Browse RUVBL2 in pancancer
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF06068 TIP49 C-terminus |
||||||||||
| Function |
Possesses single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity; hexamerization is thought to be critical for ATP hydrolysis and adjacent subunits in the ring-like structure contribute to the ATPase activity.; FUNCTION: Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AFZ from the nucleosome.; FUNCTION: Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.; FUNCTION: Plays an essential role in oncogenic transformation by MYC and also modulates transcriptional activation by the LEF1/TCF1-CTNNB1 complex. May also inhibit the transcriptional activity of ATF2.; FUNCTION: Involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway where it negatively regulates expression of ER stress response genes. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000491 small nucleolar ribonucleoprotein complex assembly GO:0000492 box C/D snoRNP assembly GO:0006310 DNA recombination GO:0006338 chromatin remodeling GO:0006403 RNA localization GO:0006457 protein folding GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0022613 ribonucleoprotein complex biogenesis GO:0022618 ribonucleoprotein complex assembly GO:0031056 regulation of histone modification GO:0031058 positive regulation of histone modification GO:0032091 negative regulation of protein binding GO:0032355 response to estradiol GO:0032392 DNA geometric change GO:0032508 DNA duplex unwinding GO:0034502 protein localization to chromosome GO:0034644 cellular response to UV GO:0035065 regulation of histone acetylation GO:0035066 positive regulation of histone acetylation GO:0043393 regulation of protein binding GO:0043543 protein acylation GO:0043967 histone H4 acetylation GO:0043968 histone H2A acetylation GO:0051098 regulation of binding GO:0051100 negative regulation of binding GO:0070199 establishment of protein localization to chromosome GO:0071103 DNA conformation change GO:0071168 protein localization to chromatin GO:0071169 establishment of protein localization to chromatin GO:0071214 cellular response to abiotic stimulus GO:0071392 cellular response to estradiol stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071478 cellular response to radiation GO:0071482 cellular response to light stimulus GO:0071733 transcriptional activation by promoter-enhancer looping GO:0071826 ribonucleoprotein complex subunit organization GO:0071898 regulation of estrogen receptor binding GO:0071899 negative regulation of estrogen receptor binding GO:0090202 gene looping GO:0090579 dsDNA loop formation GO:0090670 RNA localization to Cajal body GO:0090671 telomerase RNA localization to Cajal body GO:0090672 telomerase RNA localization GO:1900120 regulation of receptor binding GO:1900121 negative regulation of receptor binding GO:1901983 regulation of protein acetylation GO:1901985 positive regulation of protein acetylation GO:1902275 regulation of chromatin organization GO:1904872 regulation of telomerase RNA localization to Cajal body GO:1904874 positive regulation of telomerase RNA localization to Cajal body GO:1905269 positive regulation of chromatin organization GO:2000756 regulation of peptidyl-lysine acetylation GO:2000758 positive regulation of peptidyl-lysine acetylation |
| Molecular Function |
GO:0000979 RNA polymerase II core promoter sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0001046 core promoter sequence-specific DNA binding GO:0001047 core promoter binding GO:0001158 enhancer sequence-specific DNA binding GO:0003678 DNA helicase activity GO:0003682 chromatin binding GO:0004003 ATP-dependent DNA helicase activity GO:0004386 helicase activity GO:0008026 ATP-dependent helicase activity GO:0008094 DNA-dependent ATPase activity GO:0016887 ATPase activity GO:0031490 chromatin DNA binding GO:0035326 enhancer binding GO:0042623 ATPase activity, coupled GO:0043139 5'-3' DNA helicase activity GO:0043141 ATP-dependent 5'-3' DNA helicase activity GO:0043566 structure-specific DNA binding GO:0051082 unfolded protein binding GO:0070035 purine NTP-dependent helicase activity |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000791 euchromatin GO:0000812 Swr1 complex GO:0005719 nuclear euchromatin GO:0016363 nuclear matrix GO:0031011 Ino80 complex GO:0031248 protein acetyltransferase complex GO:0033202 DNA helicase complex GO:0034399 nuclear periphery GO:0034708 methyltransferase complex GO:0035097 histone methyltransferase complex GO:0035267 NuA4 histone acetyltransferase complex GO:0043189 H4/H2A histone acetyltransferase complex GO:0044454 nuclear chromosome part GO:0044665 MLL1/2 complex GO:0070603 SWI/SNF superfamily-type complex GO:0071339 MLL1 complex GO:0097255 R2TP complex GO:0097346 INO80-type complex GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-73886: Chromosome Maintenance R-HSA-180786: Extension of Telomeres R-HSA-3214847: HATs acetylate histones R-HSA-171319: Telomere Extension By Telomerase R-HSA-157579: Telomere Maintenance |
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for RUVBL2. |
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
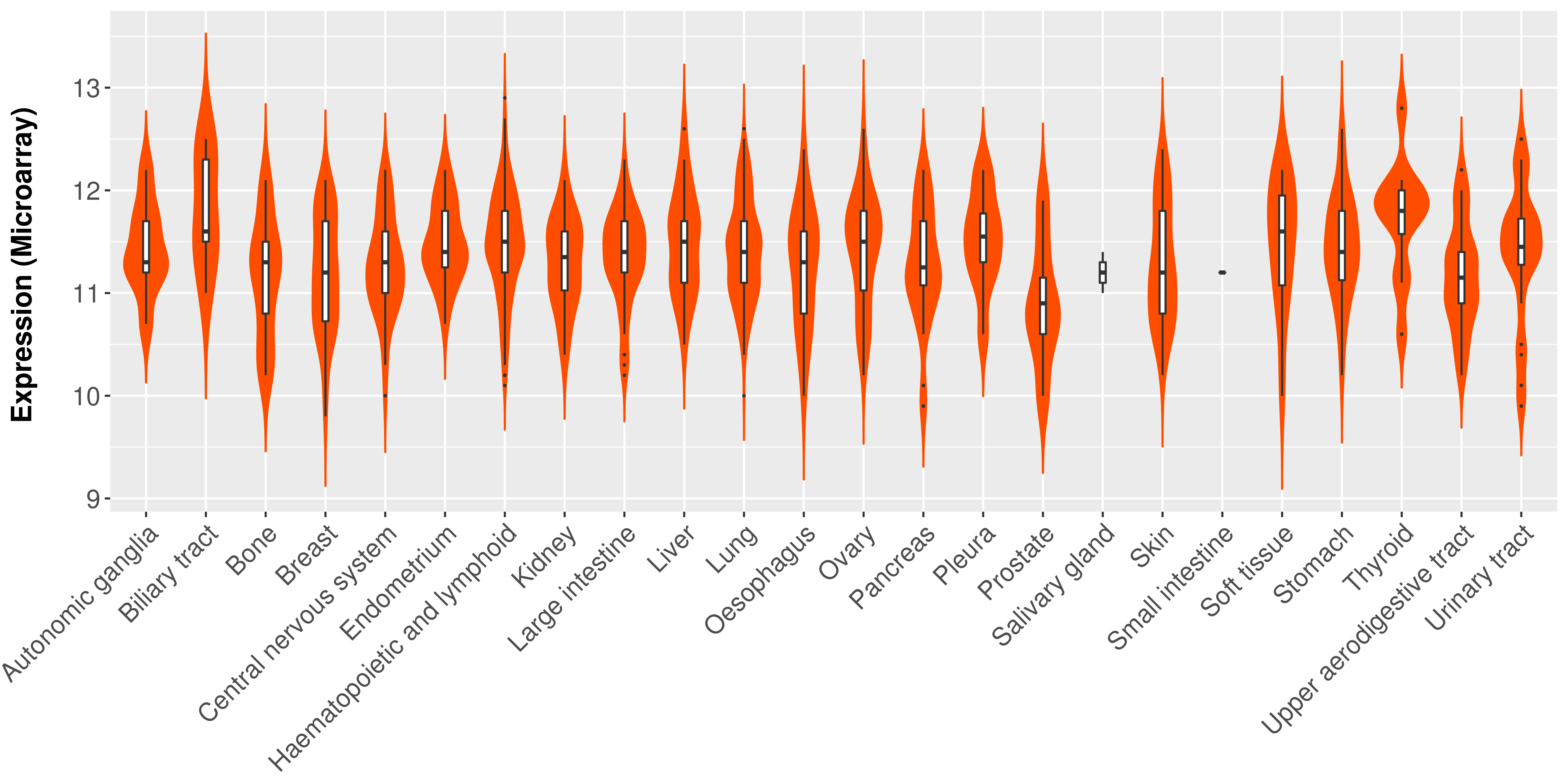
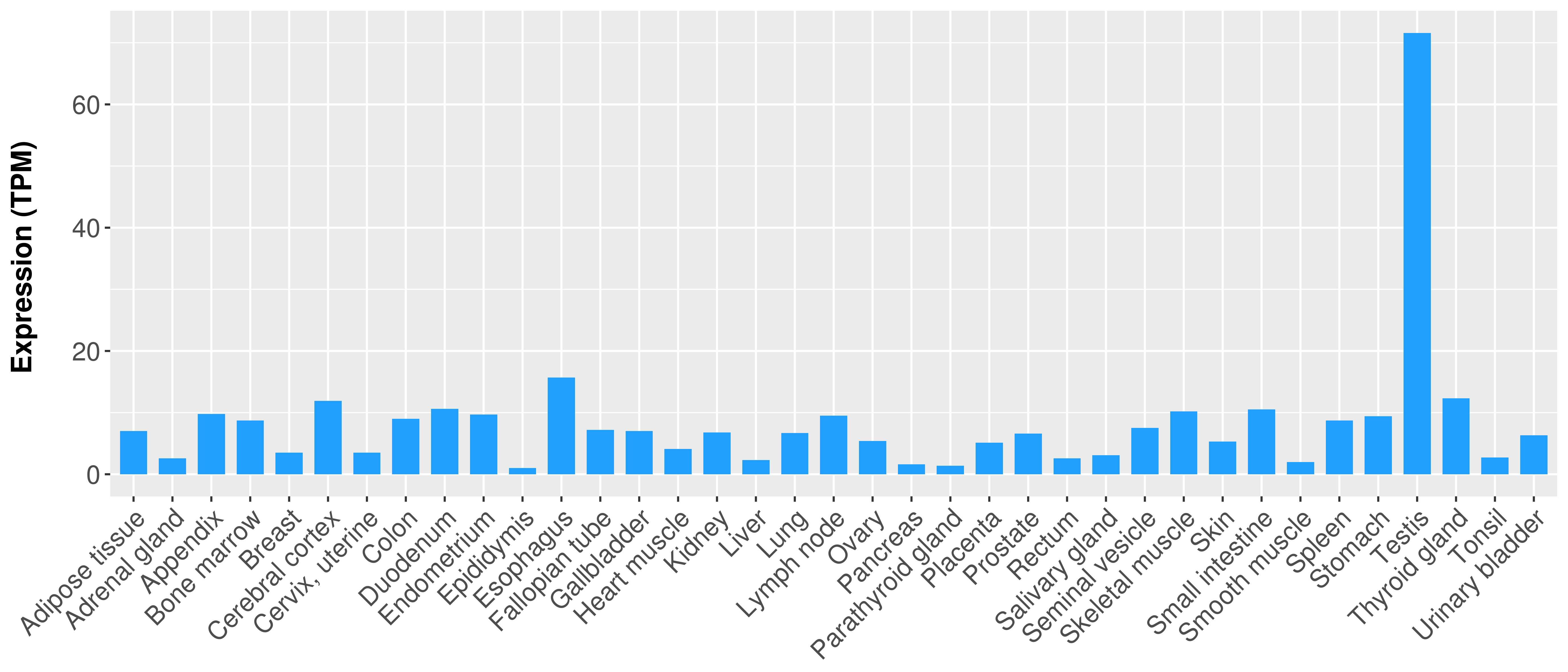
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
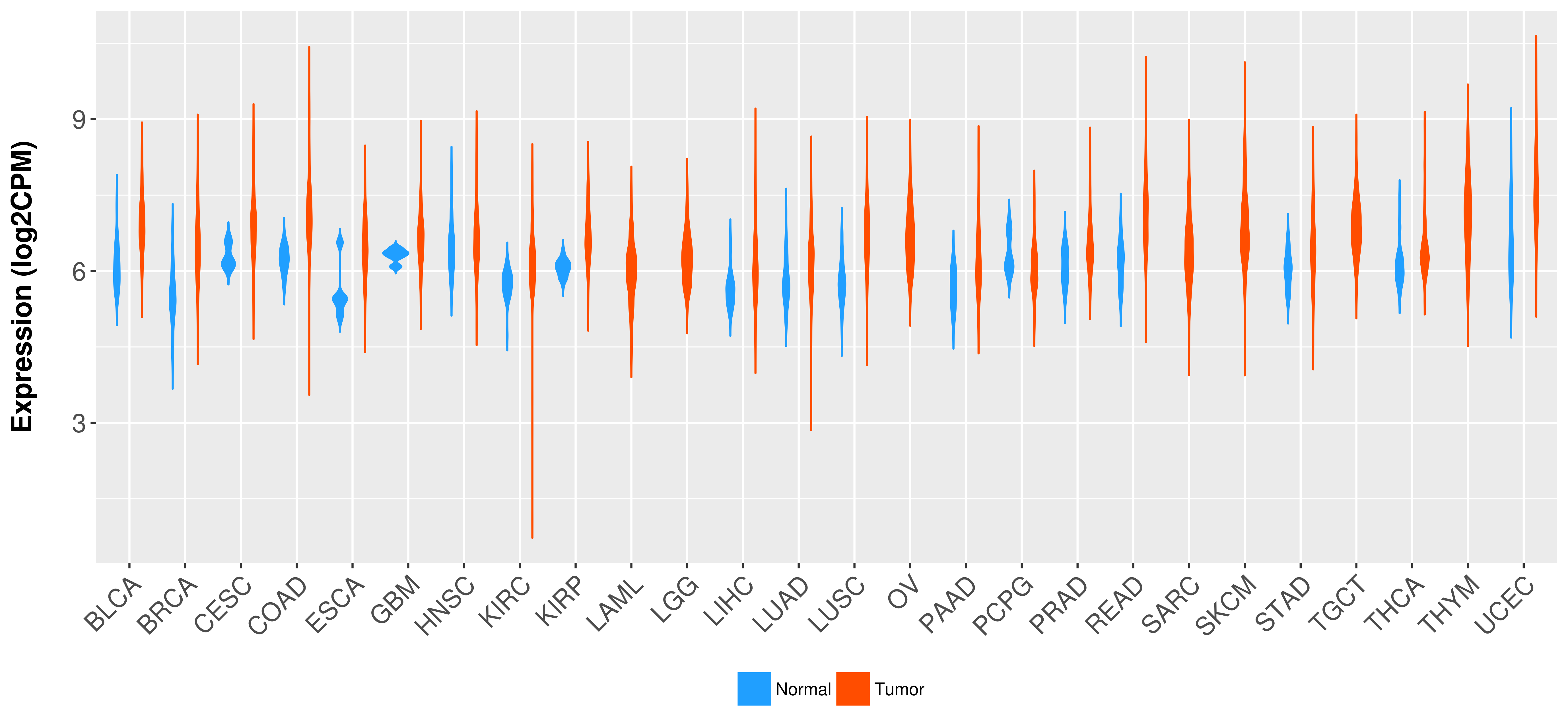
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
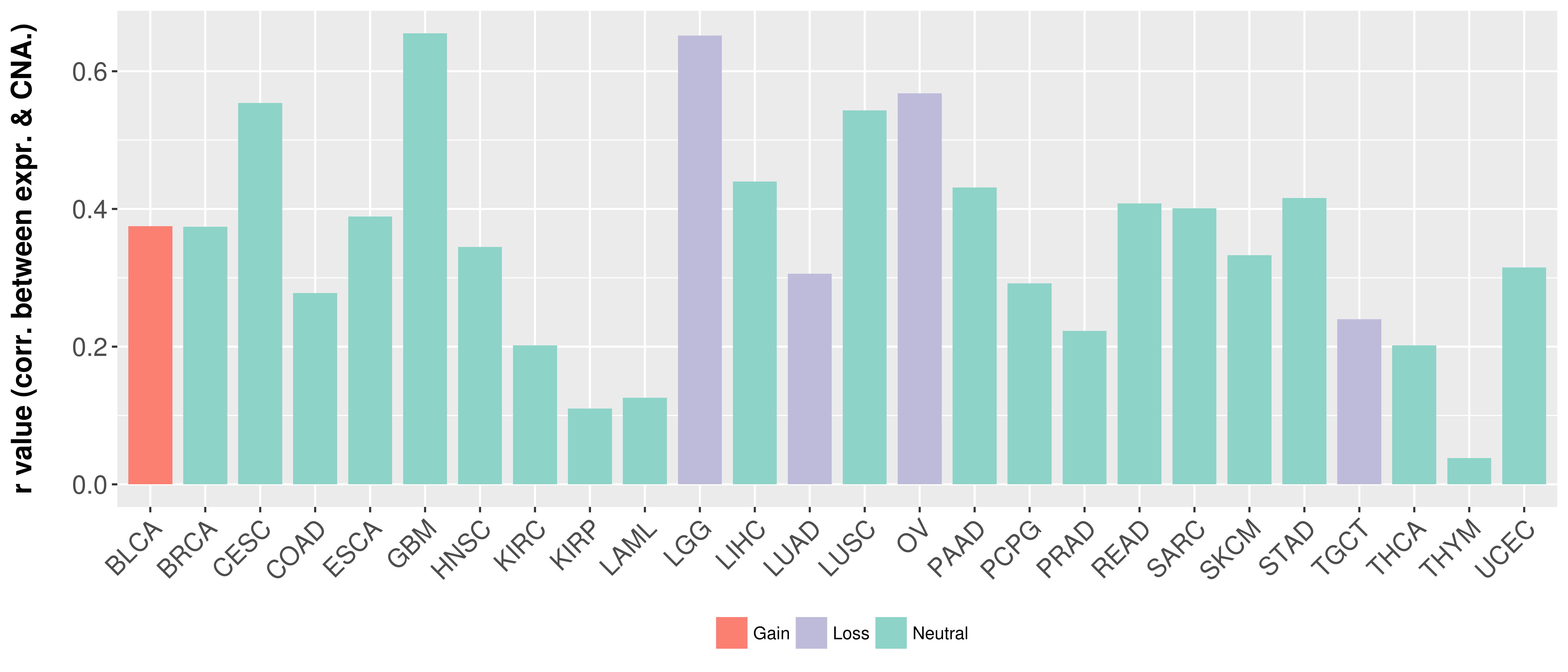
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
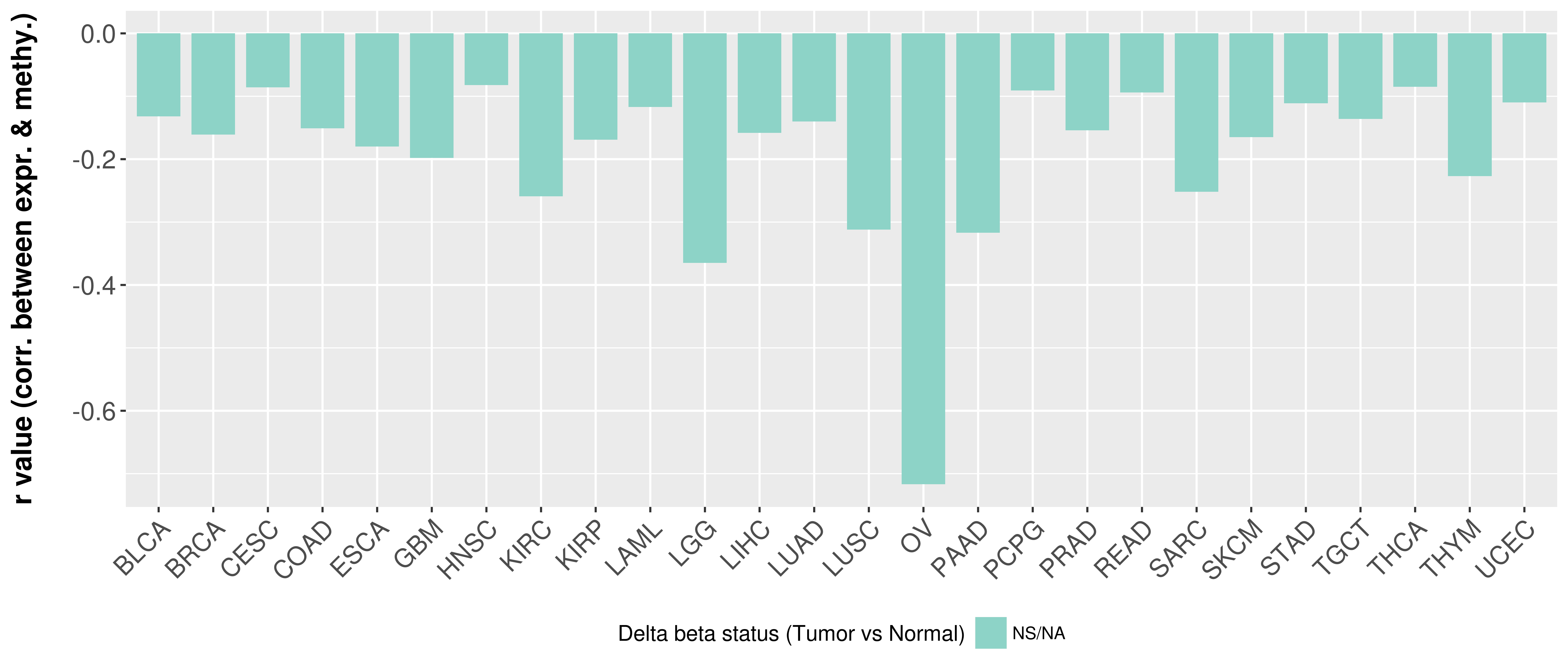
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
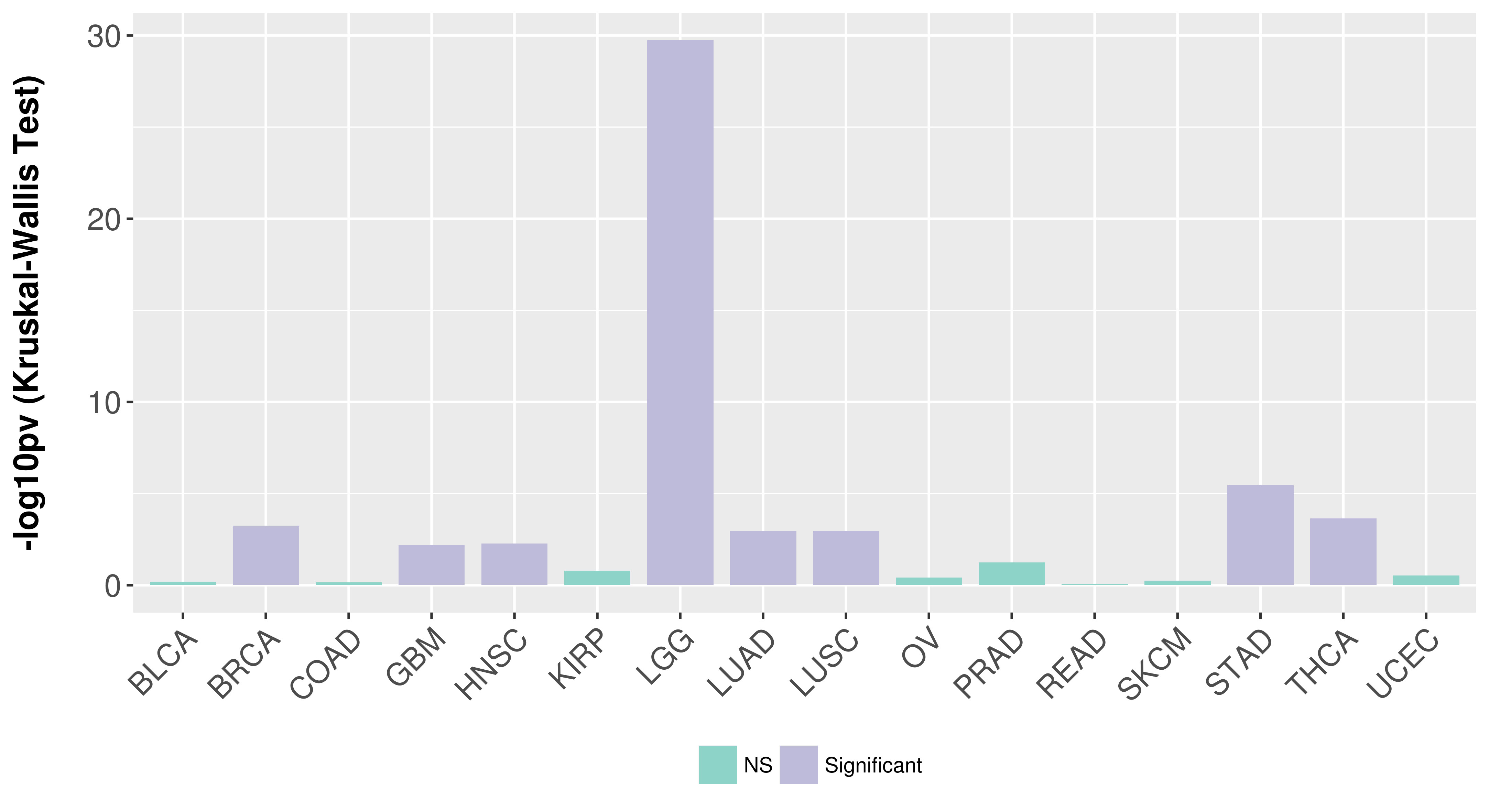
Association between expresson and subtype.
|
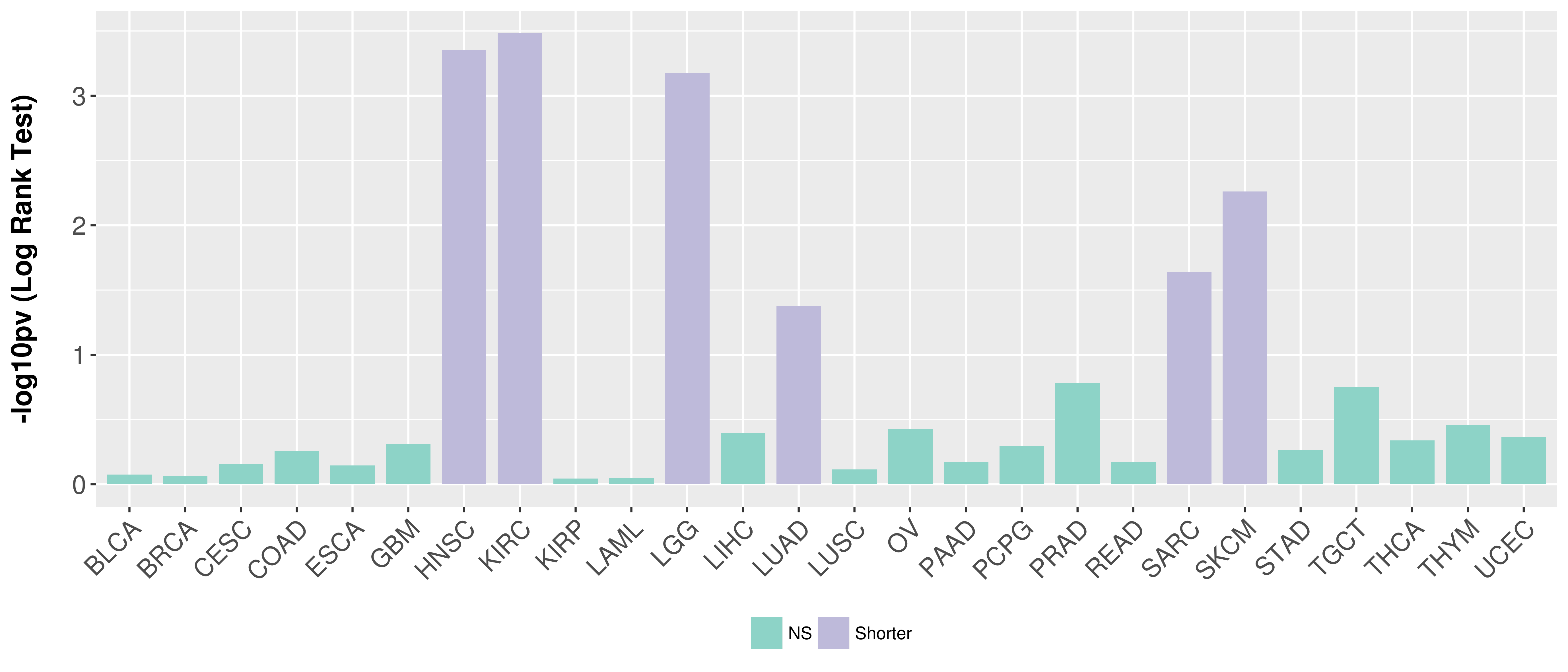
Overall survival analysis based on expression.
|
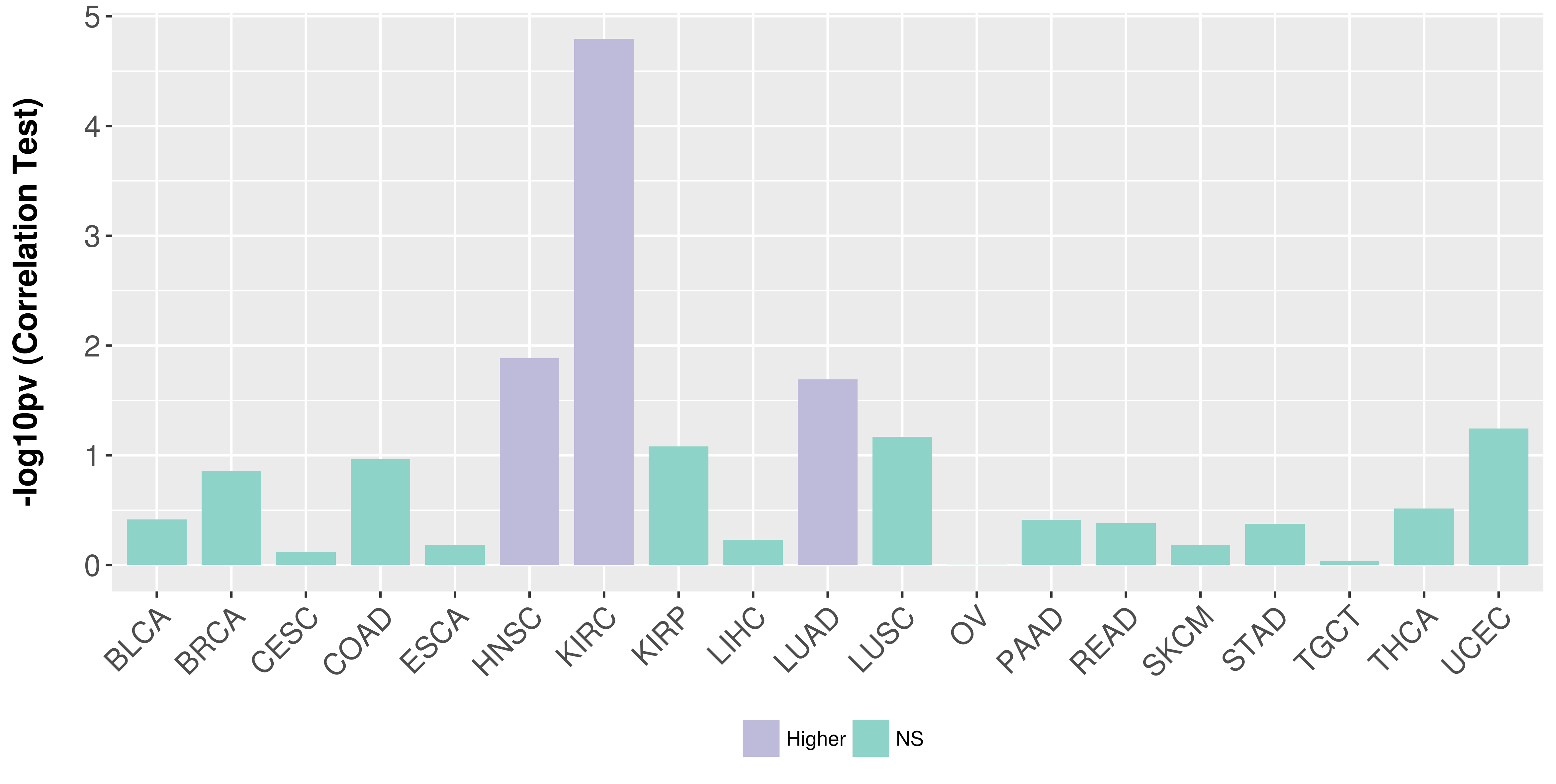
Association between expresson and stage.
|
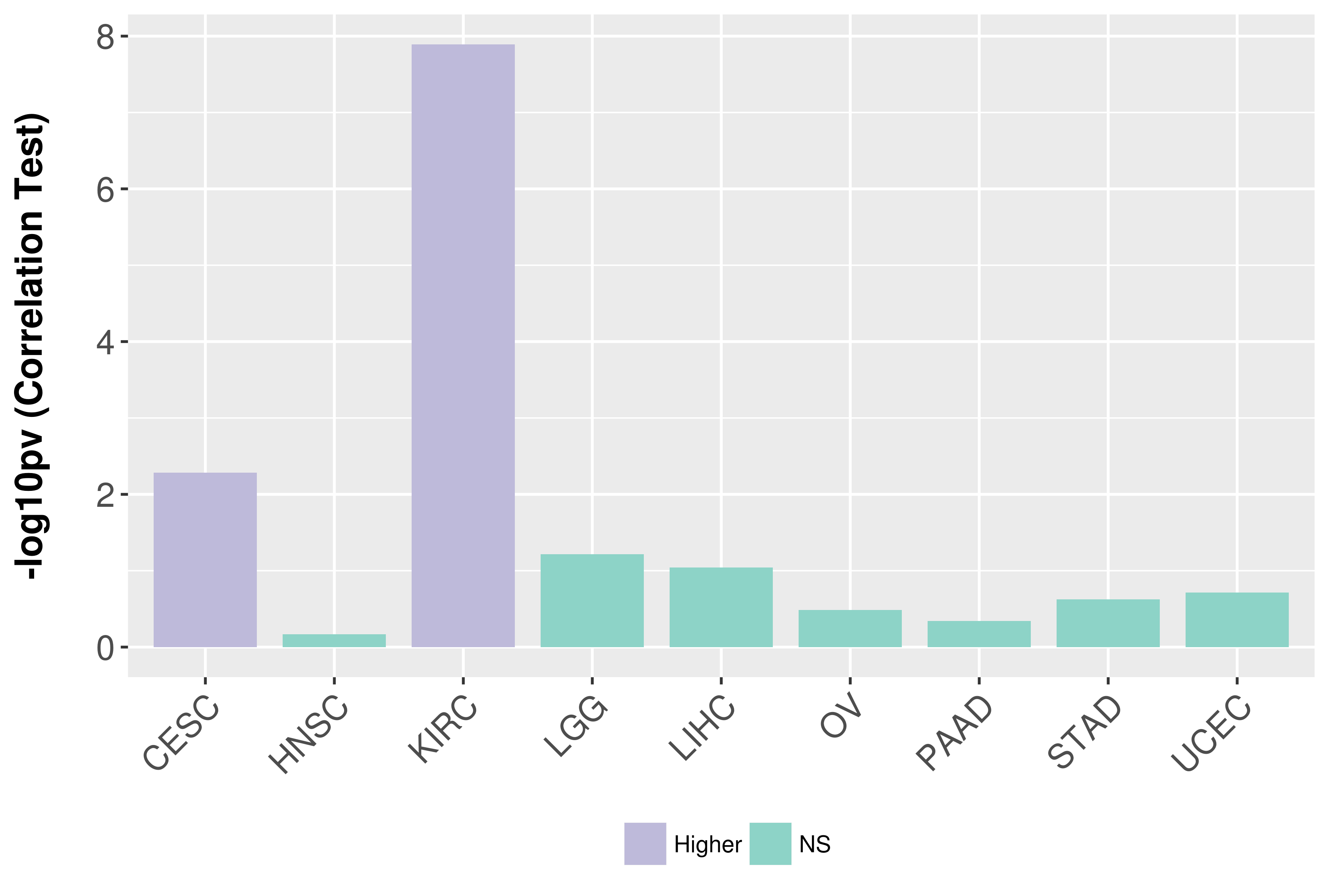
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for RUVBL2. |
| Summary | |
|---|---|
| Symbol | RUVBL2 |
| Name | RuvB like AAA ATPase 2 |
| Aliases | RVB2; TIP48; TIP49b; Reptin52; ECP51; TIH2; INO80J; reptin; RuvB (E coli homolog)-like 2; RuvB-like 2 (E. co ...... |
| Location | 19q13.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|