Browse SETMAR in pancancer
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05033 Pre-SET motif PF00856 SET domain PF01359 Transposase (partial DDE domain) |
||||||||||
| Function |
Protein derived from the fusion of a methylase with the transposase of an Hsmar1 transposon that plays a role in DNA double-strand break repair, stalled replication fork restart and DNA integration. DNA-binding protein, it is indirectly recruited to sites of DNA damage through protein-protein interactions. Has also kept a sequence-specific DNA-binding activity recognizing the 19-mer core of the 5'-terminal inverted repeats (TIRs) of the Hsmar1 element and displays a DNA nicking and end joining activity (PubMed:16332963, PubMed:16672366, PubMed:17877369, PubMed:17403897, PubMed:18263876, PubMed:22231448, PubMed:24573677, PubMed:20521842). In parallel, has a histone methyltransferase activity and methylates 'Lys-4' and 'Lys-36' of histone H3. Specifically mediates dimethylation of H3 'Lys-36' at sites of DNA double-strand break and may recruit proteins required for efficient DSB repair through non-homologous end-joining (PubMed:16332963, PubMed:21187428, PubMed:22231448). Also regulates replication fork processing, promoting replication fork restart and regulating DNA decatenation through stimulation of the topoisomerase activity of TOP2A (PubMed:18790802, PubMed:20457750). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000726 non-recombinational repair GO:0000729 DNA double-strand break processing GO:0000737 DNA catabolic process, endonucleolytic GO:0006260 DNA replication GO:0006261 DNA-dependent DNA replication GO:0006282 regulation of DNA repair GO:0006302 double-strand break repair GO:0006303 double-strand break repair via nonhomologous end joining GO:0006308 DNA catabolic process GO:0006479 protein methylation GO:0007050 cell cycle arrest GO:0007093 mitotic cell cycle checkpoint GO:0007346 regulation of mitotic cell cycle GO:0008213 protein alkylation GO:0010452 histone H3-K36 methylation GO:0010639 negative regulation of organelle organization GO:0010911 regulation of isomerase activity GO:0010912 positive regulation of isomerase activity GO:0010948 negative regulation of cell cycle process GO:0015074 DNA integration GO:0016570 histone modification GO:0016571 histone methylation GO:0018022 peptidyl-lysine methylation GO:0018027 peptidyl-lysine dimethylation GO:0018205 peptidyl-lysine modification GO:0019439 aromatic compound catabolic process GO:0031297 replication fork processing GO:0031570 DNA integrity checkpoint GO:0032259 methylation GO:0032781 positive regulation of ATPase activity GO:0033044 regulation of chromosome organization GO:0034655 nucleobase-containing compound catabolic process GO:0034968 histone lysine methylation GO:0043414 macromolecule methylation GO:0043462 regulation of ATPase activity GO:0044270 cellular nitrogen compound catabolic process GO:0044774 mitotic DNA integrity checkpoint GO:0045005 DNA-dependent DNA replication maintenance of fidelity GO:0045739 positive regulation of DNA repair GO:0045786 negative regulation of cell cycle GO:0045787 positive regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0046700 heterocycle catabolic process GO:0051052 regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051568 histone H3-K4 methylation GO:0071156 regulation of cell cycle arrest GO:0071157 negative regulation of cell cycle arrest GO:0090305 nucleic acid phosphodiester bond hydrolysis GO:0097676 histone H3-K36 dimethylation GO:1901361 organic cyclic compound catabolic process GO:2000371 regulation of DNA topoisomerase (ATP-hydrolyzing) activity GO:2000373 positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity GO:2000779 regulation of double-strand break repair GO:2000781 positive regulation of double-strand break repair GO:2001020 regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus GO:2001032 regulation of double-strand break repair via nonhomologous end joining GO:2001034 positive regulation of double-strand break repair via nonhomologous end joining GO:2001251 negative regulation of chromosome organization |
| Molecular Function |
GO:0000014 single-stranded DNA endodeoxyribonuclease activity GO:0003697 single-stranded DNA binding GO:0004518 nuclease activity GO:0004519 endonuclease activity GO:0004520 endodeoxyribonuclease activity GO:0004536 deoxyribonuclease activity GO:0008168 methyltransferase activity GO:0008170 N-methyltransferase activity GO:0008276 protein methyltransferase activity GO:0008757 S-adenosylmethionine-dependent methyltransferase activity GO:0016278 lysine N-methyltransferase activity GO:0016279 protein-lysine N-methyltransferase activity GO:0016741 transferase activity, transferring one-carbon groups GO:0018024 histone-lysine N-methyltransferase activity GO:0042054 histone methyltransferase activity GO:0042800 histone methyltransferase activity (H3-K4 specific) GO:0043566 structure-specific DNA binding GO:0044547 DNA topoisomerase binding GO:0046975 histone methyltransferase activity (H3-K36 specific) |
| Cellular Component |
GO:0000793 condensed chromosome GO:0035861 site of double-strand break |
| KEGG |
hsa00310 Lysine degradation |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
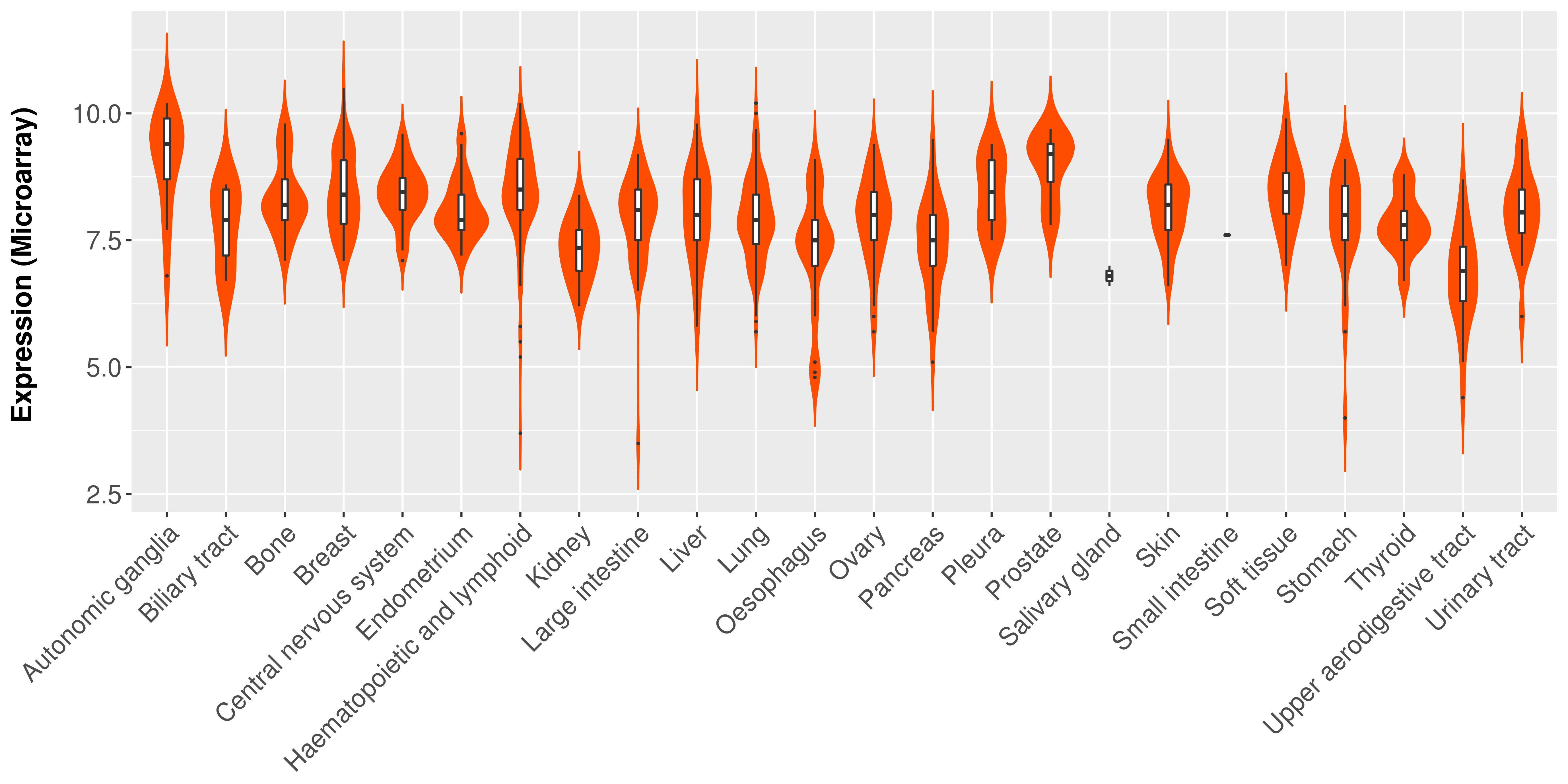
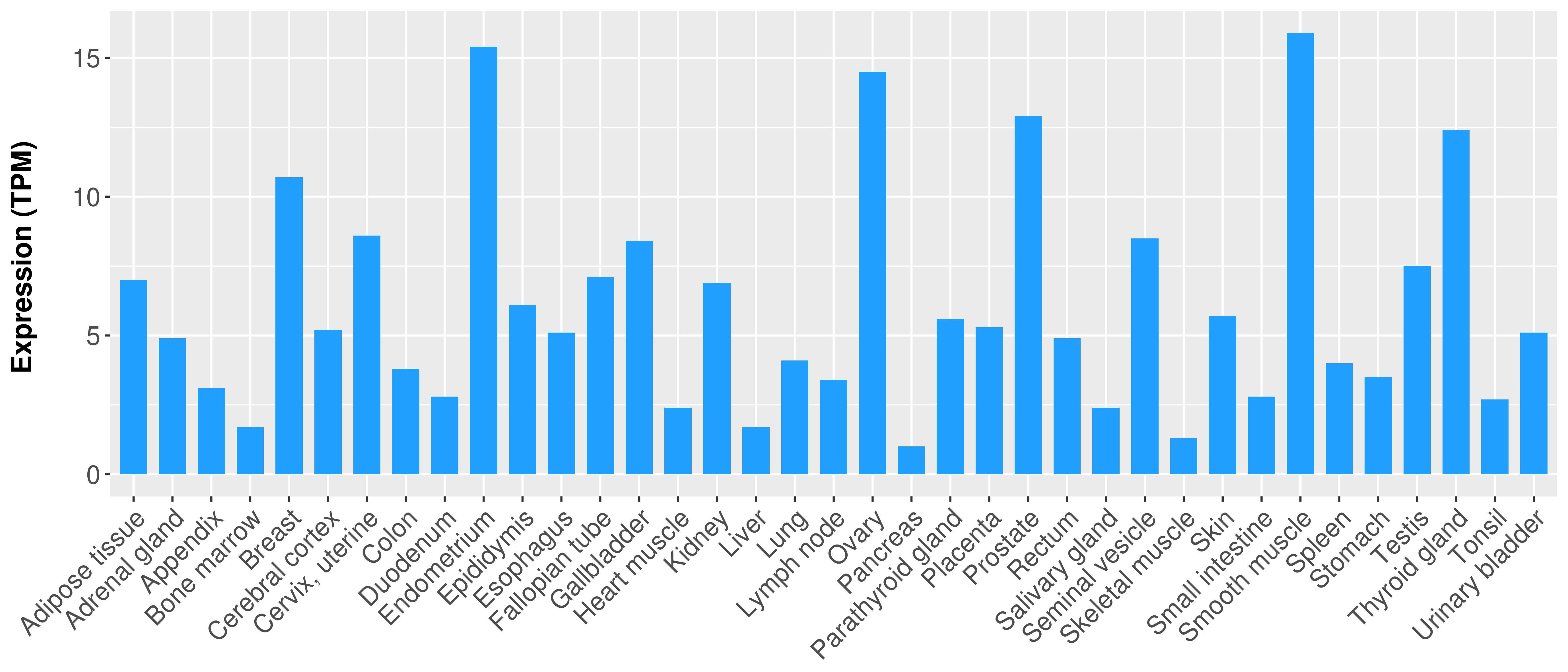
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
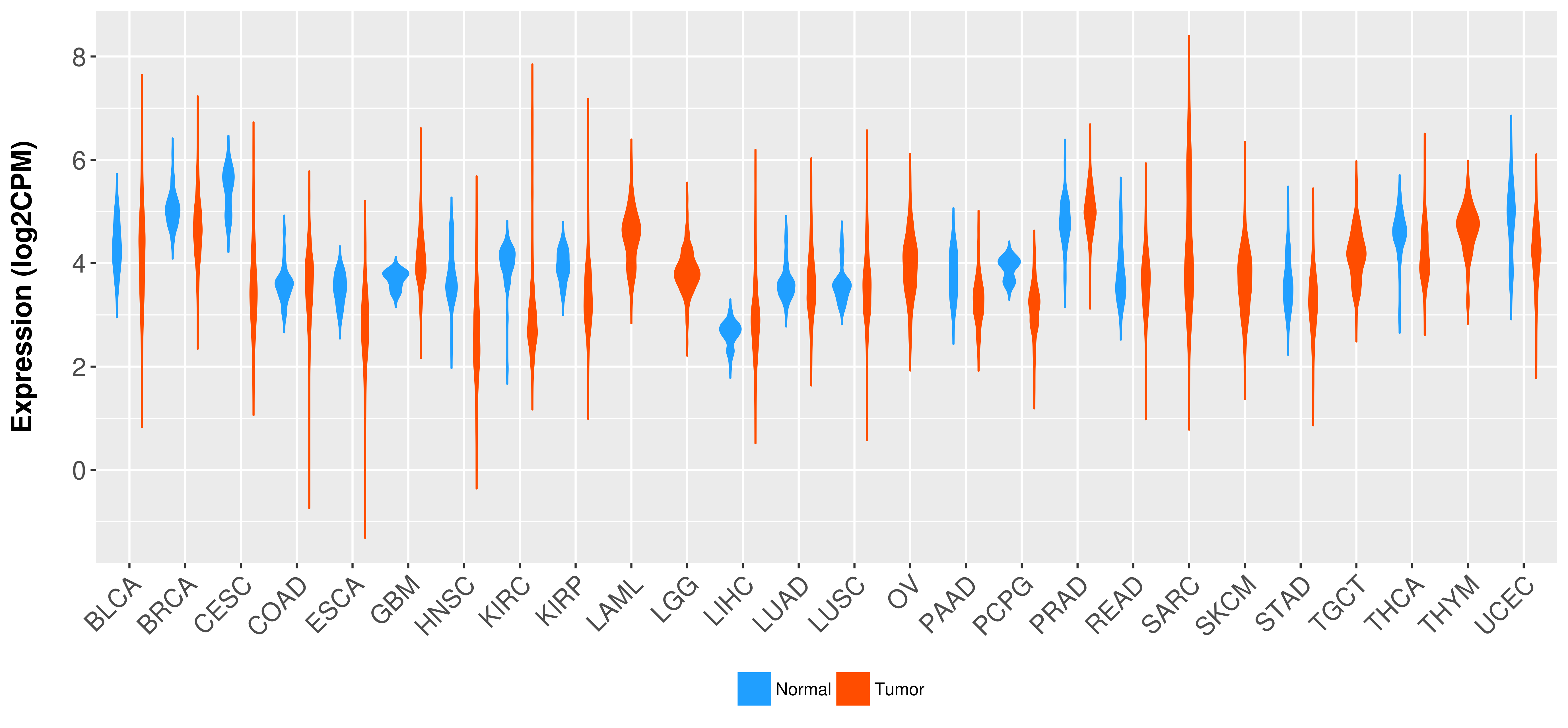
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
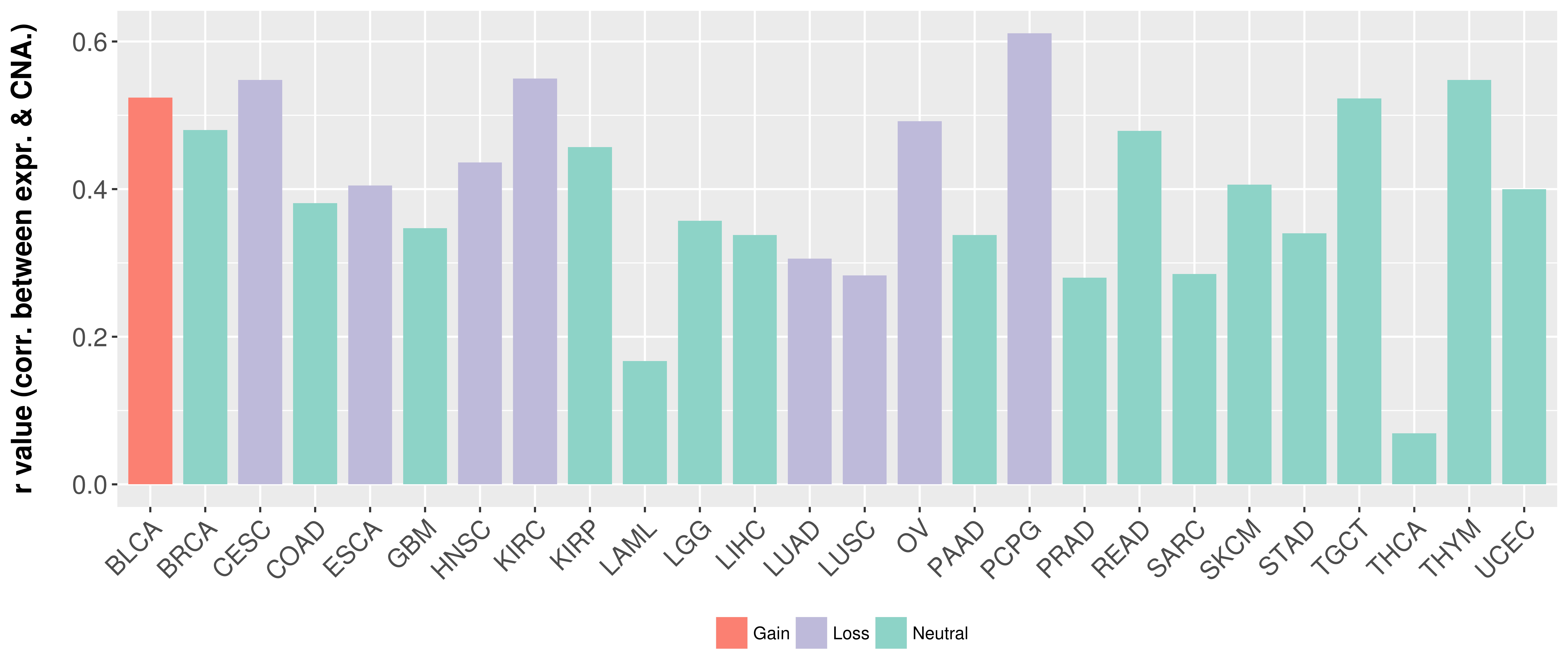
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
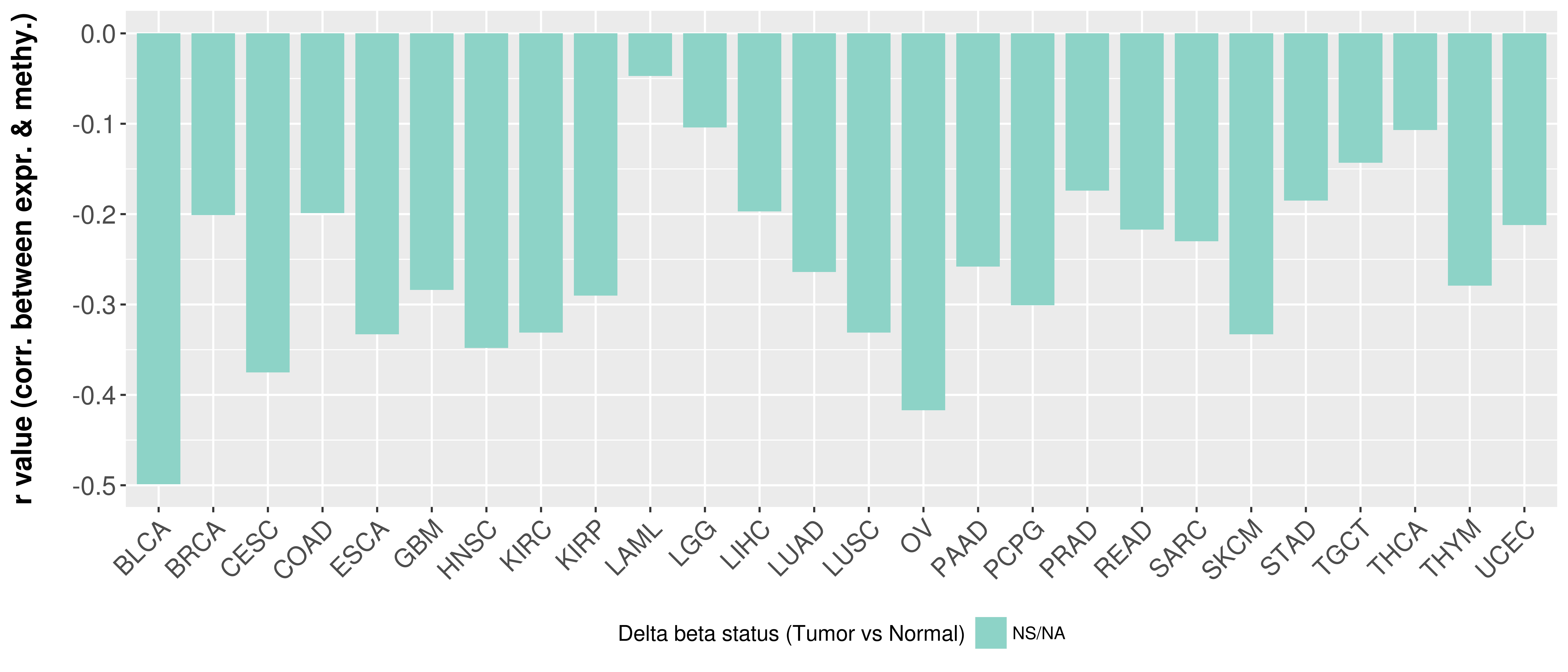
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
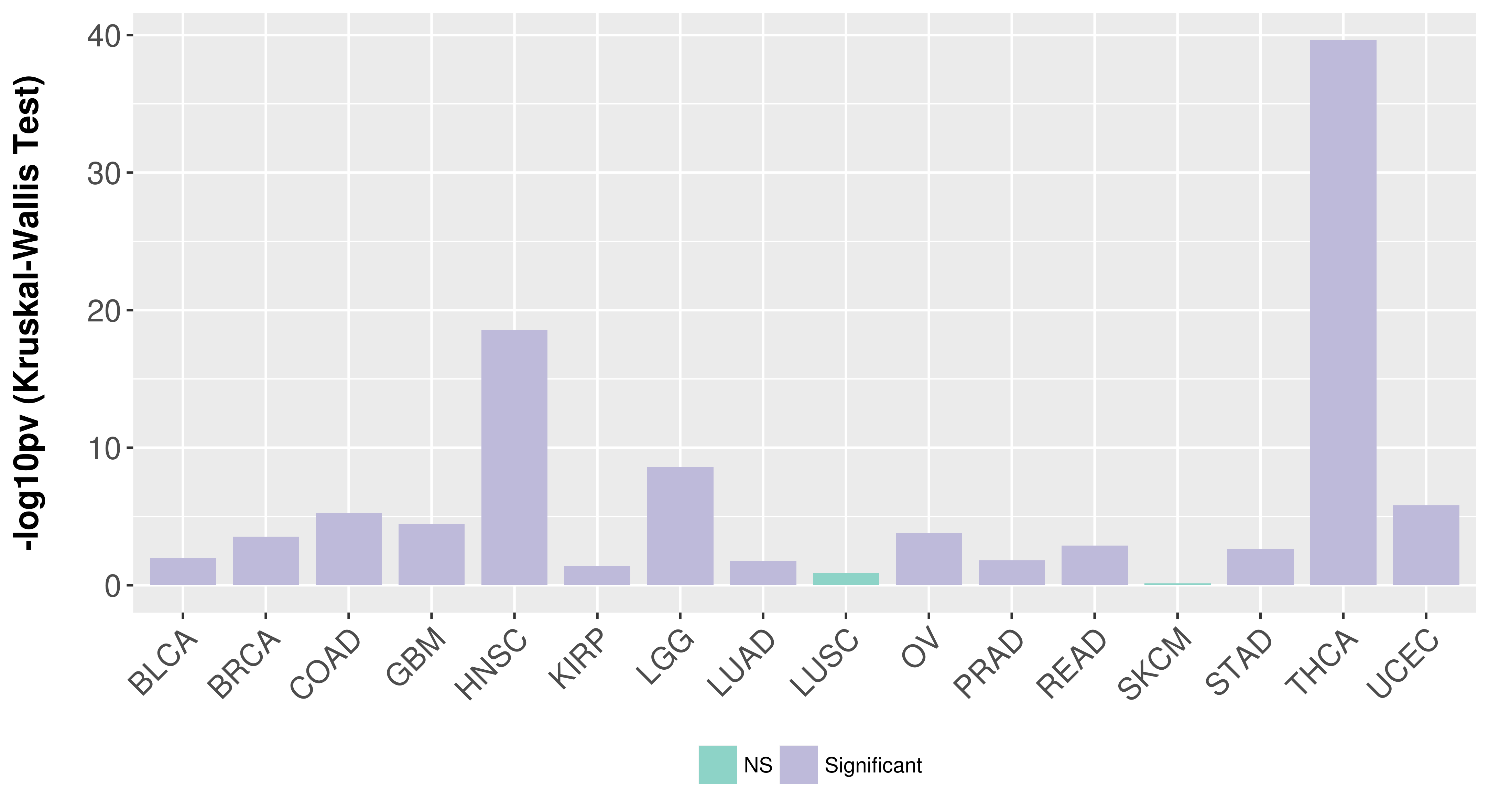
Association between expresson and subtype.
|
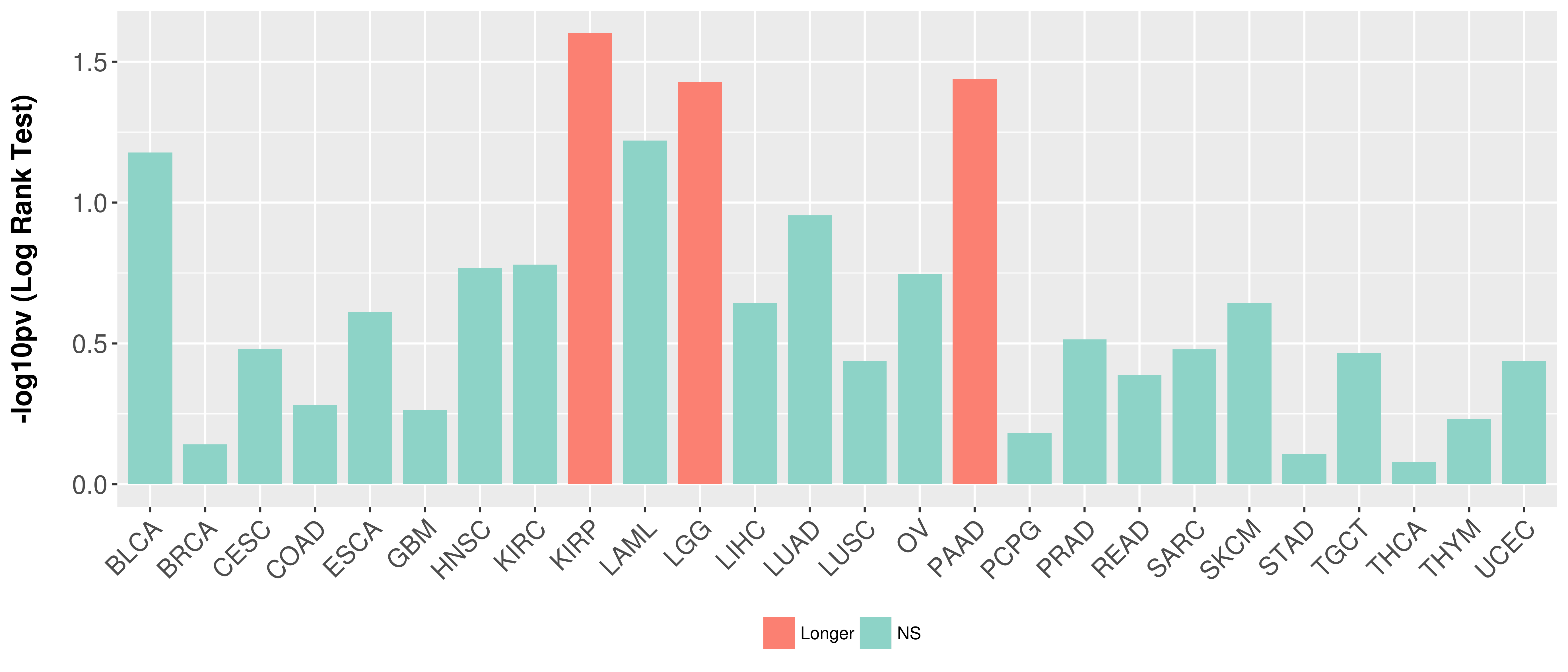
Overall survival analysis based on expression.
|
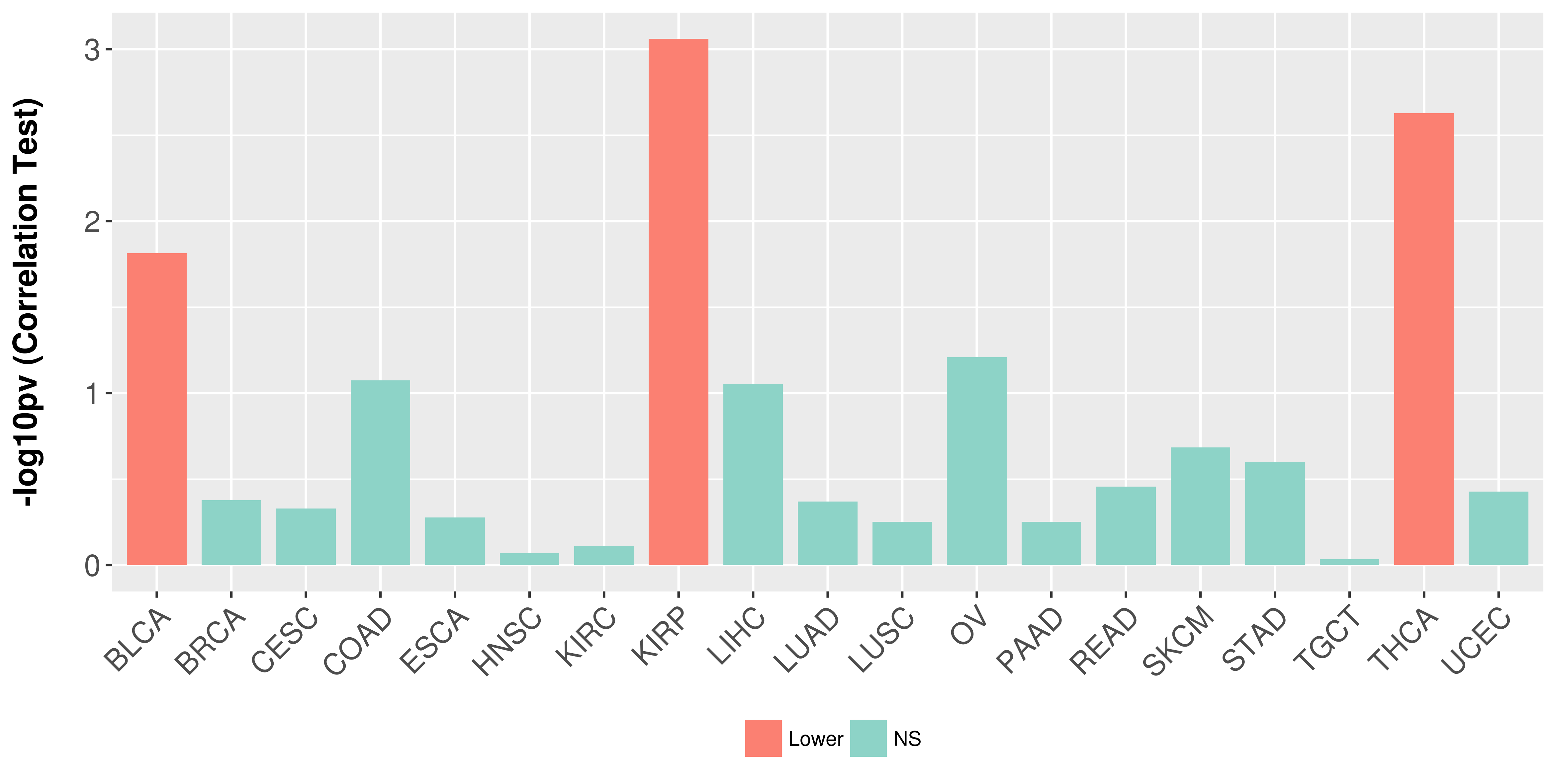
Association between expresson and stage.
|
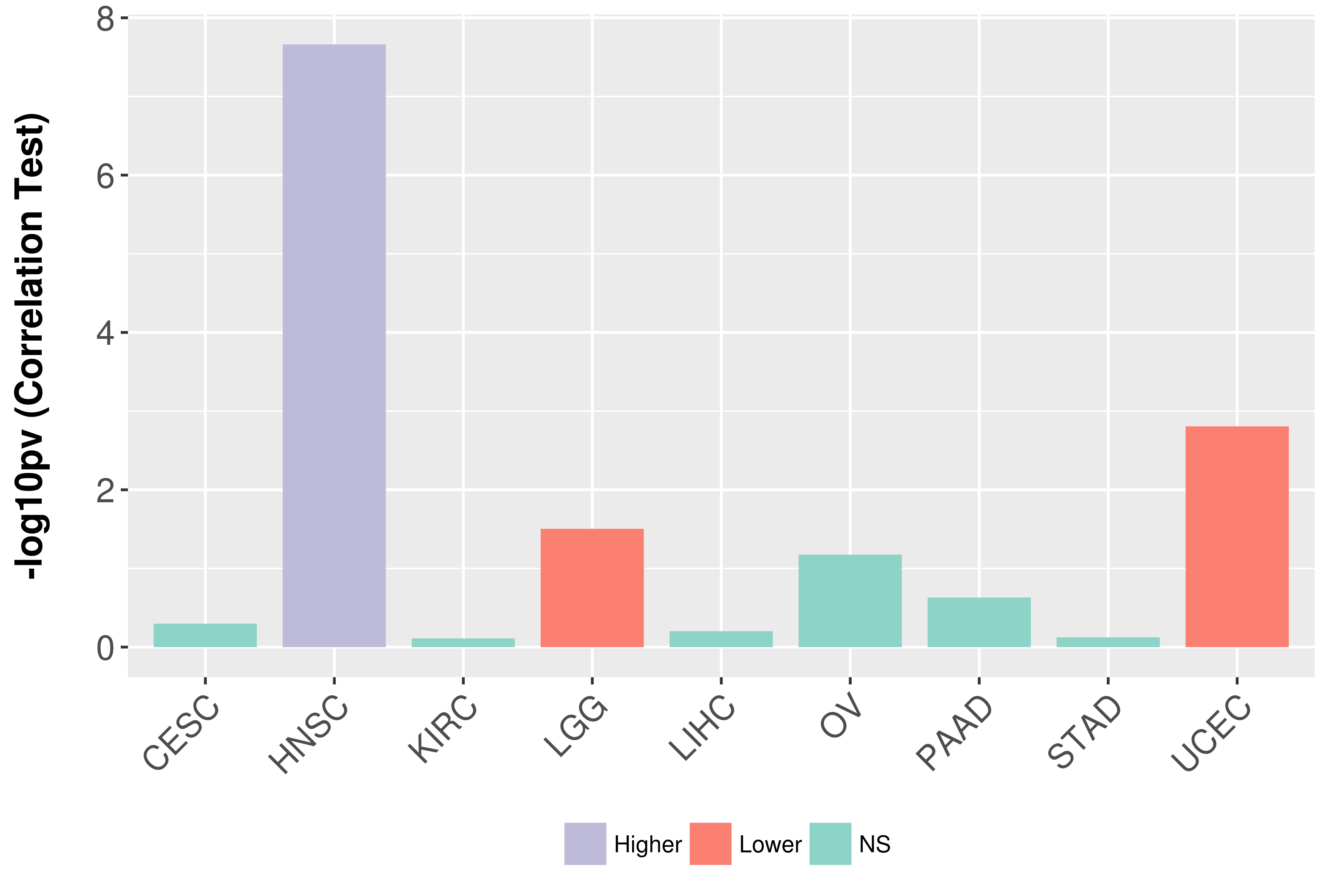
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SETMAR. |
| Summary | |
|---|---|
| Symbol | SETMAR |
| Name | SET domain and mariner transposase fusion gene |
| Aliases | metnase; HsMar1; SET domain and mariner transposase fusion gene-containing protein; SET domain and mariner t ...... |
| Location | 3p26.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SETMAR. |
| There is no record for SETMAR. |