Browse SIRT4 in pancancer
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02146 Sir2 family |
||||||||||
| Function |
Acts as NAD-dependent protein lipoamidase, ADP-ribosyl transferase and deacetylase. Catalyzes more efficiently removal of lipoyl- and biotinyl- than acetyl-lysine modifications. Inhibits the pyruvate dehydrogenase complex (PDH) activity via the enzymatic hydrolysis of the lipoamide cofactor from the E2 component, DLAT, in a phosphorylation-independent manner (PubMed:25525879). Catalyzes the transfer of ADP-ribosyl groups onto target proteins, including mitochondrial GLUD1, inhibiting GLUD1 enzyme activity. Acts as a negative regulator of mitochondrial glutamine metabolism by mediating mono ADP-ribosylation of GLUD1: expressed in response to DNA damage and negatively regulates anaplerosis by inhibiting GLUD1, leading to block metabolism of glutamine into tricarboxylic acid cycle and promoting cell cycle arrest (PubMed:16959573, PubMed:17715127). In response to mTORC1 signal, SIRT4 expression is repressed, promoting anaplerosis and cell proliferation. Acts as a tumor suppressor (PubMed:23562301, PubMed:23663782). Also acts as a NAD-dependent protein deacetylase: mediates deacetylation of 'Lys-471' of MLYCD, inhibiting its activity, thereby acting as a regulator of lipid homeostasis (By similarity). Controls fatty acid oxidation by inhibiting PPARA transcriptional activation. Impairs SIRT1:PPARA interaction probably through the regulation of NAD(+) levels (PubMed:24043310). Down-regulates insulin secretion. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000820 regulation of glutamine family amino acid metabolic process GO:0001666 response to hypoxia GO:0002790 peptide secretion GO:0002791 regulation of peptide secretion GO:0002792 negative regulation of peptide secretion GO:0006342 chromatin silencing GO:0006471 protein ADP-ribosylation GO:0006476 protein deacetylation GO:0006486 protein glycosylation GO:0006520 cellular amino acid metabolic process GO:0006541 glutamine metabolic process GO:0006626 protein targeting to mitochondrion GO:0006627 protein processing involved in protein targeting to mitochondrion GO:0006631 fatty acid metabolic process GO:0006839 mitochondrial transport GO:0009064 glutamine family amino acid metabolic process GO:0009100 glycoprotein metabolic process GO:0009101 glycoprotein biosynthetic process GO:0009306 protein secretion GO:0009914 hormone transport GO:0010565 regulation of cellular ketone metabolic process GO:0010639 negative regulation of organelle organization GO:0010656 negative regulation of muscle cell apoptotic process GO:0010657 muscle cell apoptotic process GO:0010658 striated muscle cell apoptotic process GO:0010659 cardiac muscle cell apoptotic process GO:0010660 regulation of muscle cell apoptotic process GO:0010662 regulation of striated muscle cell apoptotic process GO:0010664 negative regulation of striated muscle cell apoptotic process GO:0010665 regulation of cardiac muscle cell apoptotic process GO:0010667 negative regulation of cardiac muscle cell apoptotic process GO:0010817 regulation of hormone levels GO:0010821 regulation of mitochondrion organization GO:0010823 negative regulation of mitochondrion organization GO:0010955 negative regulation of protein processing GO:0015833 peptide transport GO:0016458 gene silencing GO:0016485 protein processing GO:0018205 peptidyl-lysine modification GO:0019216 regulation of lipid metabolic process GO:0019217 regulation of fatty acid metabolic process GO:0019395 fatty acid oxidation GO:0023061 signal release GO:0030072 peptide hormone secretion GO:0030073 insulin secretion GO:0030258 lipid modification GO:0032386 regulation of intracellular transport GO:0032387 negative regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0034440 lipid oxidation GO:0034982 mitochondrial protein processing GO:0034983 peptidyl-lysine deacetylation GO:0035601 protein deacylation GO:0036293 response to decreased oxygen levels GO:0036294 cellular response to decreased oxygen levels GO:0040029 regulation of gene expression, epigenetic GO:0042180 cellular ketone metabolic process GO:0042886 amide transport GO:0043413 macromolecule glycosylation GO:0045814 negative regulation of gene expression, epigenetic GO:0045833 negative regulation of lipid metabolic process GO:0045834 positive regulation of lipid metabolic process GO:0045861 negative regulation of proteolysis GO:0045922 negative regulation of fatty acid metabolic process GO:0046320 regulation of fatty acid oxidation GO:0046322 negative regulation of fatty acid oxidation GO:0046676 negative regulation of insulin secretion GO:0046879 hormone secretion GO:0046883 regulation of hormone secretion GO:0046888 negative regulation of hormone secretion GO:0046889 positive regulation of lipid biosynthetic process GO:0046890 regulation of lipid biosynthetic process GO:0050708 regulation of protein secretion GO:0050709 negative regulation of protein secretion GO:0050796 regulation of insulin secretion GO:0051048 negative regulation of secretion GO:0051051 negative regulation of transport GO:0051224 negative regulation of protein transport GO:0051341 regulation of oxidoreductase activity GO:0051604 protein maturation GO:0070085 glycosylation GO:0070482 response to oxygen levels GO:0070585 protein localization to mitochondrion GO:0070613 regulation of protein processing GO:0071453 cellular response to oxygen levels GO:0071456 cellular response to hypoxia GO:0072350 tricarboxylic acid metabolic process GO:0072655 establishment of protein localization to mitochondrion GO:0090087 regulation of peptide transport GO:0090276 regulation of peptide hormone secretion GO:0090278 negative regulation of peptide hormone secretion GO:0090317 negative regulation of intracellular protein transport GO:0098732 macromolecule deacylation GO:1901605 alpha-amino acid metabolic process GO:1903214 regulation of protein targeting to mitochondrion GO:1903215 negative regulation of protein targeting to mitochondrion GO:1903216 regulation of protein processing involved in protein targeting to mitochondrion GO:1903217 negative regulation of protein processing involved in protein targeting to mitochondrion GO:1903317 regulation of protein maturation GO:1903318 negative regulation of protein maturation GO:1903531 negative regulation of secretion by cell GO:1903533 regulation of protein targeting GO:1903747 regulation of establishment of protein localization to mitochondrion GO:1903748 negative regulation of establishment of protein localization to mitochondrion GO:1903828 negative regulation of cellular protein localization GO:1904182 regulation of pyruvate dehydrogenase activity GO:1904950 negative regulation of establishment of protein localization |
| Molecular Function |
GO:0003950 NAD+ ADP-ribosyltransferase activity GO:0016757 transferase activity, transferring glycosyl groups GO:0016763 transferase activity, transferring pentosyl groups GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0047708 biotinidase activity GO:0048037 cofactor binding GO:0050662 coenzyme binding GO:0051287 NAD binding GO:0061690 lipoamidase activity GO:0070403 NAD+ binding |
| Cellular Component |
GO:0005743 mitochondrial inner membrane GO:0005759 mitochondrial matrix |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SIRT4. |
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
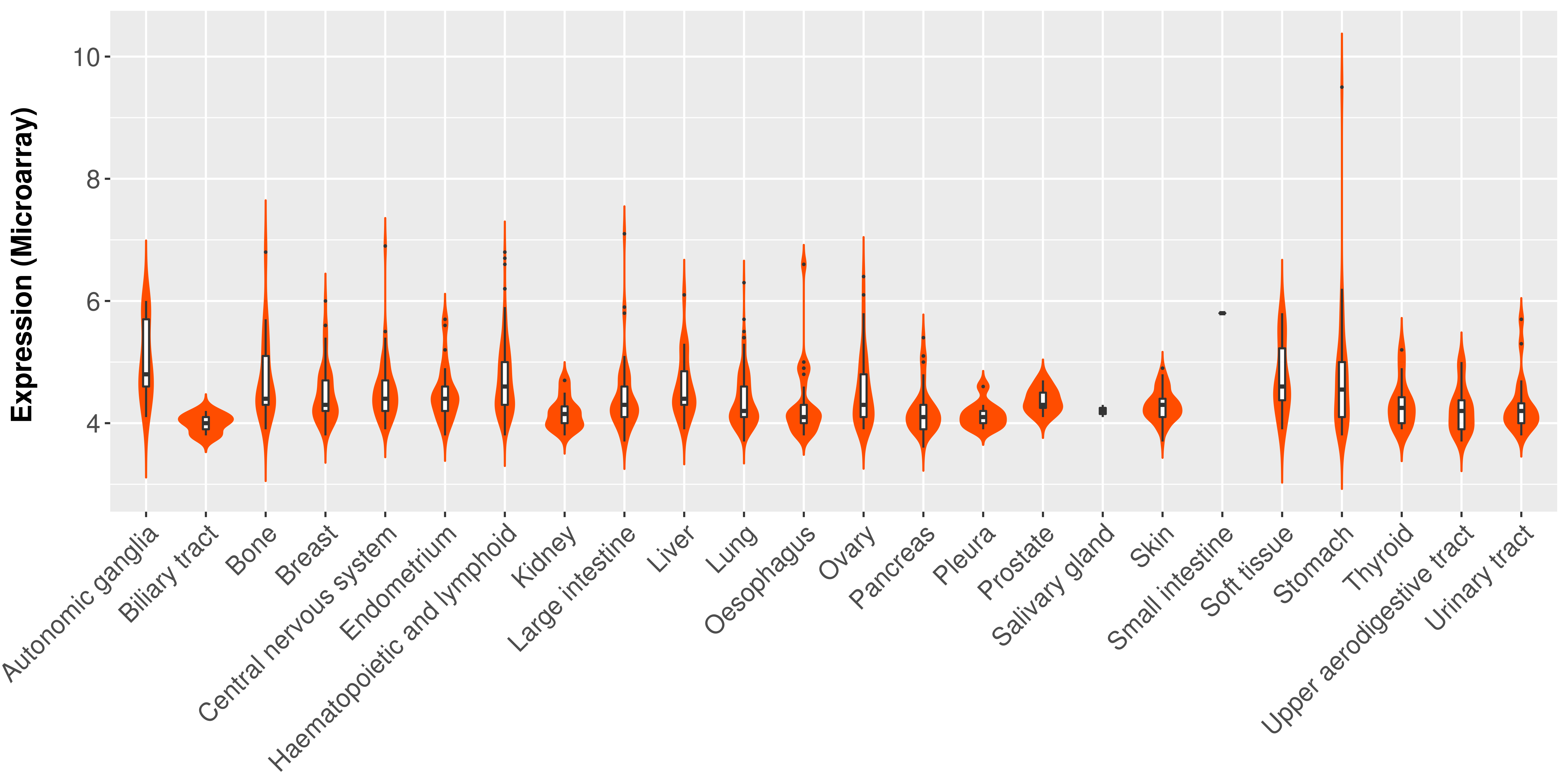
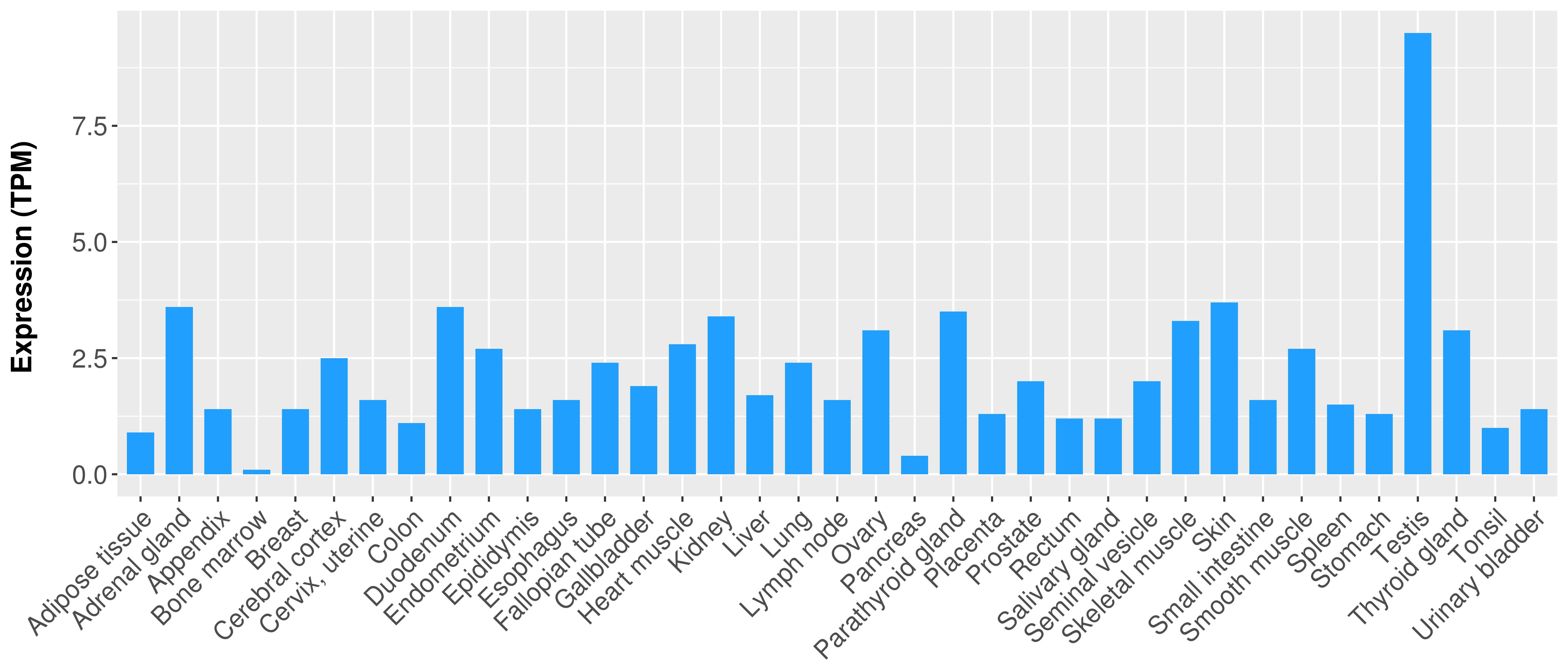
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
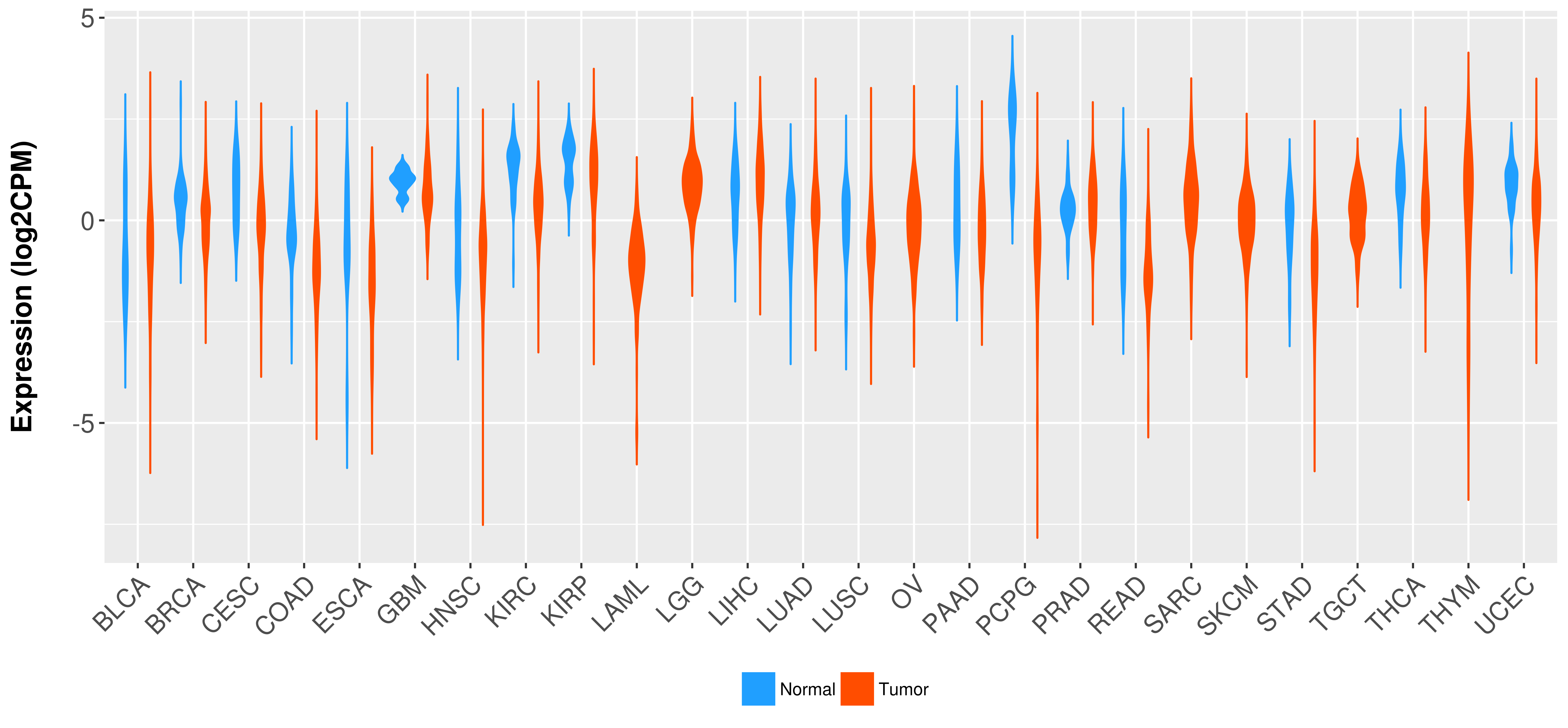
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
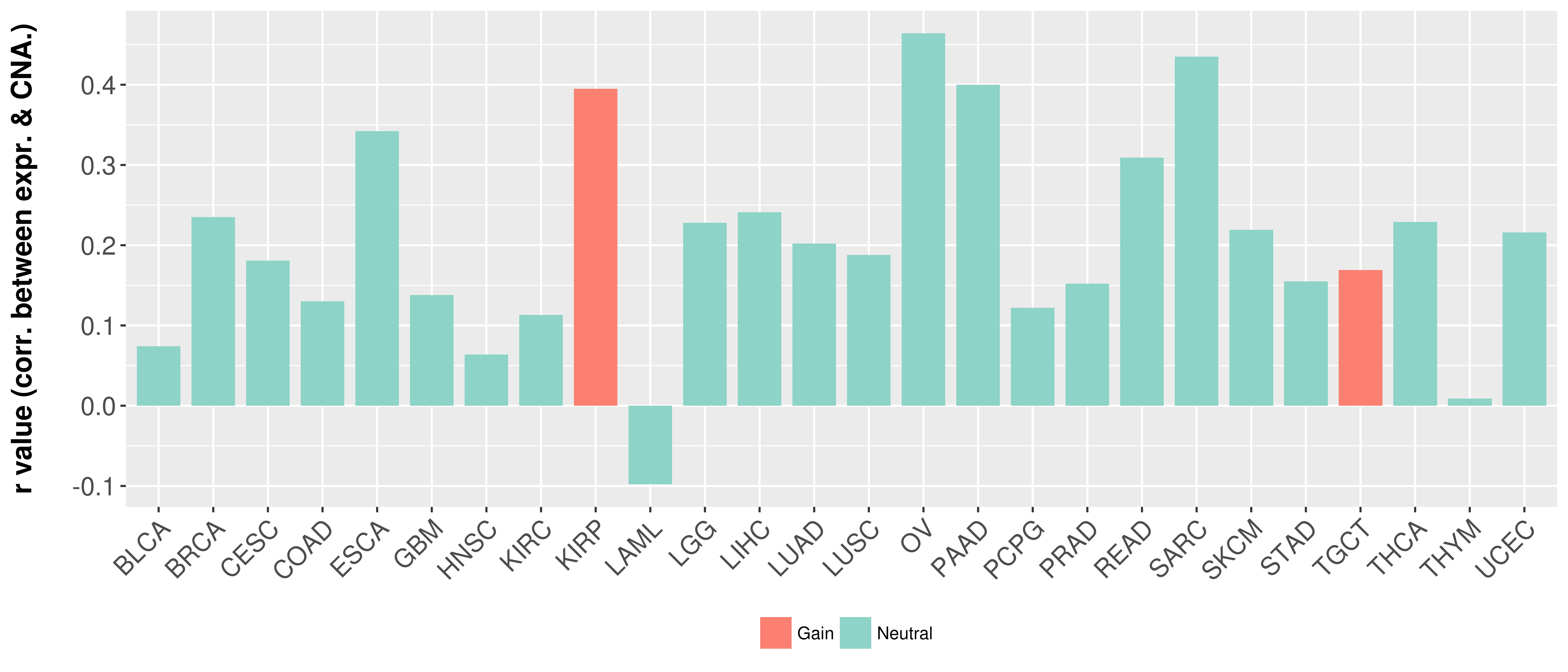
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |

Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
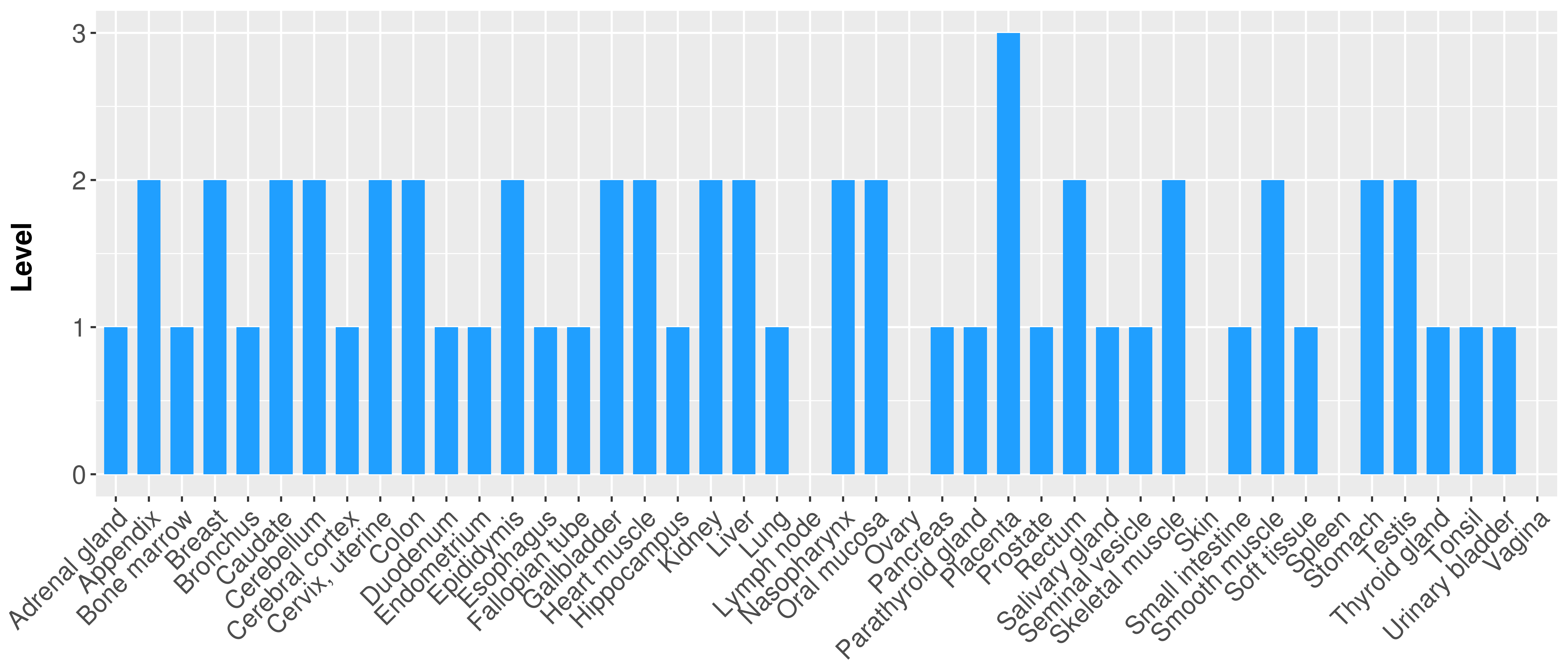
|
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
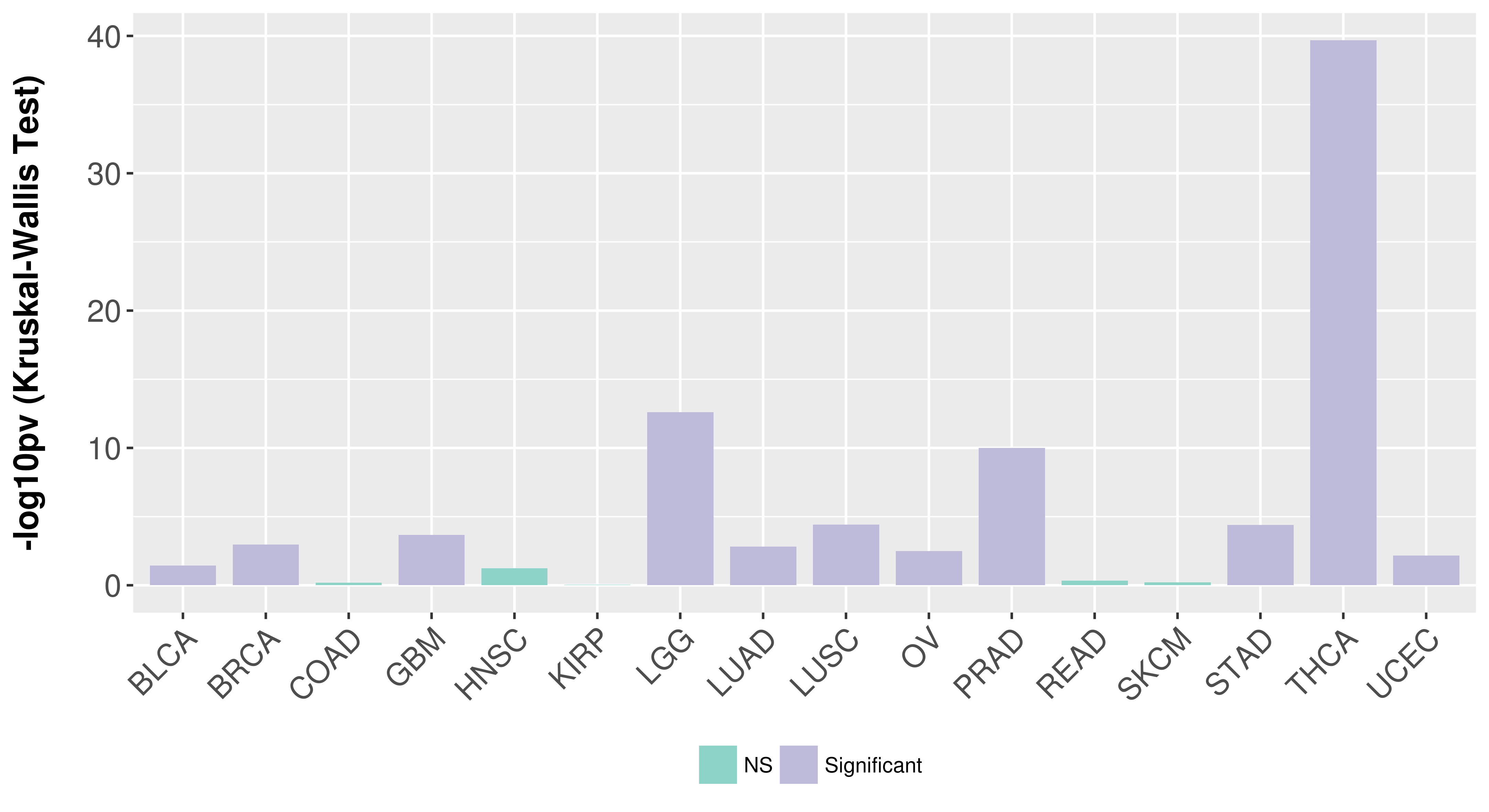
Association between expresson and subtype.
|
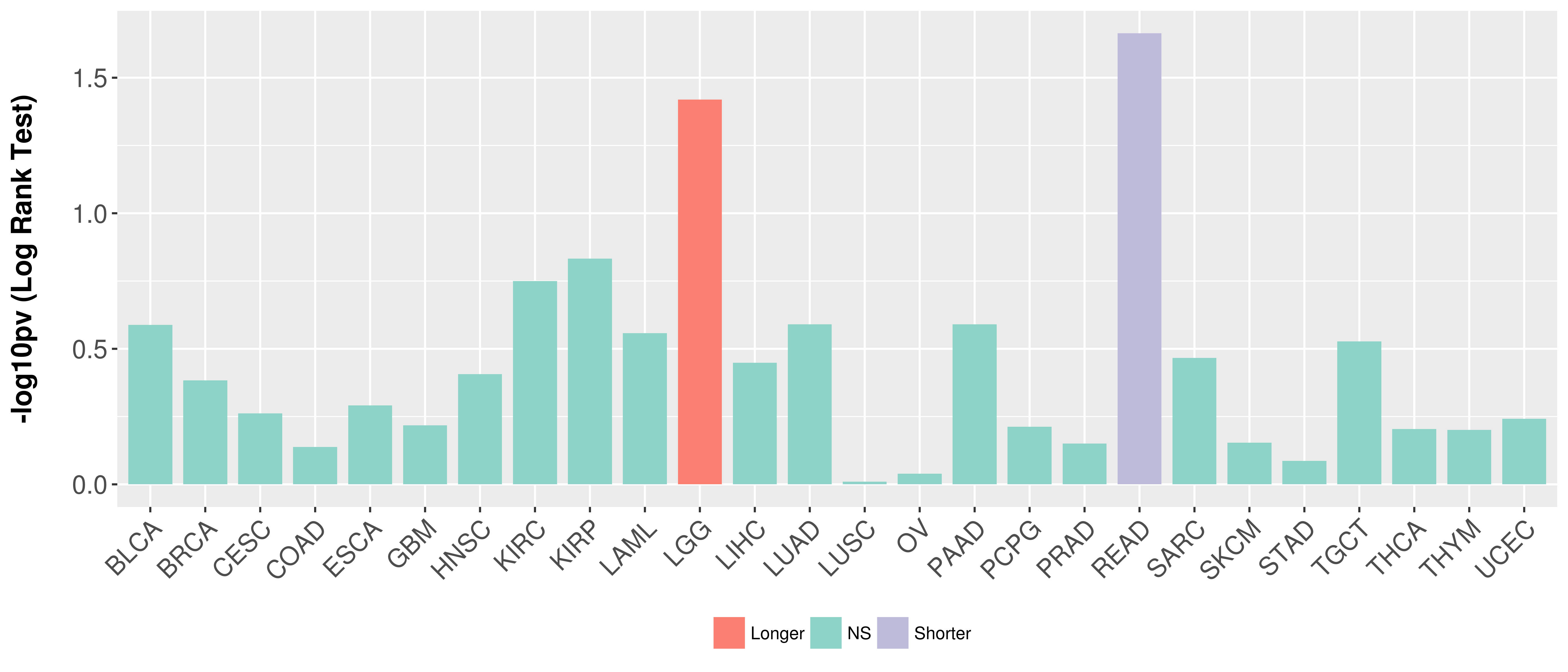
Overall survival analysis based on expression.
|
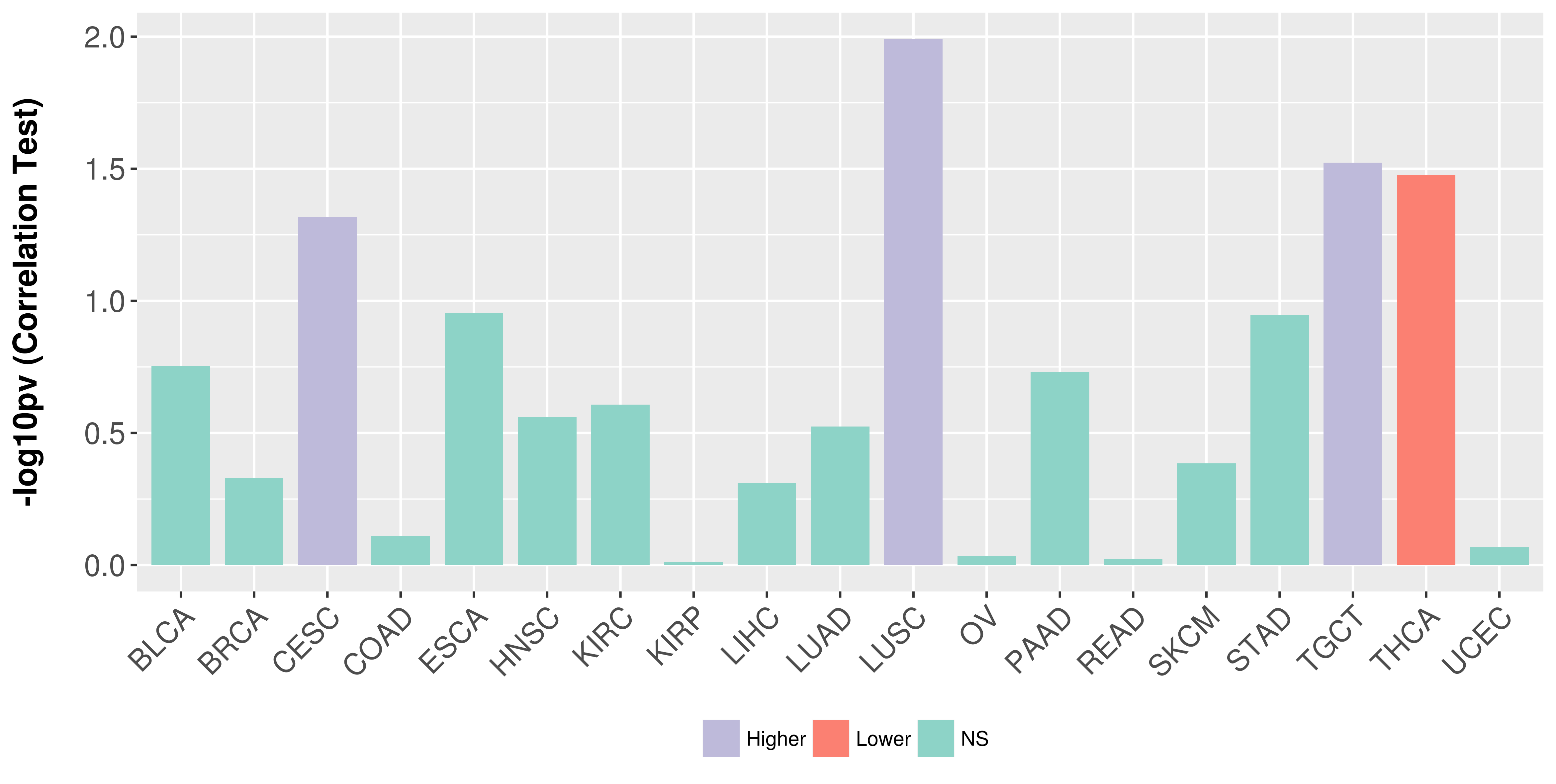
Association between expresson and stage.
|
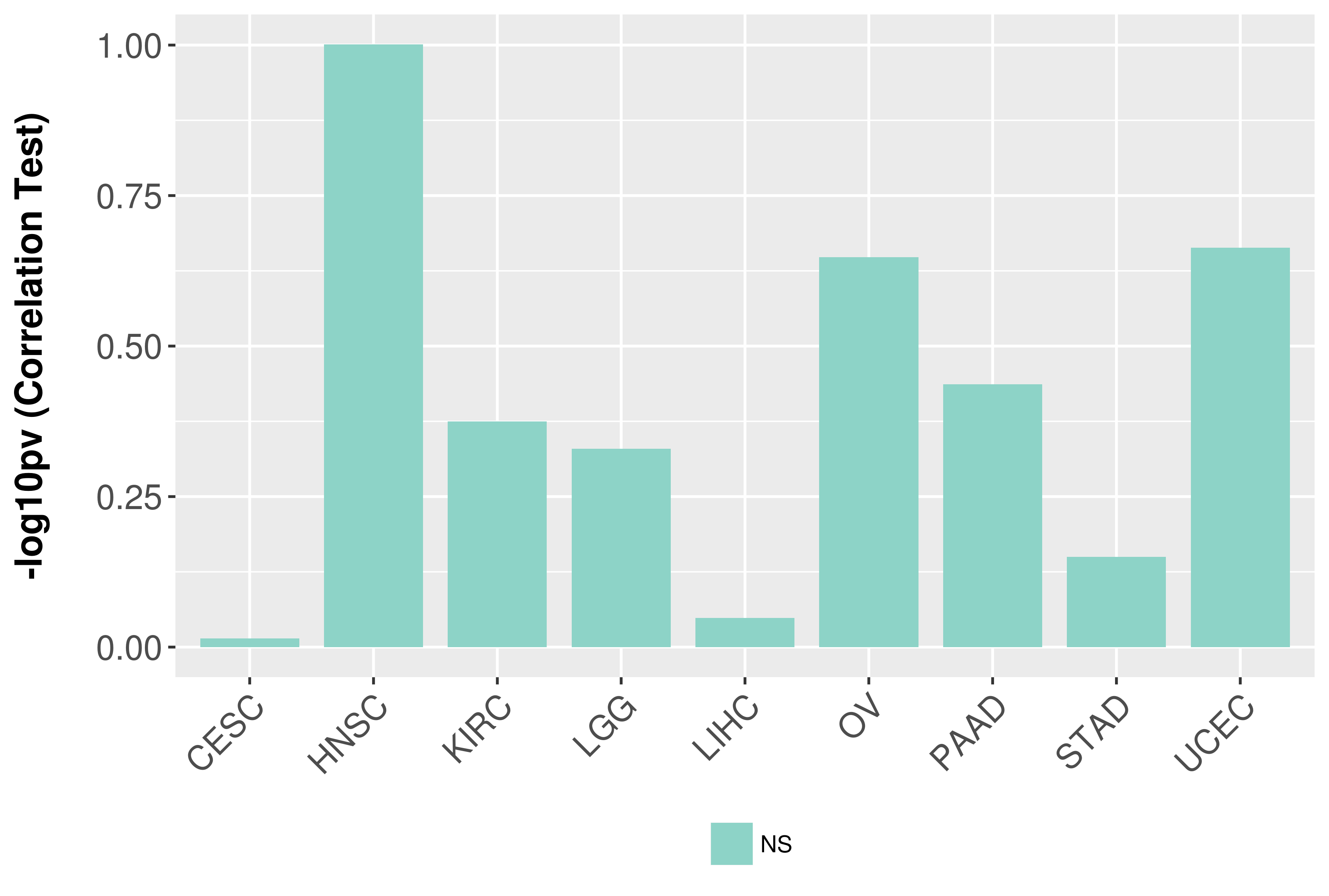
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SIRT4. |
| Summary | |
|---|---|
| Symbol | SIRT4 |
| Name | sirtuin 4 |
| Aliases | SIR2L4; sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 4; sirtuin (silent mat ...... |
| Location | 12q24.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|