Browse RBBP4 in pancancer
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF12265 Histone-binding protein RBBP4 or subunit C of CAF1 complex PF00400 WD domain |
||||||||||
| Function |
Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA. Component of several complexes which regulate chromatin metabolism. These include the chromatin assembly factor 1 (CAF-1) complex, which is required for chromatin assembly following DNA replication and DNA repair; the core histone deacetylase (HDAC) complex, which promotes histone deacetylation and consequent transcriptional repression; the nucleosome remodeling and histone deacetylase complex (the NuRD complex), which promotes transcriptional repression by histone deacetylation and nucleosome remodeling; the PRC2/EED-EZH2 complex, which promotes repression of homeotic genes during development; and the NURF (nucleosome remodeling factor) complex. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006260 DNA replication GO:0006323 DNA packaging GO:0006333 chromatin assembly or disassembly GO:0006334 nucleosome assembly GO:0006335 DNA replication-dependent nucleosome assembly GO:0006336 DNA replication-independent nucleosome assembly GO:0006338 chromatin remodeling GO:0006476 protein deacetylation GO:0016570 histone modification GO:0016575 histone deacetylation GO:0031055 chromatin remodeling at centromere GO:0031497 chromatin assembly GO:0034080 CENP-A containing nucleosome assembly GO:0034508 centromere complex assembly GO:0034723 DNA replication-dependent nucleosome organization GO:0034724 DNA replication-independent nucleosome organization GO:0034728 nucleosome organization GO:0035601 protein deacylation GO:0040029 regulation of gene expression, epigenetic GO:0043044 ATP-dependent chromatin remodeling GO:0043434 response to peptide hormone GO:0043486 histone exchange GO:0045814 negative regulation of gene expression, epigenetic GO:0060416 response to growth hormone GO:0061641 CENP-A containing chromatin organization GO:0065004 protein-DNA complex assembly GO:0071103 DNA conformation change GO:0071824 protein-DNA complex subunit organization GO:0072331 signal transduction by p53 class mediator GO:0098732 macromolecule deacylation GO:1901652 response to peptide GO:1901796 regulation of signal transduction by p53 class mediator |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001158 enhancer sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0003682 chromatin binding GO:0004407 histone deacetylase activity GO:0008094 DNA-dependent ATPase activity GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0016887 ATPase activity GO:0019213 deacetylase activity GO:0031490 chromatin DNA binding GO:0031491 nucleosome binding GO:0031492 nucleosomal DNA binding GO:0033558 protein deacetylase activity GO:0035326 enhancer binding GO:0042393 histone binding GO:0042623 ATPase activity, coupled GO:0042826 histone deacetylase binding GO:0043566 structure-specific DNA binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016580 Sin3 complex GO:0016581 NuRD complex GO:0016589 NURF complex GO:0017053 transcriptional repressor complex GO:0031010 ISWI-type complex GO:0031519 PcG protein complex GO:0033186 CAF-1 complex GO:0034708 methyltransferase complex GO:0035097 histone methyltransferase complex GO:0035098 ESC/E(Z) complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0070822 Sin3-type complex GO:0090545 CHD-type complex GO:0090568 nuclear transcriptional repressor complex |
| KEGG | - |
| Reactome |
R-HSA-5619507: Activation of HOX genes during differentiation R-HSA-5617472: Activation of anterior HOX genes in hindbrain development during early embryogenesis R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-2559583: Cellular Senescence R-HSA-2262752: Cellular responses to stress R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-73886: Chromosome Maintenance R-HSA-606279: Deposition of new CENPA-containing nucleosomes at the centromere R-HSA-1266738: Developmental Biology R-HSA-427389: ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression R-HSA-212165: Epigenetic regulation of gene expression R-HSA-1538133: G0 and Early G1 R-HSA-69275: G2/M Transition R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214815: HDACs deacetylate histones R-HSA-453279: Mitotic G1-G1/S phases R-HSA-453274: Mitotic G2-G2/M phases R-HSA-774815: Nucleosome assembly R-HSA-2559580: Oxidative Stress Induced Senescence R-HSA-3214841: PKMTs methylate histone lysines R-HSA-212300: PRC2 methylates histones and DNA R-HSA-156711: Polo-like kinase mediated events R-HSA-5250913: Positive epigenetic regulation of rRNA expression R-HSA-73854: RNA Polymerase I Promoter Clearance R-HSA-73864: RNA Polymerase I Transcription R-HSA-73762: RNA Polymerase I Transcription Initiation R-HSA-504046: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804758: Regulation of TP53 Activity through Acetylation R-HSA-3700989: Transcriptional Regulation by TP53 |
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for RBBP4. |
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
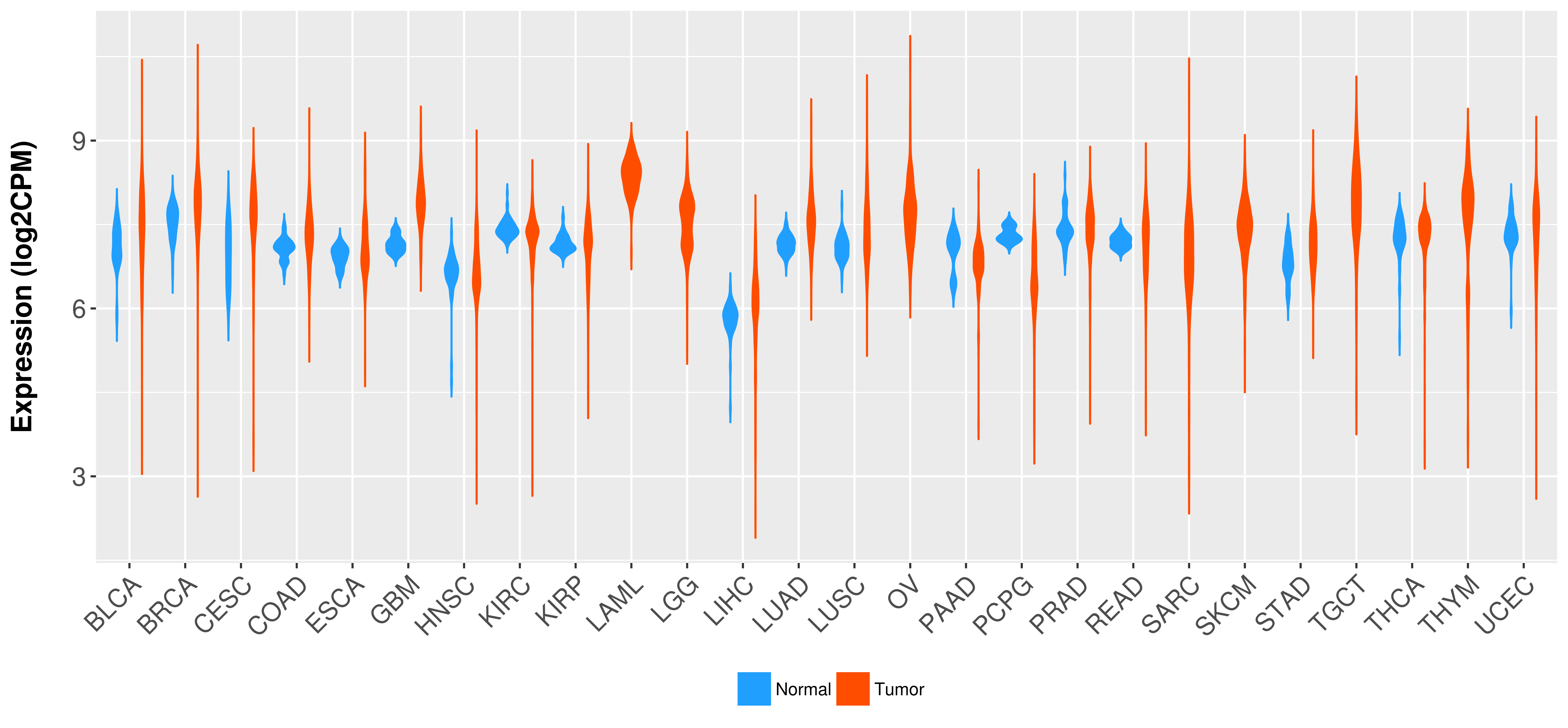
Differential expression analysis for cancers with more than 10 normal samples
|
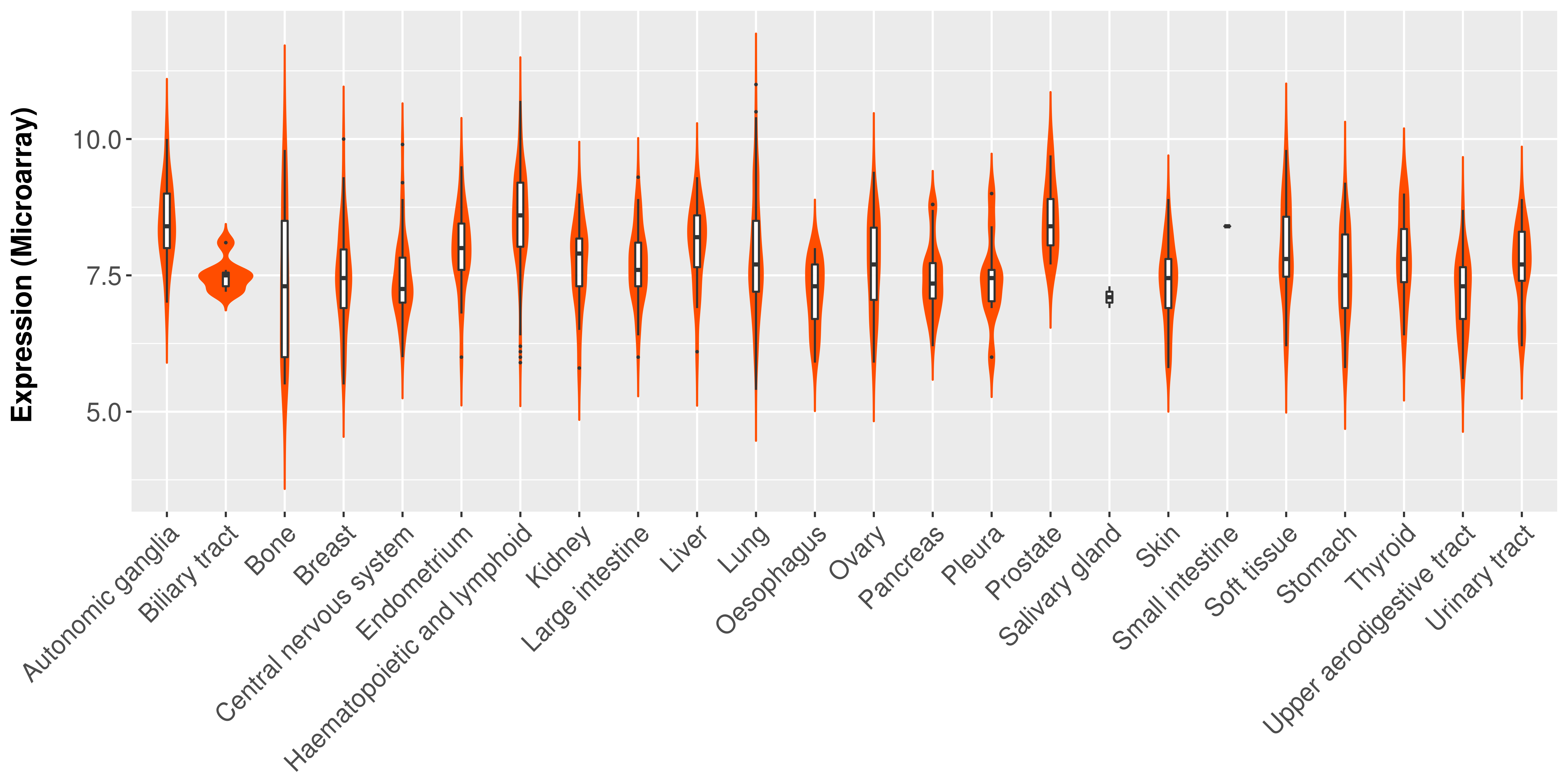
|
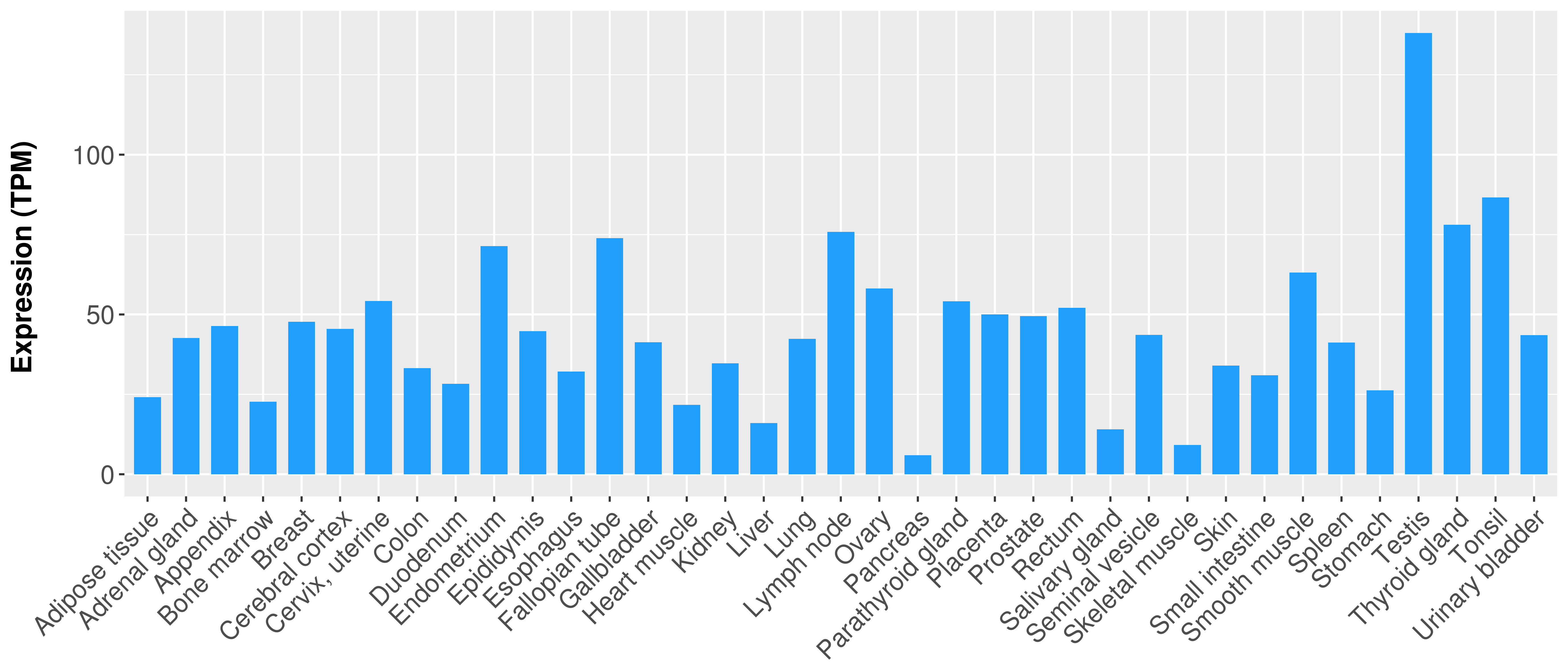
|
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
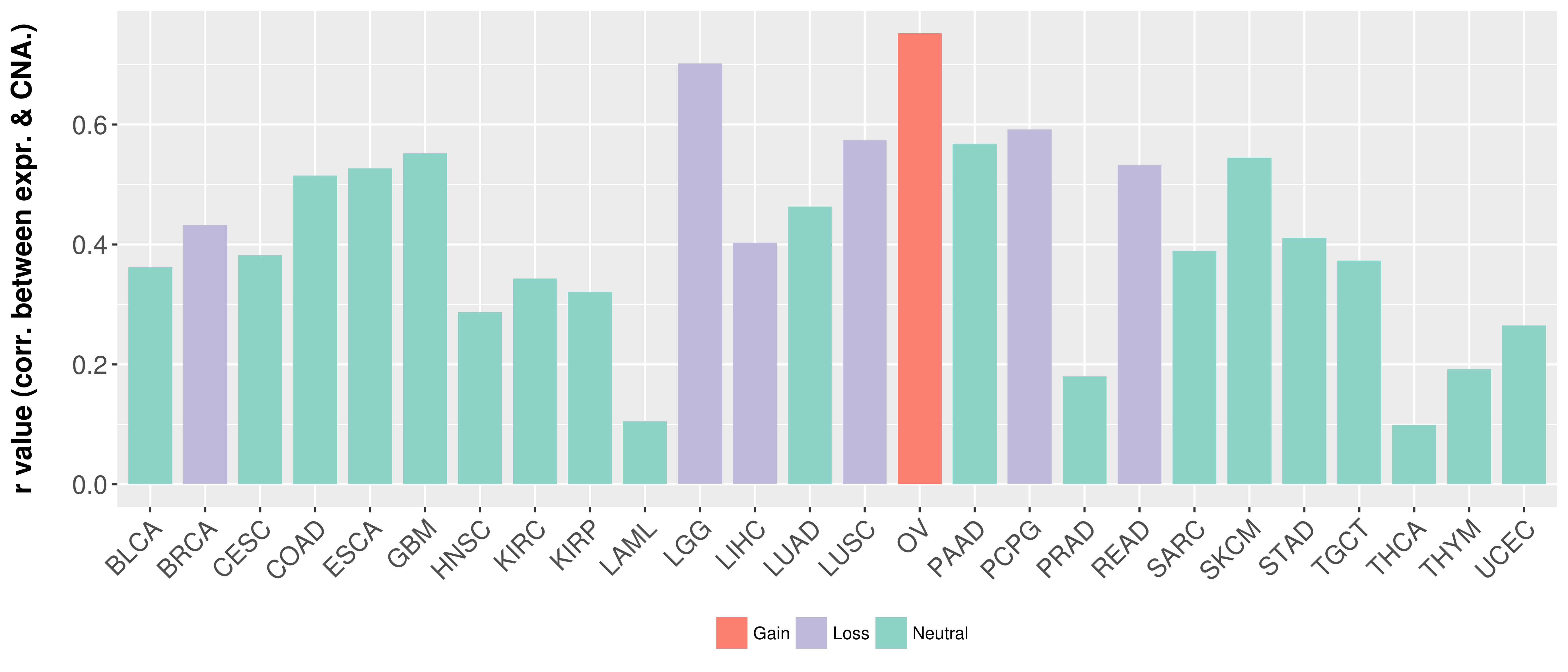
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
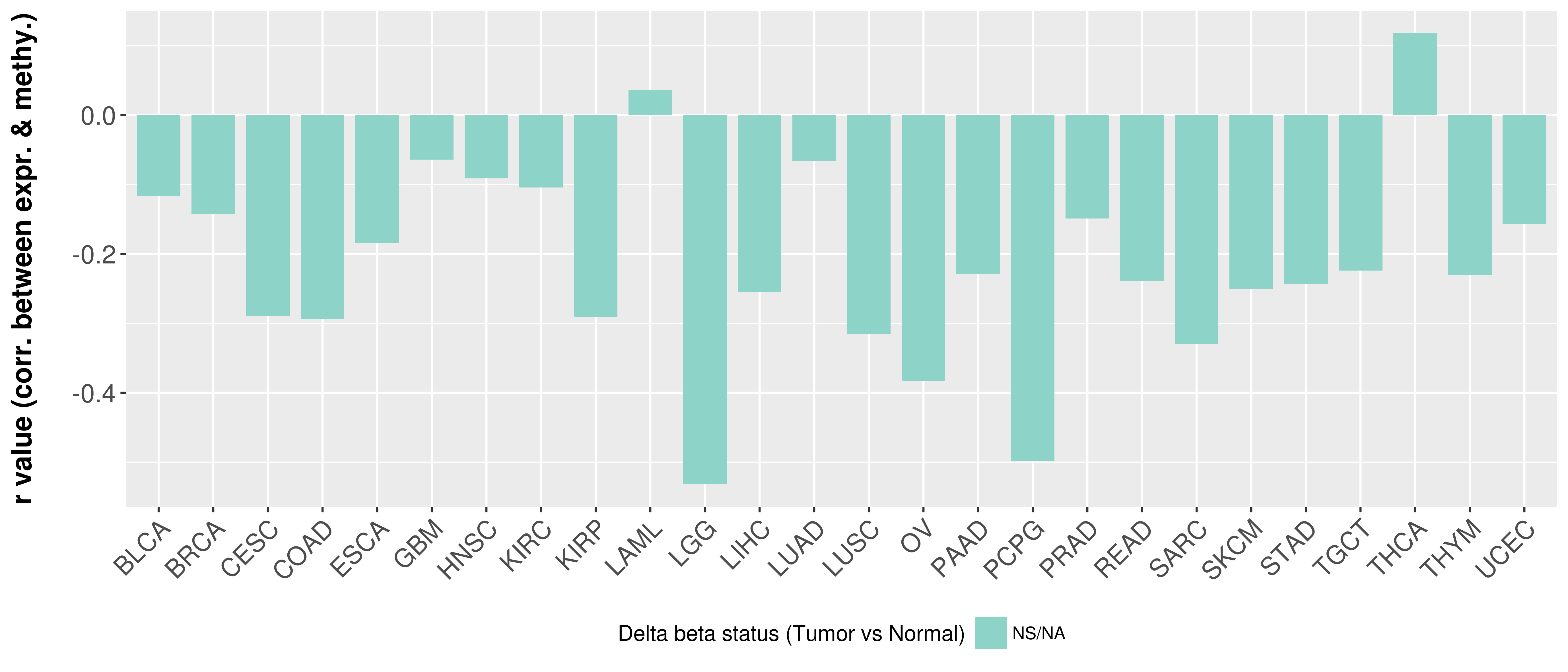
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
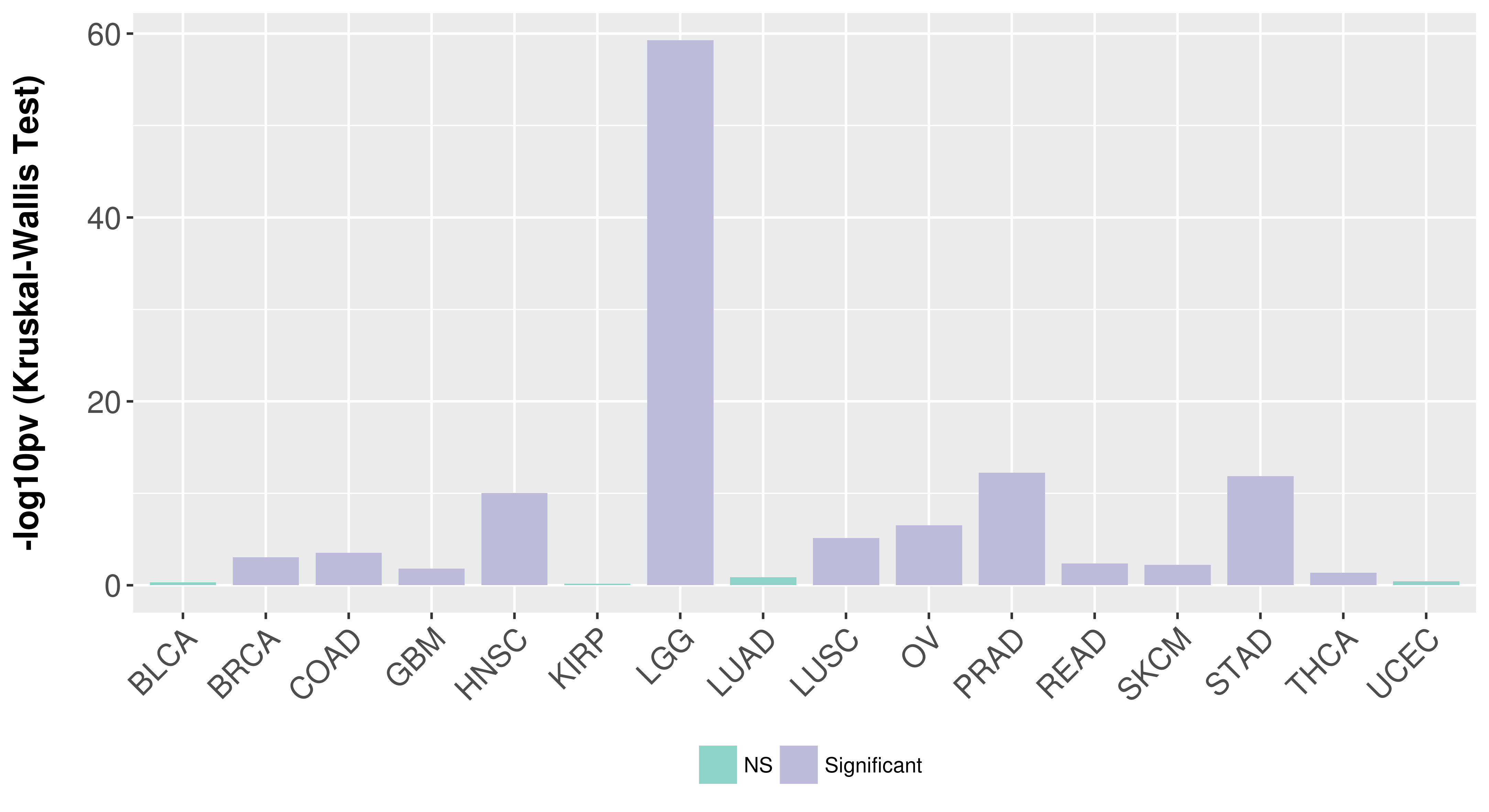
Association between expresson and subtype.
|
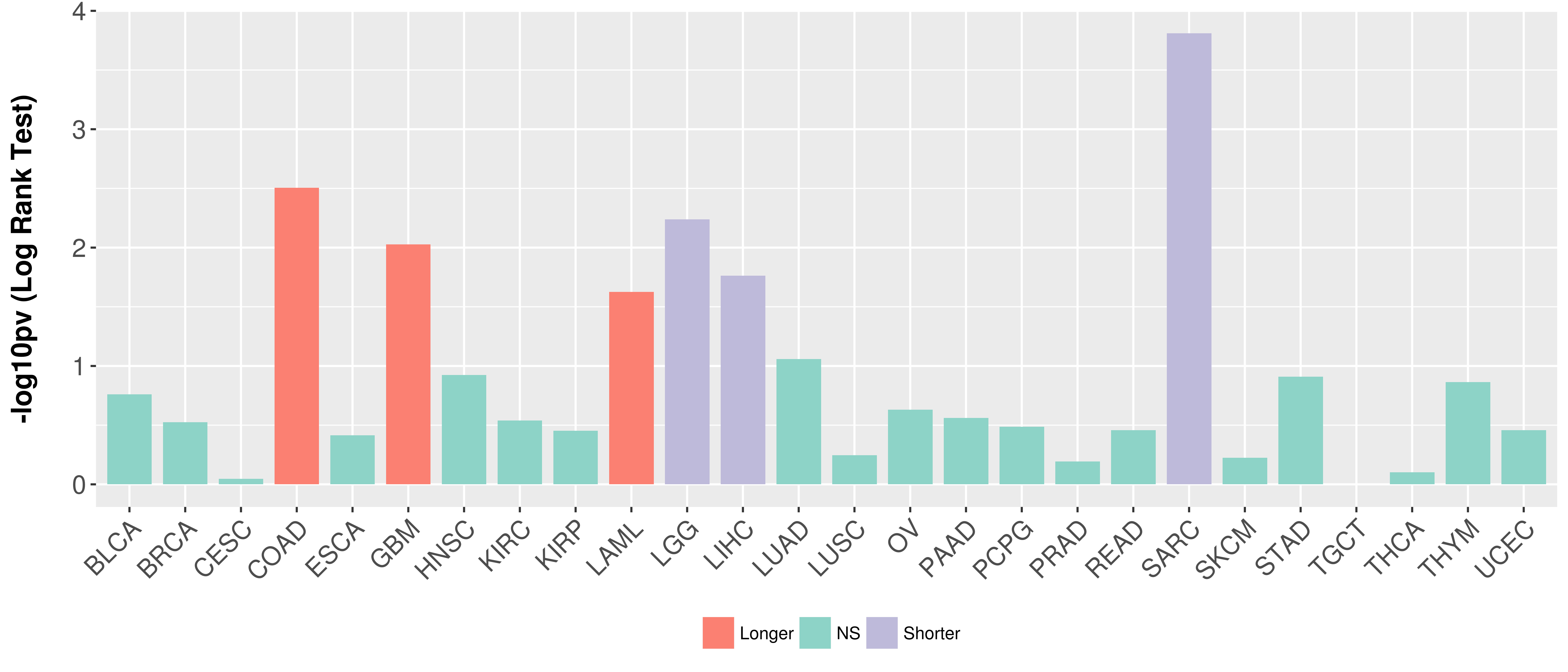
Overall survival analysis based on expression.
|
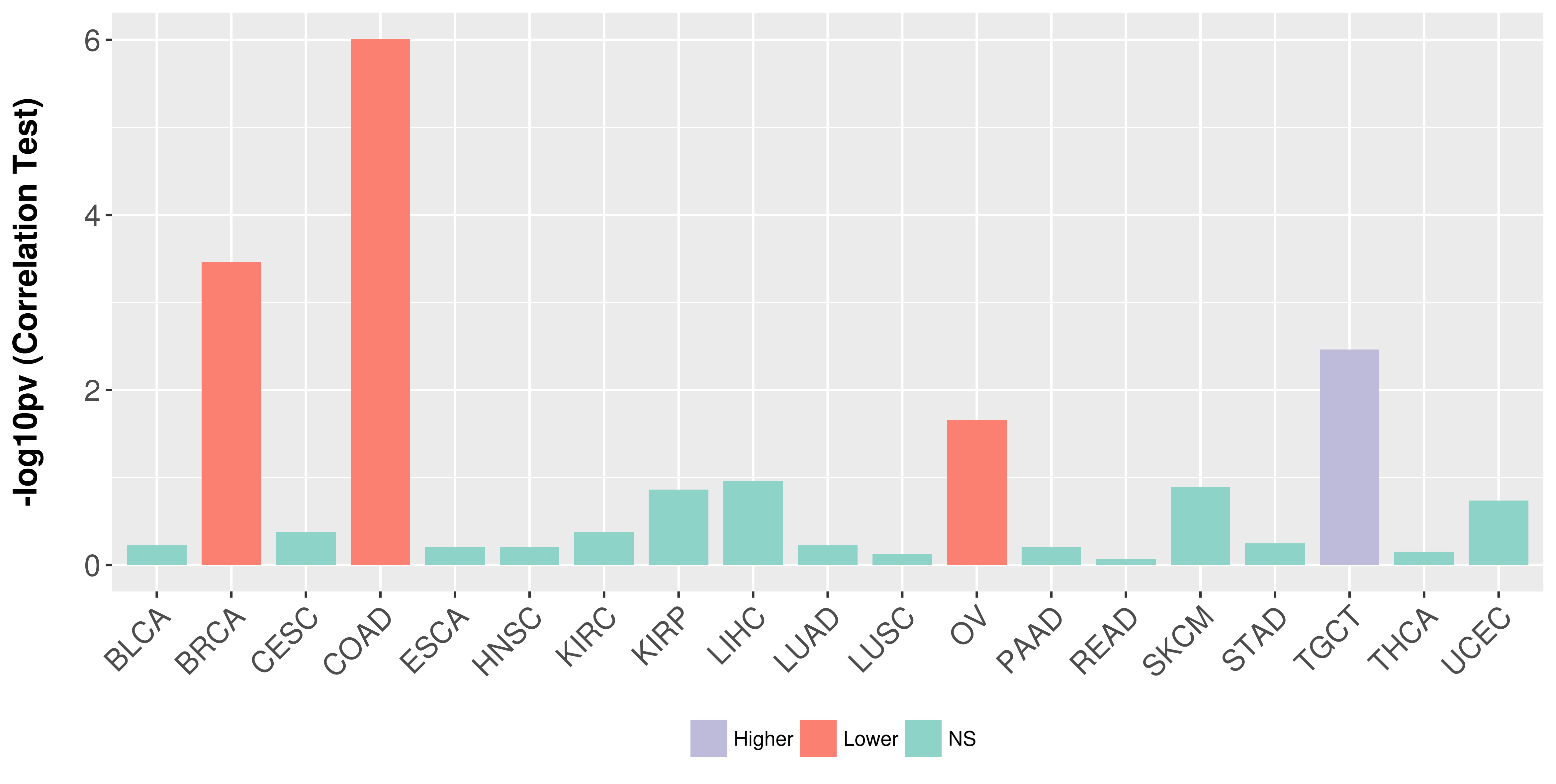
Association between expresson and stage.
|
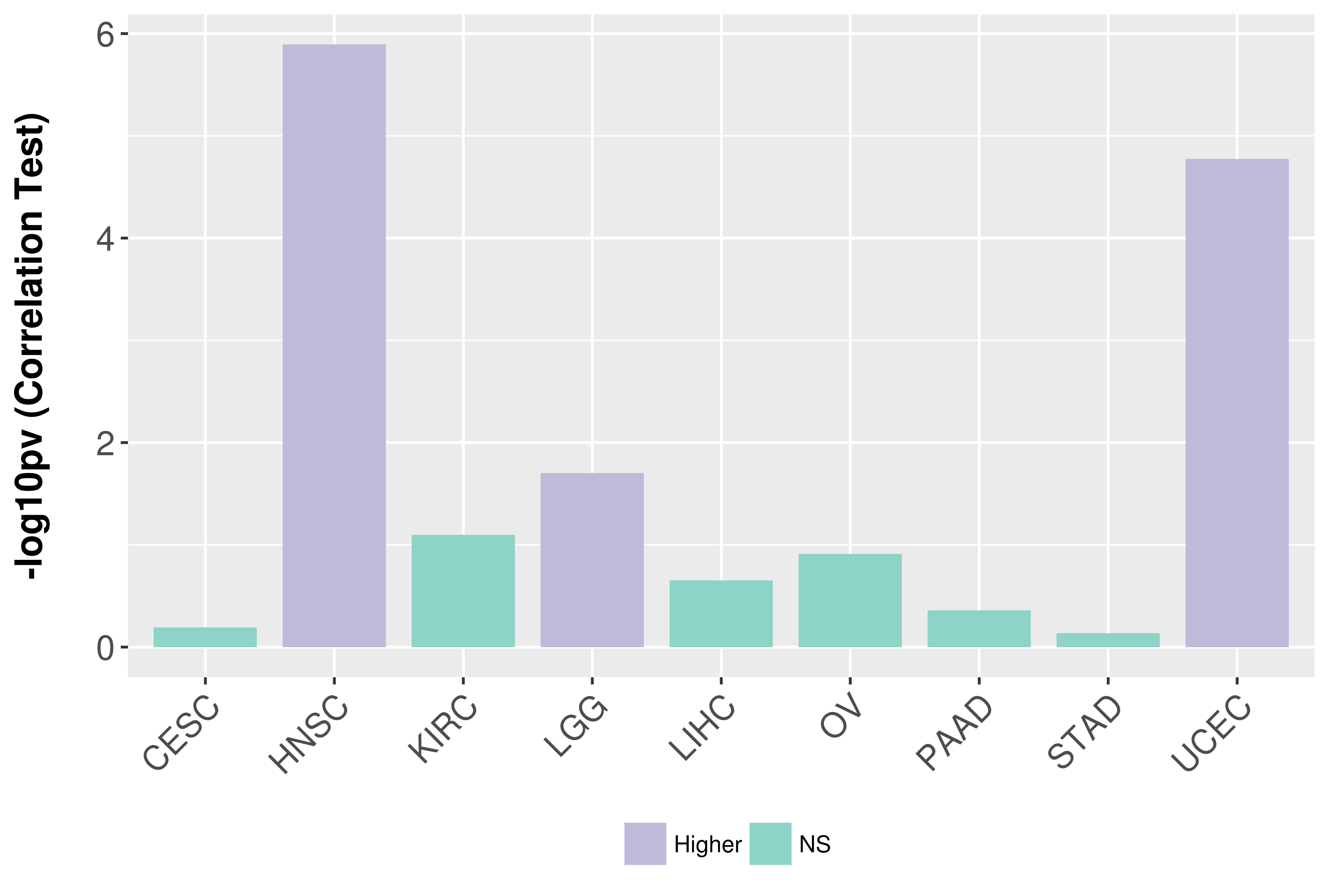
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for RBBP4. |
| Summary | |
|---|---|
| Symbol | RBBP4 |
| Name | RB binding protein 4, chromatin remodeling factor |
| Aliases | RbAp48; NURF55; lin-53; retinoblastoma-binding protein 4; CAF-1 subunit C; CAF-I 48 kDa subunit; CAF-I p48; ...... |
| Location | 1p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for RBBP4. |