathway GO:0060161 positive regulation of dopamine receptor signaling pathway GO:0061008 hepaticobiliary system development GO:0070371 ERK1 and ERK2 cascade GO:0070372 regulation of ERK1 and ERK2 cascade GO:0071826 ribonucleoprotein complex subunit organization GO:0072331 signal transduction by p53 class mediator GO:0090161 Golgi ribbon formation GO:0097421 liver regeneration GO:1901796 regulation of signal transduction by p53 class mediator GO:1904990 regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway GO:1904992 positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway |
| Molecular Function |
GO:0001046 core promoter sequence-specific DNA binding GO:0001047 core promoter binding GO:0003714 transcription corepressor activity GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008327 methyl-CpG binding GO:0008469 histone-arginine N-methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016273 arginine N-methyltransferase activity GO:0016274 protein-arginine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0035243 protein-arginine omega-N symmetric methyltransferase activity GO:0042054 histone methyltransferase activity GO:0043021 ribonucleoprotein complex binding GO:0044020 histone methyltransferase activity (H4-R3 specific) GO:0046982 protein heterodimerization activity |
| Cellular Component |
GO:0034708 methyltransferase complex GO:0034709 methylosome GO:0035097 histone methyltransferase complex |
| KEGG |
hsa03013 RNA transport |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-194441: Metabolism of non-coding RNA R-HSA-3214858: RMTs methylate histone arginines R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804760: Regulation of TP53 Activity through Methylation R-HSA-3700989: Transcriptional Regulation by TP53 R-HSA-191859: snRNP Assembly |
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for PRMT5. |
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
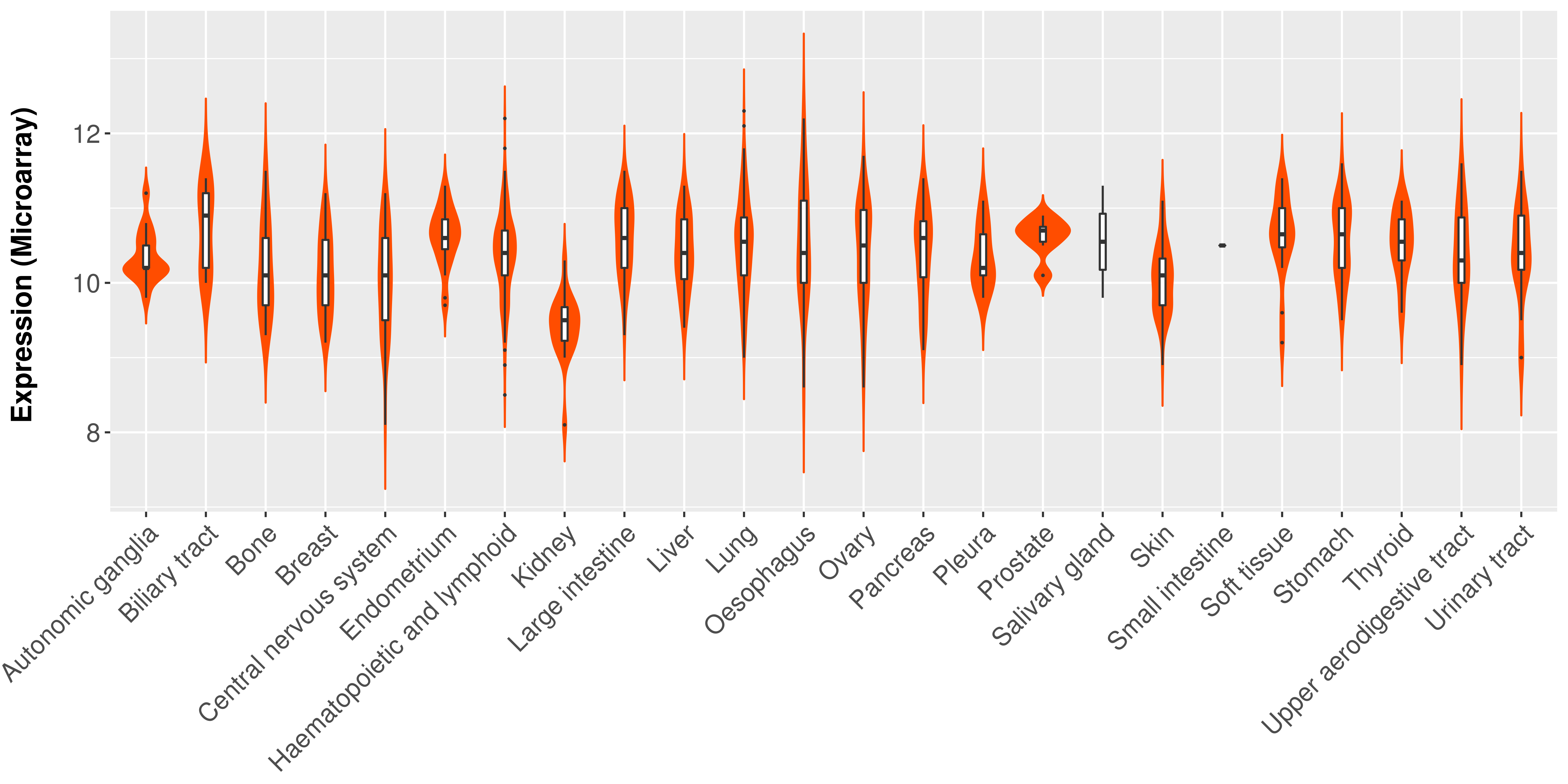
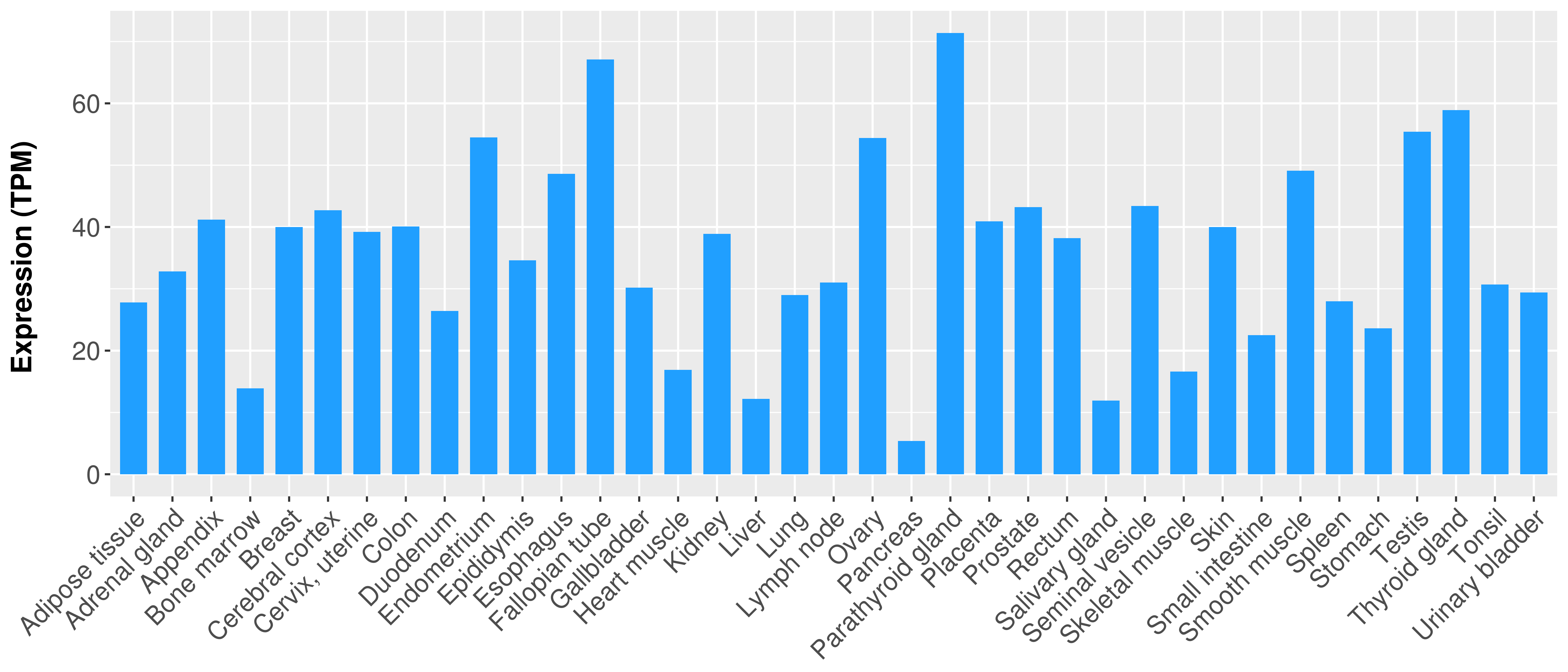
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
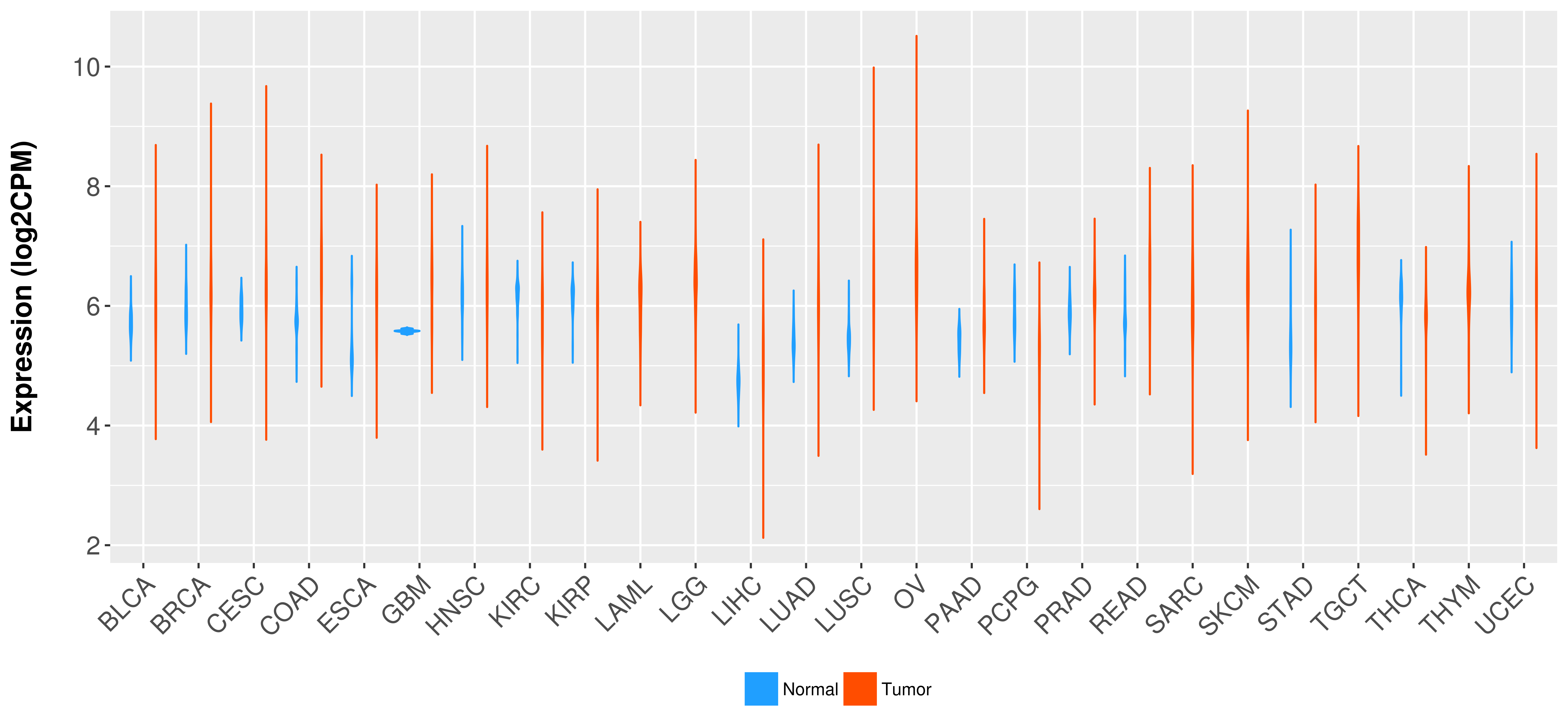
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
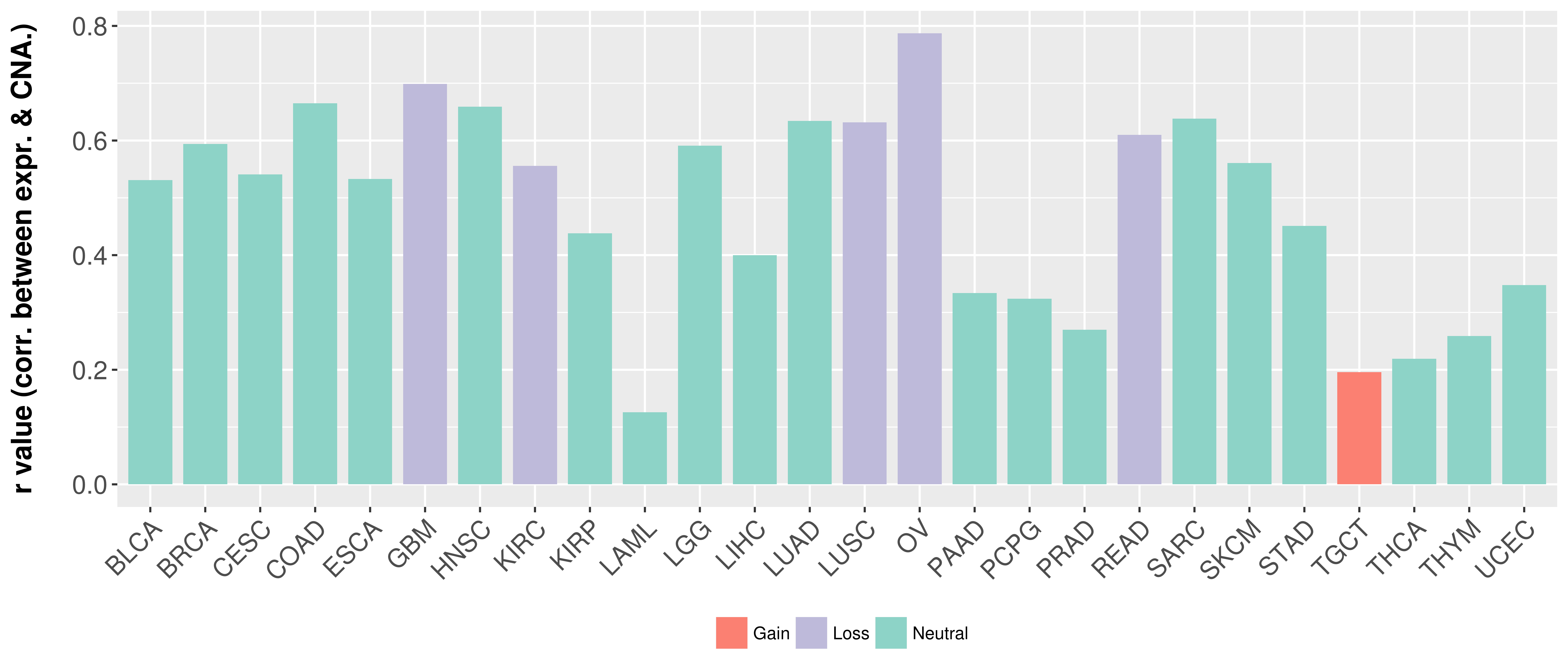
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
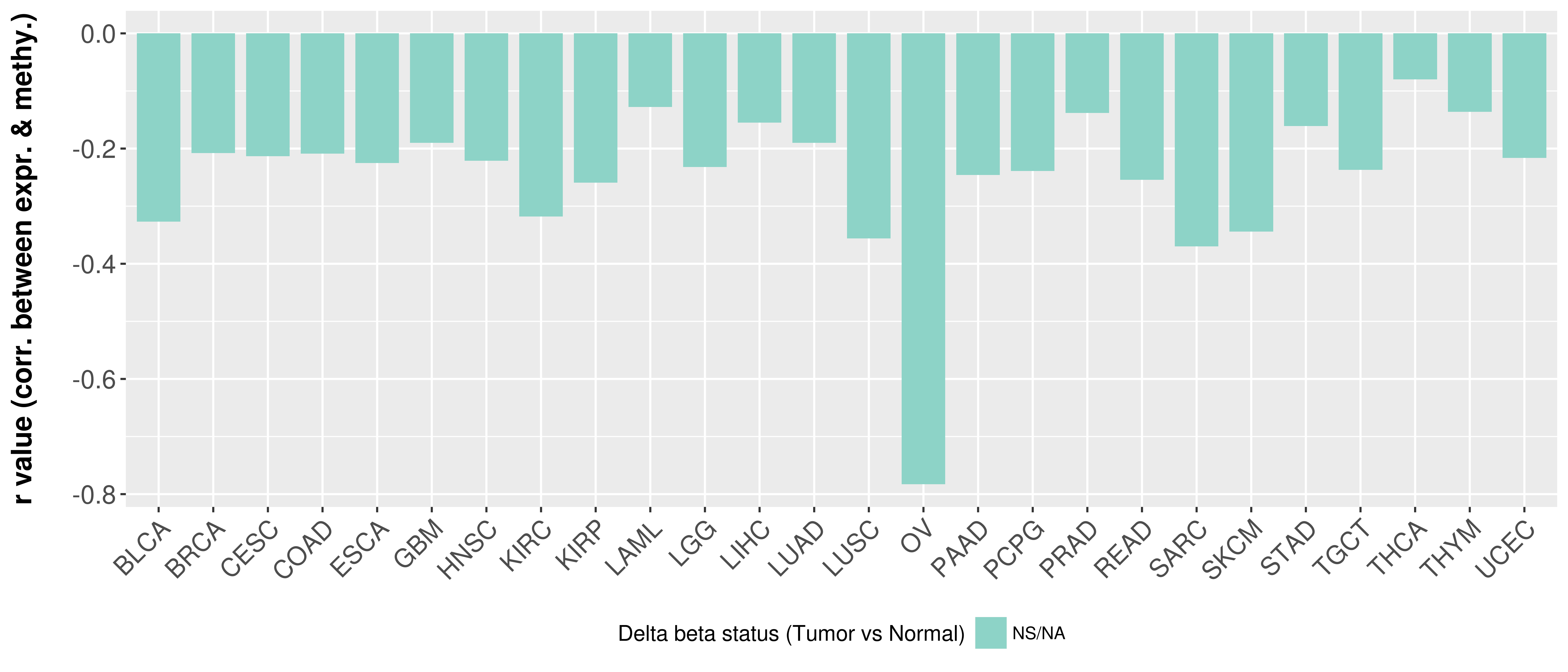
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
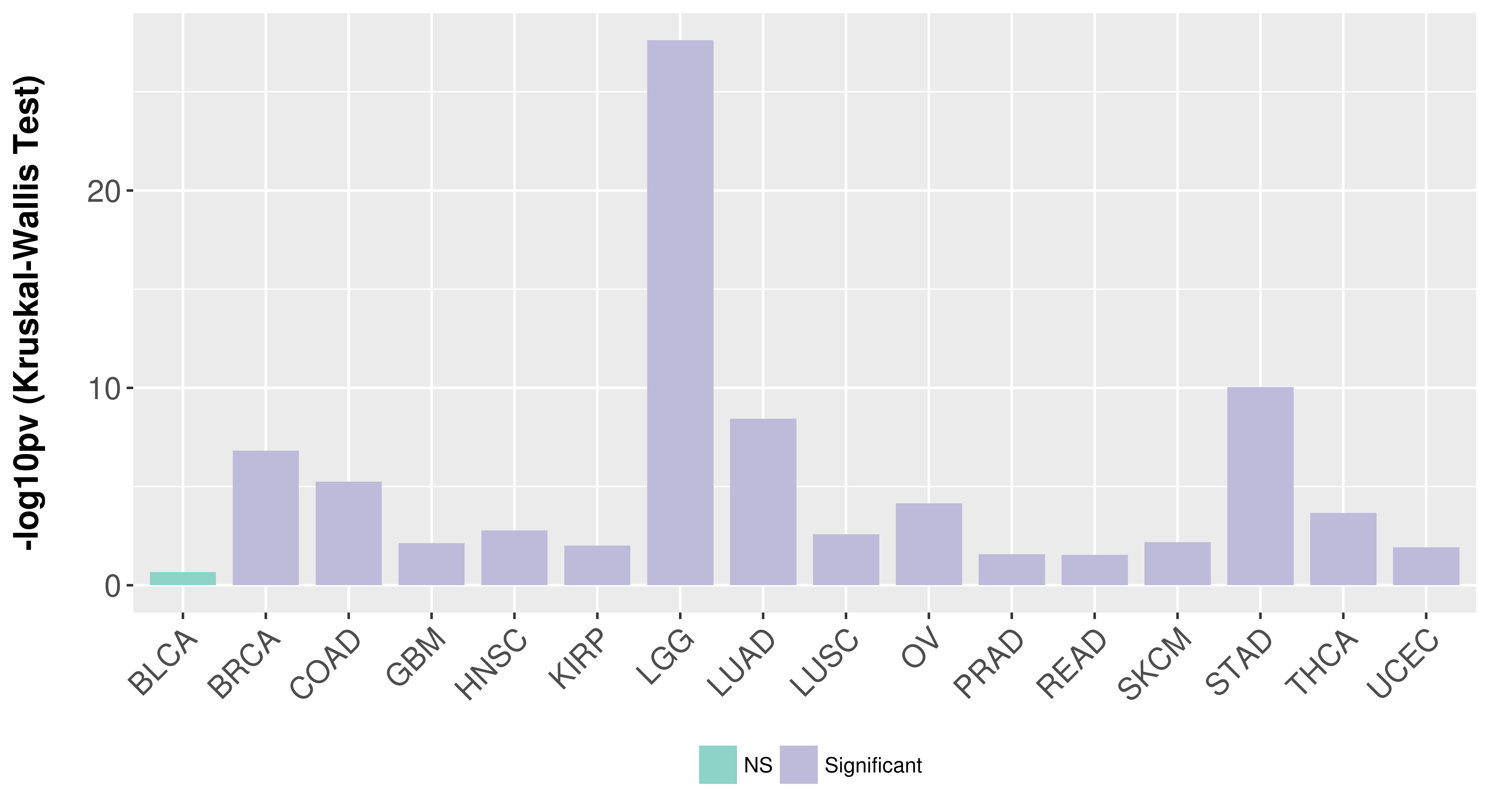
Association between expresson and subtype.
|
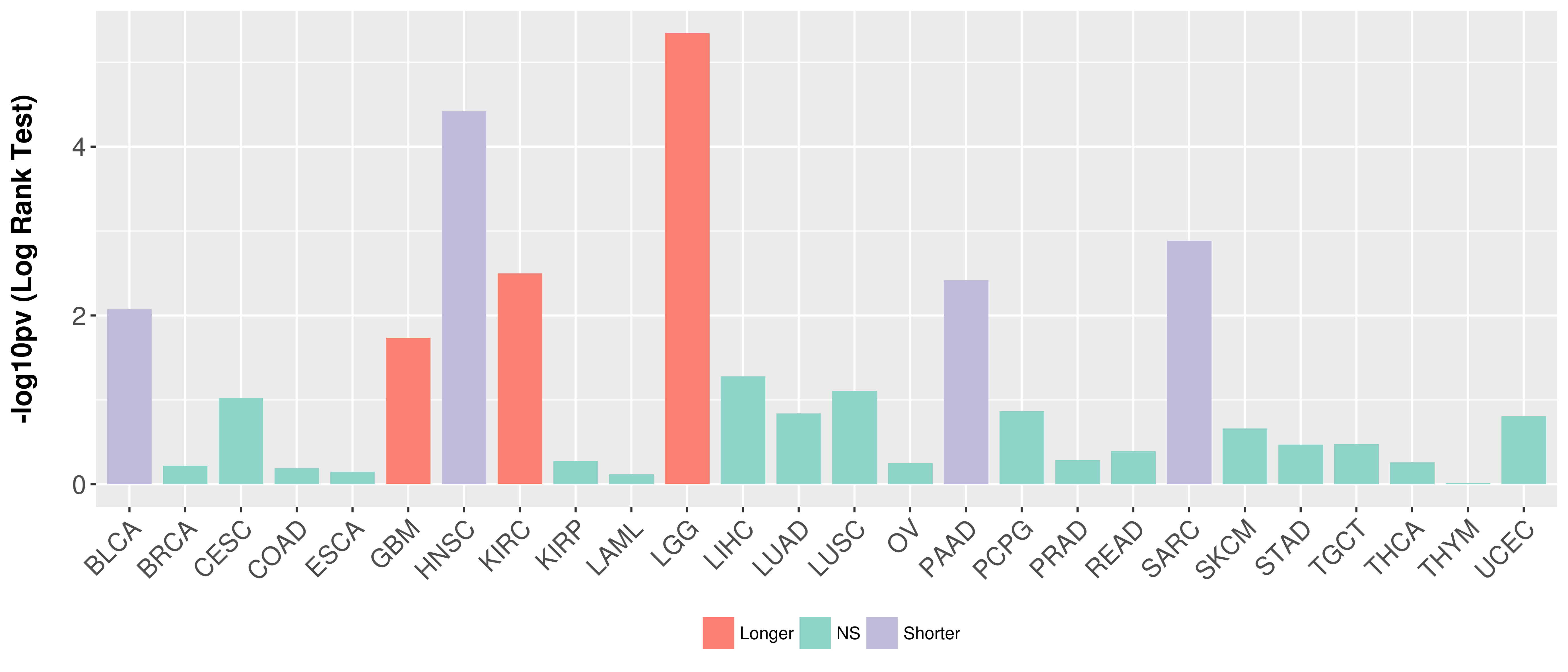
Overall survival analysis based on expression.
|
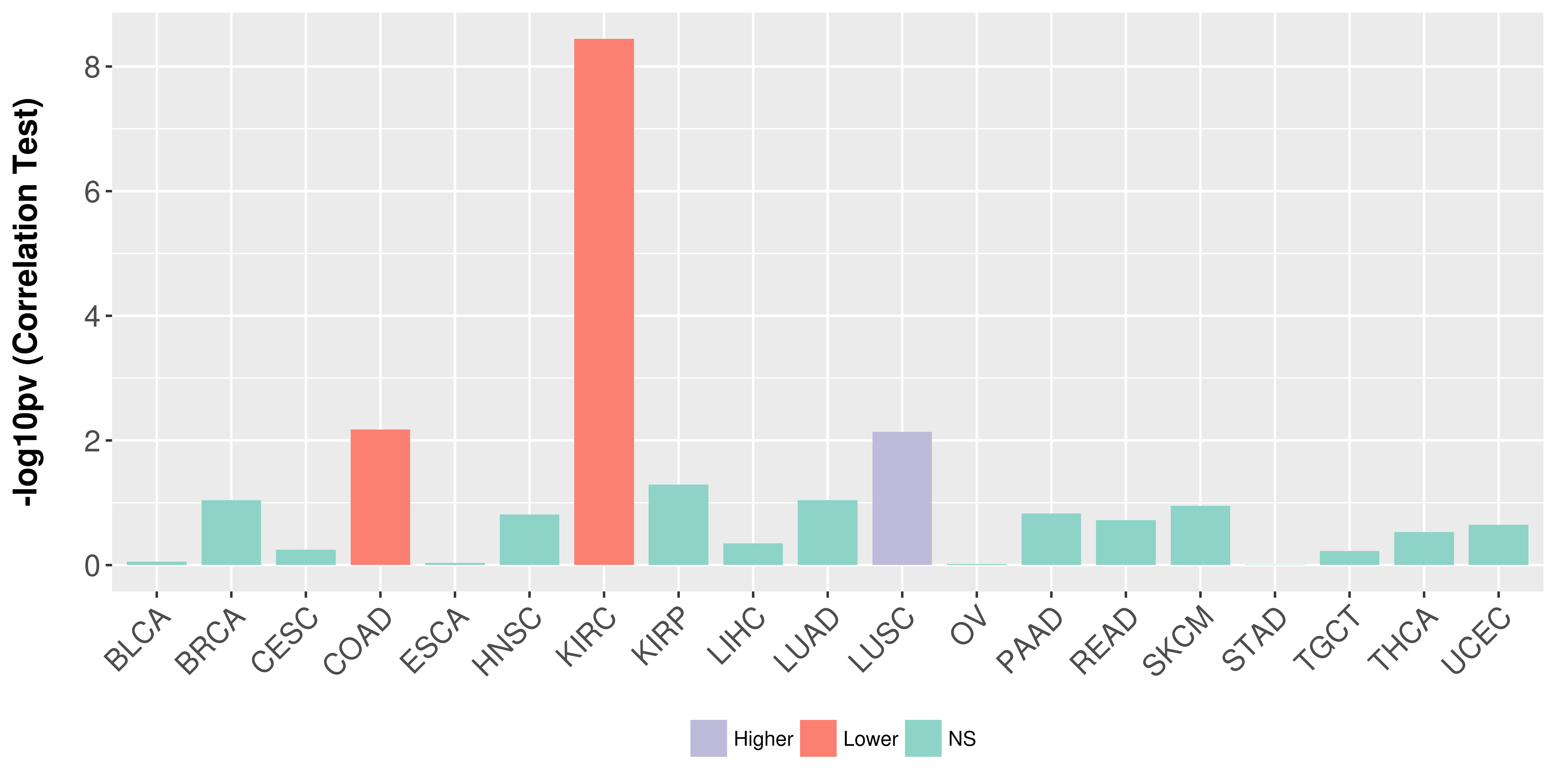
Association between expresson and stage.
|
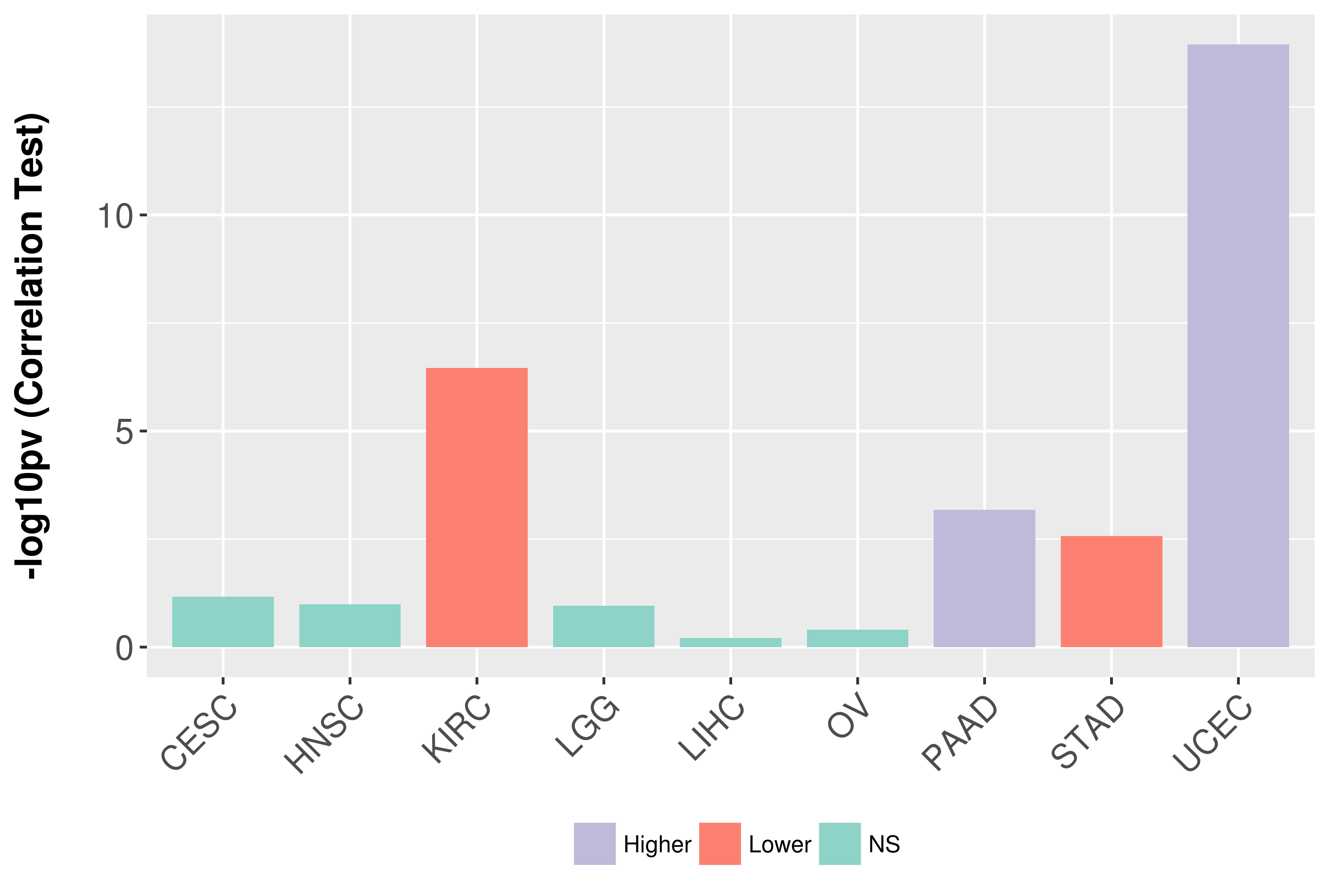
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for PRMT5. |
| Summary | |
|---|---|
| Symbol | PRMT5 |
| Name | protein arginine methyltransferase 5 |
| Aliases | SKB1Hs; HRMT1L5; SKB1; skb1 (S. pombe) homolog; SKB1 homolog (S. pombe); IBP72; JBP1; 72 kDa ICln-binding pr ...... |
| Location | 14q11.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|