Browse SETD2 in pancancer
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00856 SET domain PF08236 SRI (Set2 Rpb1 interacting) domain PF00397 WW domain |
||||||||||
| Function |
Histone methyltransferase that specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using dimethylated 'Lys-36' (H3K36me2) as substrate. Represents the main enzyme generating H3K36me3, a specific tag for epigenetic transcriptional activation. Plays a role in chromatin structure modulation during elongation by coordinating recruitment of the FACT complex and by interacting with hyperphosphorylated POLR2A. Acts as a key regulator of DNA mismatch repair in G1 and early S phase by generating H3K36me3, a mark required to recruit MSH6 subunit of the MutS alpha complex: early recruitment of the MutS alpha complex to chromatin to be replicated allows a quick identification of mismatch DNA to initiate the mismatch repair reaction. H3K36me3 also plays an essential role in the maintenance of a heterochromatic state, by recruiting DNA methyltransferase DNMT3A. H3K36me3 is also enhanced in intron-containing genes, suggesting that SETD2 recruitment is enhanced by splicing and that splicing is coupled to recruitment of elongating RNA polymerase. Required during angiogenesis. Recruited to the promoters of adenovirus 12 E1A gene in case of infection, possibly leading to regulate its expression. Trimethylates 'Lys-40' of alpha-tubulins such as TUBA1B (alpha-TubK40me3) (PubMed:27518565). Alpha-TubK40me3 is required for normal mitosis and cytokinesis and is proposed to be a specific tag in cytoskeletal remodeling (PubMed:27518565). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001501 skeletal system development GO:0001525 angiogenesis GO:0001570 vasculogenesis GO:0001701 in utero embryonic development GO:0001763 morphogenesis of a branching structure GO:0001838 embryonic epithelial tube formation GO:0001841 neural tube formation GO:0001843 neural tube closure GO:0001890 placenta development GO:0001892 embryonic placenta development GO:0006298 mismatch repair GO:0006354 DNA-templated transcription, elongation GO:0006368 transcription elongation from RNA polymerase II promoter GO:0006403 RNA localization GO:0006405 RNA export from nucleus GO:0006406 mRNA export from nucleus GO:0006479 protein methylation GO:0006913 nucleocytoplasmic transport GO:0007498 mesoderm development GO:0007507 heart development GO:0008213 protein alkylation GO:0010452 histone H3-K36 methylation GO:0010793 regulation of mRNA export from nucleus GO:0014020 primary neural tube formation GO:0015931 nucleobase-containing compound transport GO:0016331 morphogenesis of embryonic epithelium GO:0016570 histone modification GO:0016571 histone methylation GO:0018022 peptidyl-lysine methylation GO:0018023 peptidyl-lysine trimethylation GO:0018027 peptidyl-lysine dimethylation GO:0018205 peptidyl-lysine modification GO:0021915 neural tube development GO:0030900 forebrain development GO:0032239 regulation of nucleobase-containing compound transport GO:0032259 methylation GO:0032386 regulation of intracellular transport GO:0034728 nucleosome organization GO:0034968 histone lysine methylation GO:0035148 tube formation GO:0035239 tube morphogenesis GO:0035441 cell migration involved in vasculogenesis GO:0043414 macromolecule methylation GO:0046822 regulation of nucleocytoplasmic transport GO:0046831 regulation of RNA export from nucleus GO:0048332 mesoderm morphogenesis GO:0048514 blood vessel morphogenesis GO:0048562 embryonic organ morphogenesis GO:0048568 embryonic organ development GO:0048608 reproductive structure development GO:0048701 embryonic cranial skeleton morphogenesis GO:0048704 embryonic skeletal system morphogenesis GO:0048705 skeletal system morphogenesis GO:0048706 embryonic skeletal system development GO:0048863 stem cell differentiation GO:0048864 stem cell development GO:0050657 nucleic acid transport GO:0050658 RNA transport GO:0051028 mRNA transport GO:0051168 nuclear export GO:0051169 nuclear transport GO:0051236 establishment of RNA localization GO:0060039 pericardium development GO:0060562 epithelial tube morphogenesis GO:0060606 tube closure GO:0060669 embryonic placenta morphogenesis GO:0060976 coronary vasculature development GO:0060977 coronary vasculature morphogenesis GO:0061458 reproductive system development GO:0071166 ribonucleoprotein complex localization GO:0071426 ribonucleoprotein complex export from nucleus GO:0071427 mRNA-containing ribonucleoprotein complex export from nucleus GO:0071824 protein-DNA complex subunit organization GO:0072175 epithelial tube formation GO:0097198 histone H3-K36 trimethylation GO:0097676 histone H3-K36 dimethylation GO:1904888 cranial skeletal system development GO:2000197 regulation of ribonucleoprotein complex localization |
| Molecular Function |
GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018024 histone-lysine N-methyltransferase activity GO:0042054 histone methyltransferase activity GO:0046975 histone methyltransferase activity (H3-K36 specific) |
| Cellular Component | - |
| KEGG |
hsa00310 Lysine degradation |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-3214841: PKMTs methylate histone lysines |
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SETD2. |
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
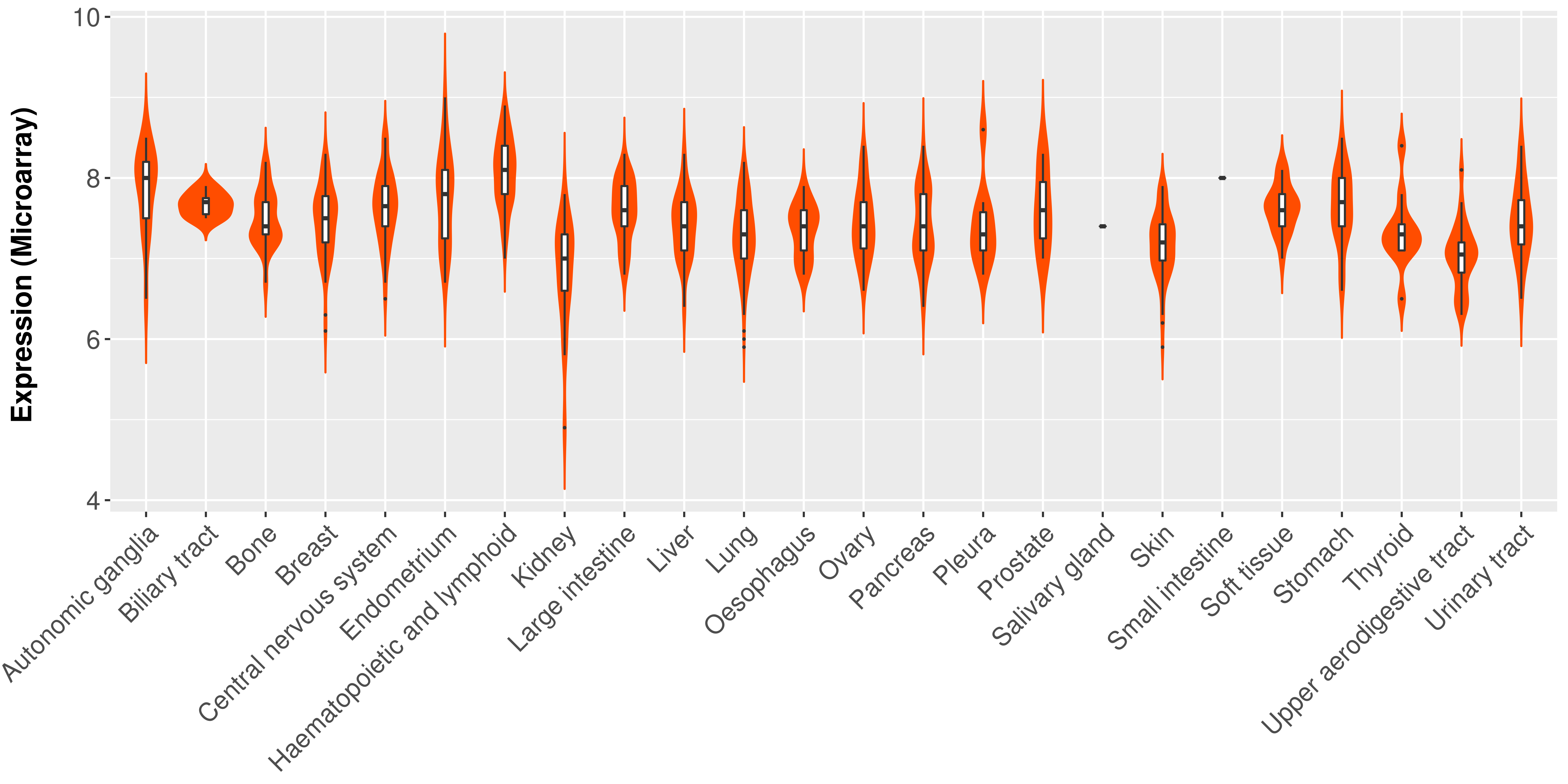
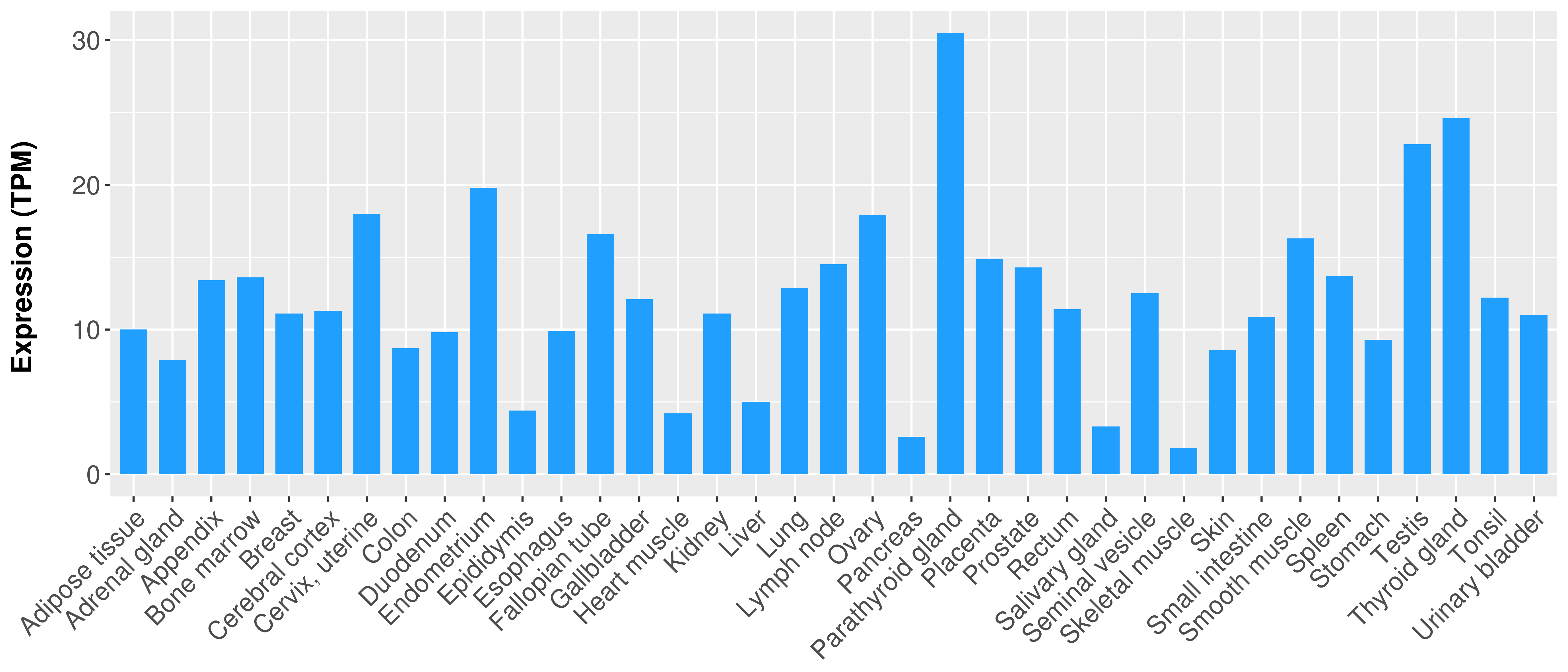
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
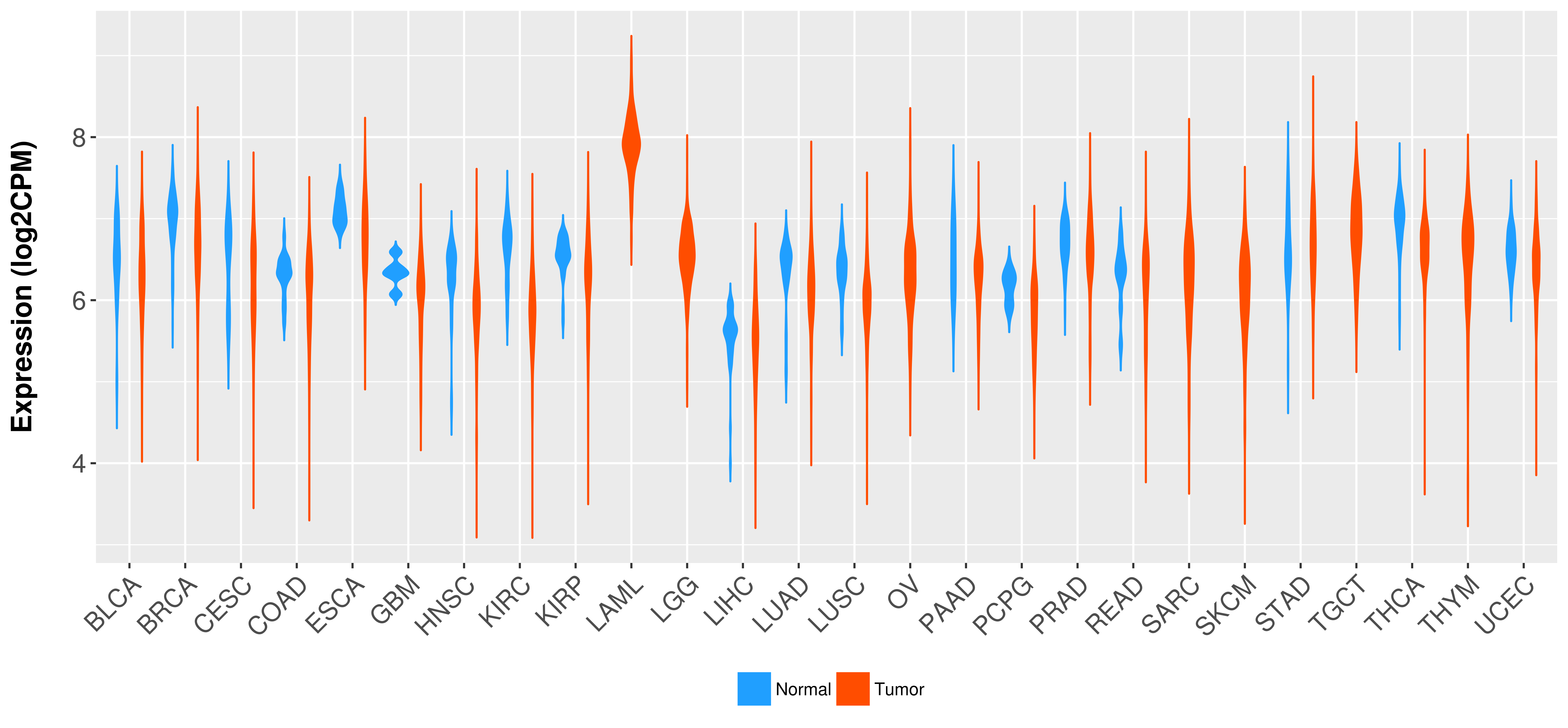
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
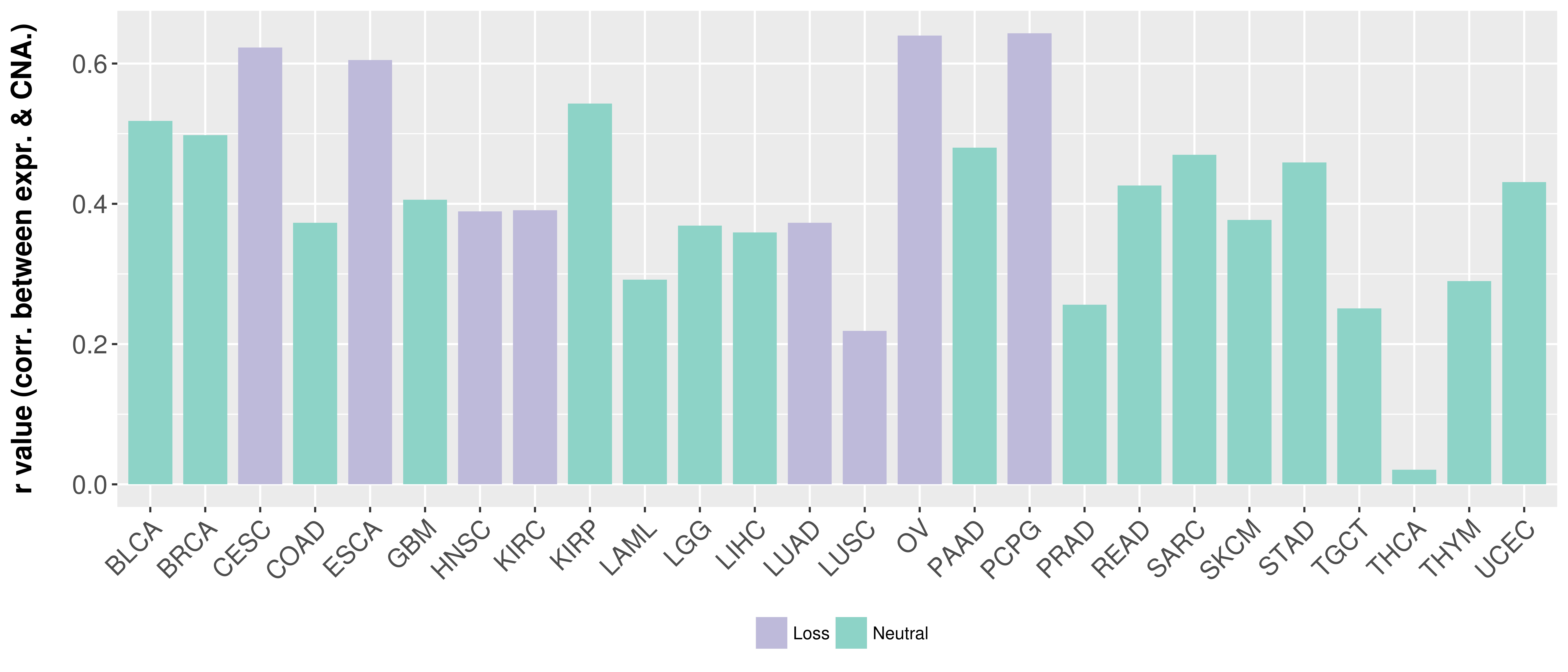
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
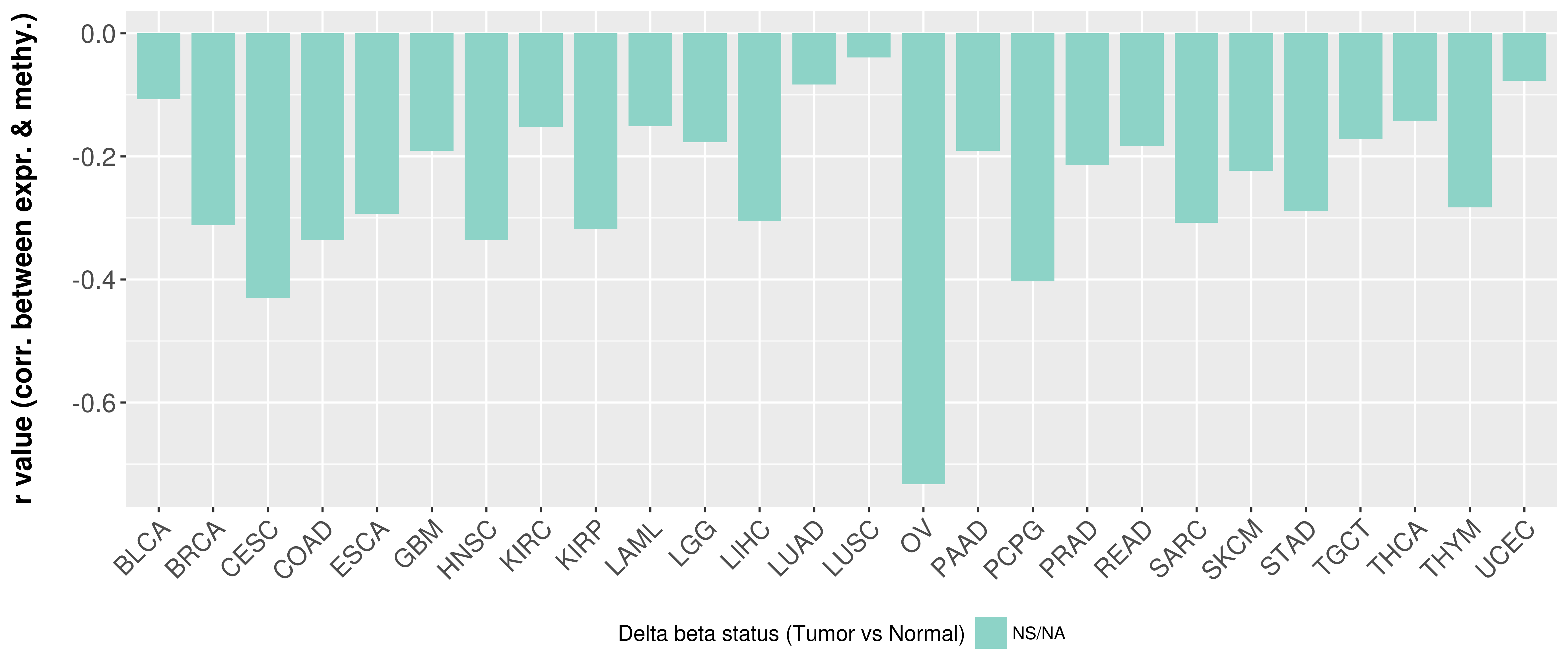
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
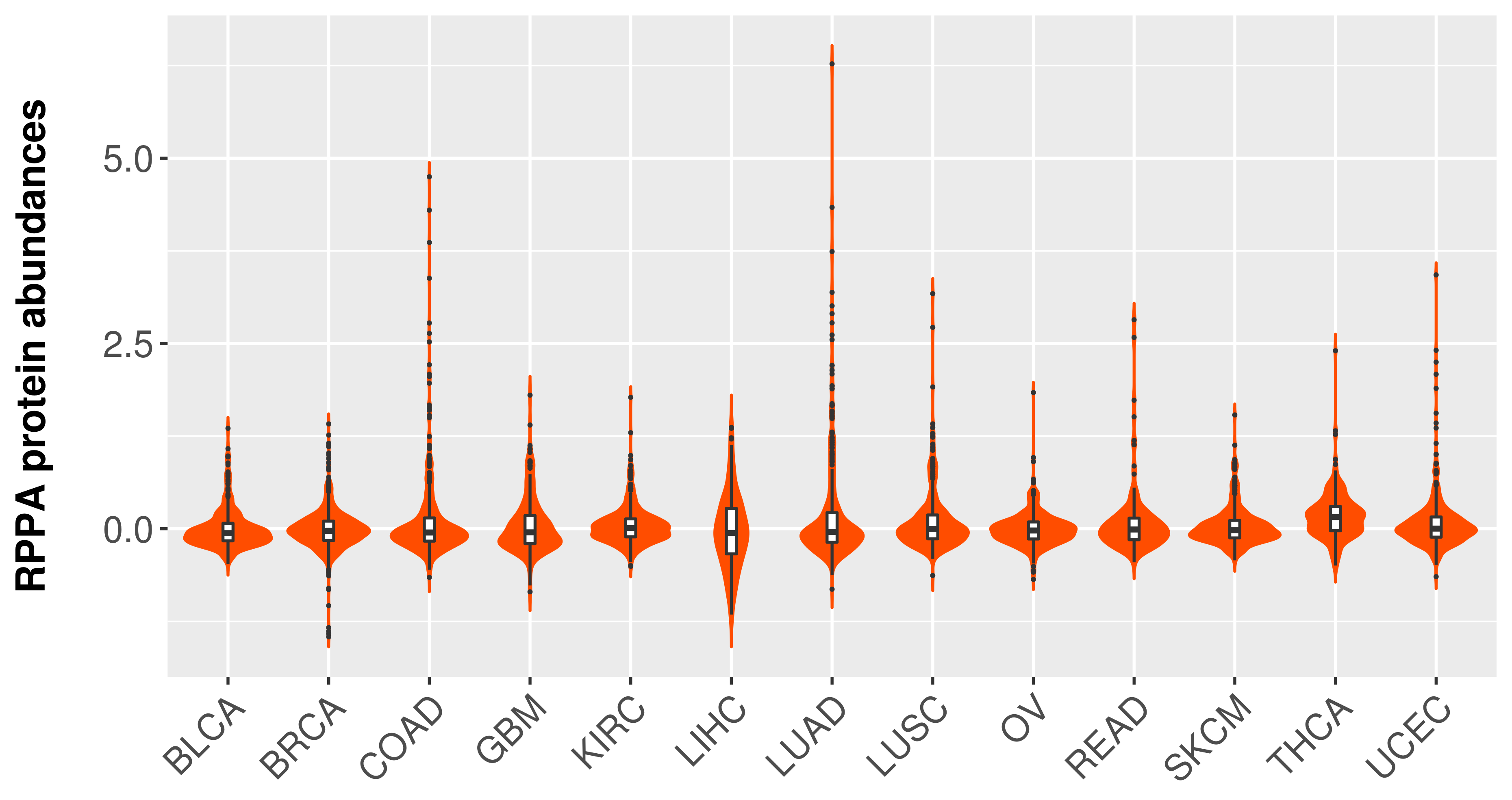
|
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
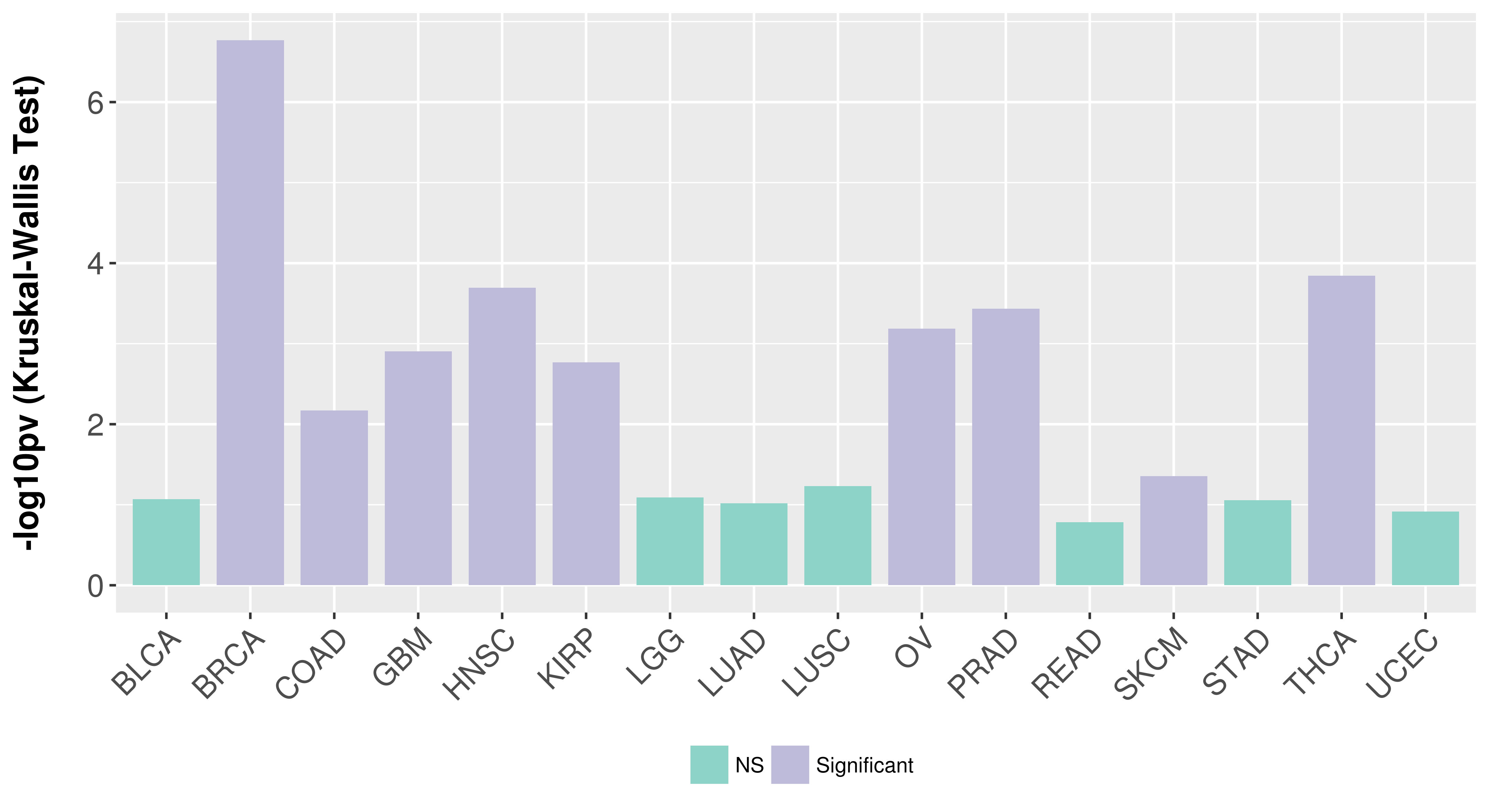
Association between expresson and subtype.
|
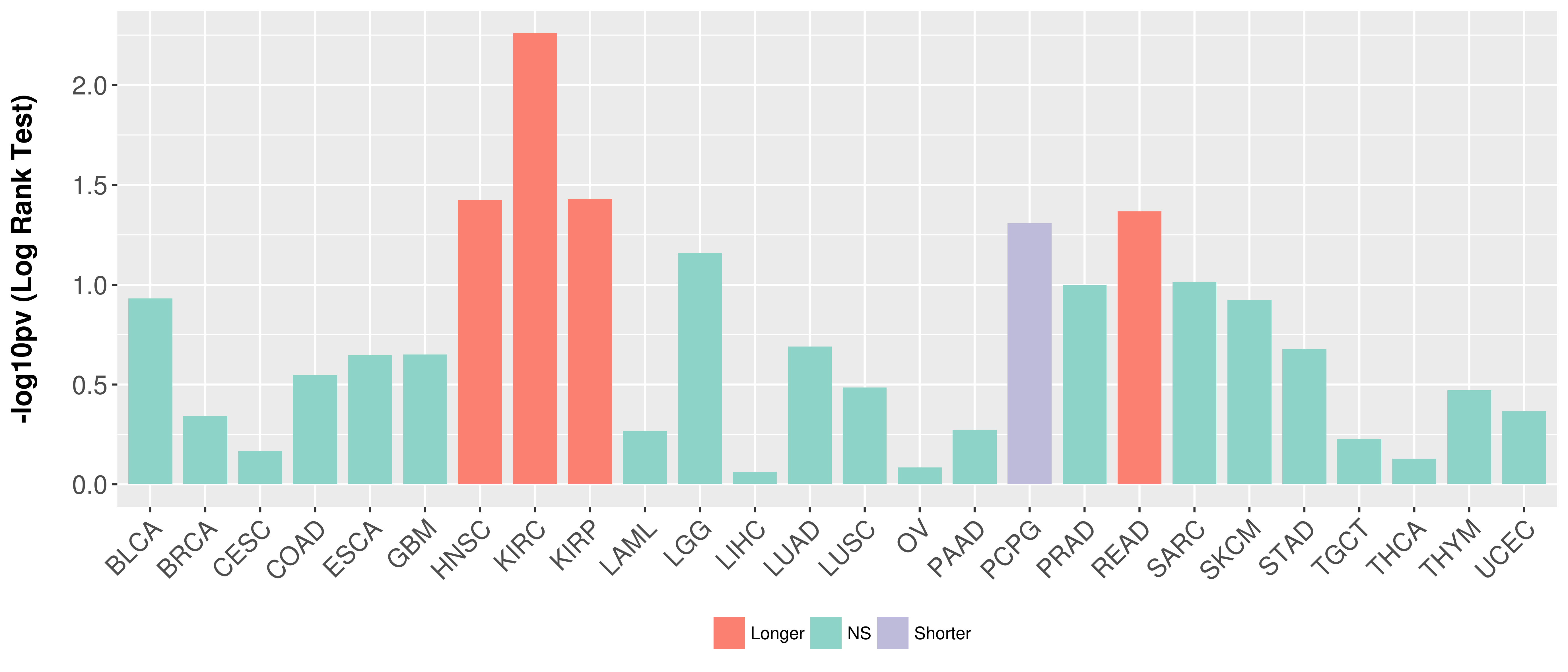
Overall survival analysis based on expression.
|
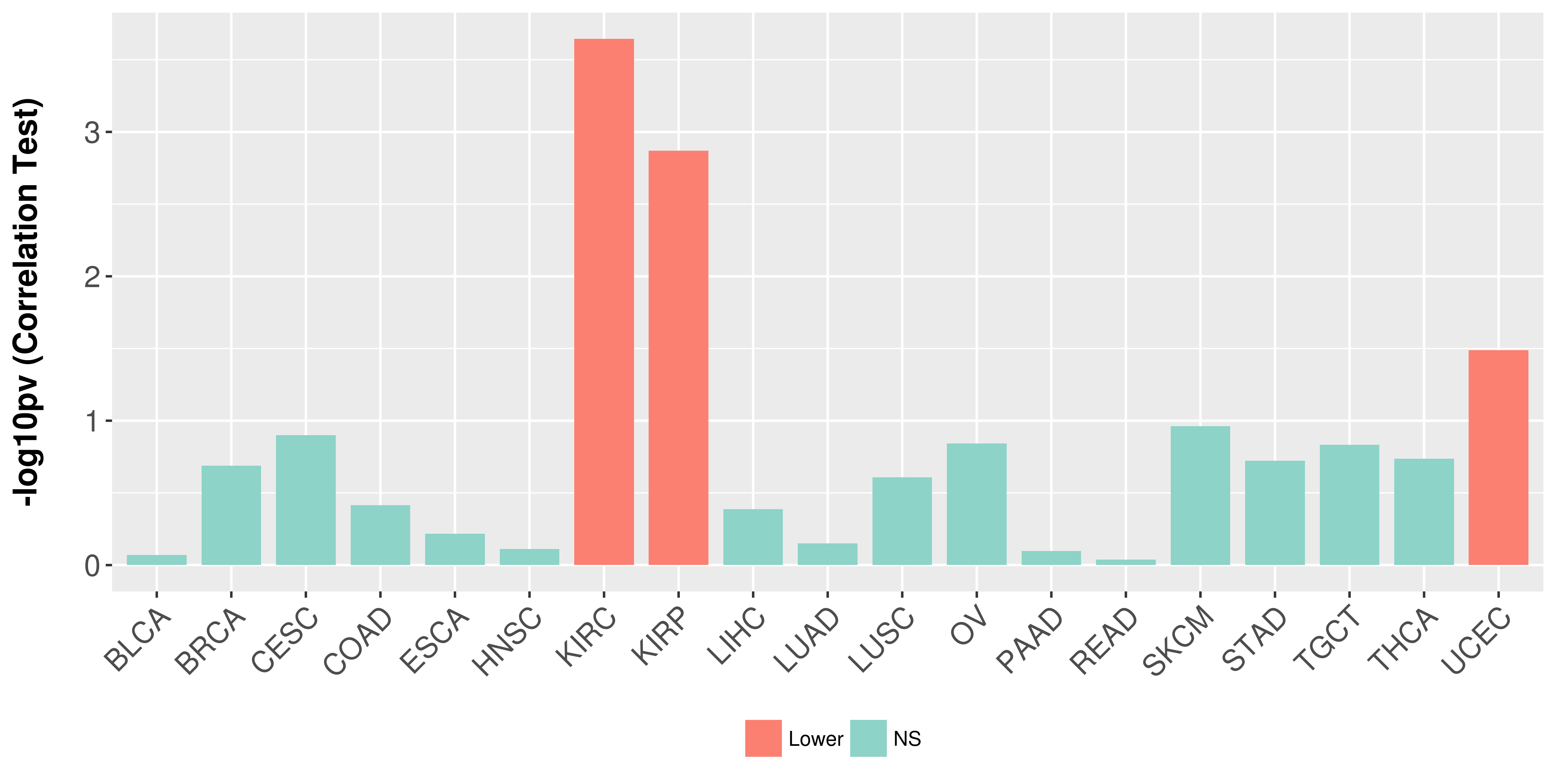
Association between expresson and stage.
|
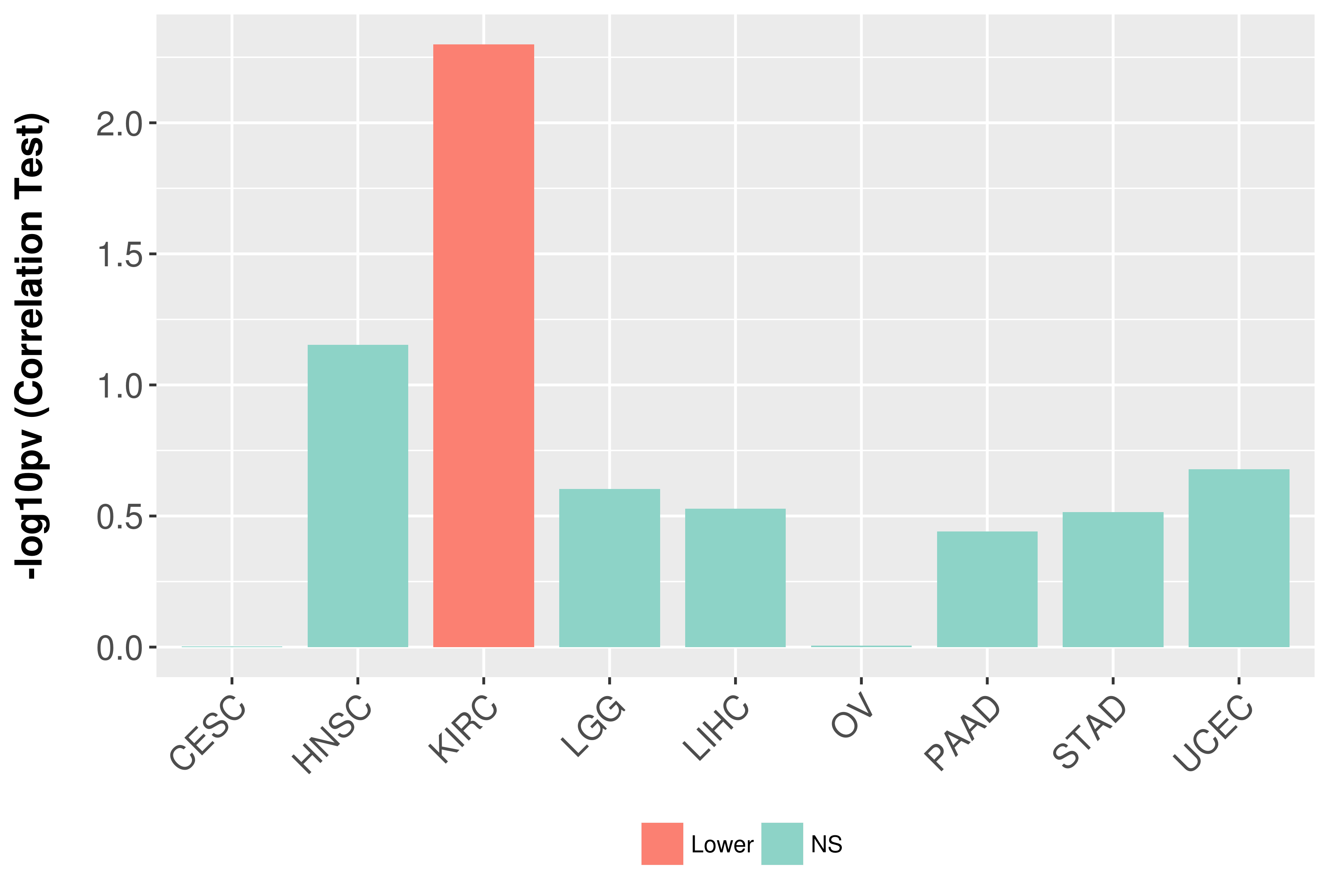
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SETD2. |
| Summary | |
|---|---|
| Symbol | SETD2 |
| Name | SET domain containing 2 |
| Aliases | HYPB; HIF-1; KIAA1732; FLJ23184; KMT3A; HBP231; HSPC069; SET2; p231HBP; huntingtin yeast partner B; huntingt ...... |
| Location | 3p21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SETD2. |