Browse SIRT6 in pancancer
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02146 Sir2 family |
||||||||||
| Function |
NAD-dependent protein deacetylase. Has deacetylase activity towards histone H3K9Ac and H3K56Ac. Modulates acetylation of histone H3 in telomeric chromatin during the S-phase of the cell cycle. Deacetylates histone H3K9Ac at NF-kappa-B target promoters and may down-regulate the expression of a subset of NF-kappa-B target genes. Acts as a corepressor of the transcription factor HIF1A to control the expression of multiple glycolytic genes to regulate glucose homeostasis. Required for genomic stability. Regulates the production of TNF protein. Has a role in the regulation of life span (By similarity). Deacetylation of nucleosomes interferes with RELA binding to target DNA. May be required for the association of WRN with telomeres during S-phase and for normal telomere maintenance. Required for genomic stability. Required for normal IGF1 serum levels and normal glucose homeostasis. Modulates cellular senescence and apoptosis. On DNA damage, promotes DNA end resection via deacetylation of RBBP8. Has very weak deacetylase activity and can bind NAD(+) in the absence of acetylated substrate. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000018 regulation of DNA recombination GO:0000724 double-strand break repair via homologous recombination GO:0000725 recombinational repair GO:0003007 heart morphogenesis GO:0003241 growth involved in heart morphogenesis GO:0003245 cardiac muscle tissue growth involved in heart morphogenesis GO:0003247 post-embryonic cardiac muscle cell growth involved in heart morphogenesis GO:0006090 pyruvate metabolic process GO:0006091 generation of precursor metabolites and energy GO:0006096 glycolytic process GO:0006109 regulation of carbohydrate metabolic process GO:0006110 regulation of glycolytic process GO:0006140 regulation of nucleotide metabolic process GO:0006165 nucleoside diphosphate phosphorylation GO:0006282 regulation of DNA repair GO:0006284 base-excision repair GO:0006302 double-strand break repair GO:0006310 DNA recombination GO:0006471 protein ADP-ribosylation GO:0006476 protein deacetylation GO:0006486 protein glycosylation GO:0006732 coenzyme metabolic process GO:0006733 oxidoreduction coenzyme metabolic process GO:0006757 ATP generation from ADP GO:0007507 heart development GO:0007517 muscle organ development GO:0008643 carbohydrate transport GO:0008645 hexose transport GO:0009100 glycoprotein metabolic process GO:0009101 glycoprotein biosynthetic process GO:0009116 nucleoside metabolic process GO:0009118 regulation of nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009123 nucleoside monophosphate metabolic process GO:0009126 purine nucleoside monophosphate metabolic process GO:0009132 nucleoside diphosphate metabolic process GO:0009135 purine nucleoside diphosphate metabolic process GO:0009141 nucleoside triphosphate metabolic process GO:0009144 purine nucleoside triphosphate metabolic process GO:0009150 purine ribonucleotide metabolic process GO:0009161 ribonucleoside monophosphate metabolic process GO:0009167 purine ribonucleoside monophosphate metabolic process GO:0009179 purine ribonucleoside diphosphate metabolic process GO:0009185 ribonucleoside diphosphate metabolic process GO:0009199 ribonucleoside triphosphate metabolic process GO:0009205 purine ribonucleoside triphosphate metabolic process GO:0009894 regulation of catabolic process GO:0009895 negative regulation of catabolic process GO:0009991 response to extracellular stimulus GO:0010569 regulation of double-strand break repair via homologous recombination GO:0010675 regulation of cellular carbohydrate metabolic process GO:0010677 negative regulation of cellular carbohydrate metabolic process GO:0010827 regulation of glucose transport GO:0010829 negative regulation of glucose transport GO:0014706 striated muscle tissue development GO:0015749 monosaccharide transport GO:0015758 glucose transport GO:0016049 cell growth GO:0016052 carbohydrate catabolic process GO:0016570 histone modification GO:0016575 histone deacetylation GO:0019362 pyridine nucleotide metabolic process GO:0031647 regulation of protein stability GO:0031648 protein destabilization GO:0031667 response to nutrient levels GO:0033500 carbohydrate homeostasis GO:0035265 organ growth GO:0035601 protein deacylation GO:0042278 purine nucleoside metabolic process GO:0042326 negative regulation of phosphorylation GO:0042593 glucose homeostasis GO:0043413 macromolecule glycosylation GO:0043467 regulation of generation of precursor metabolites and energy GO:0043470 regulation of carbohydrate catabolic process GO:0044262 cellular carbohydrate metabolic process GO:0044723 single-organism carbohydrate metabolic process GO:0044724 single-organism carbohydrate catabolic process GO:0045820 negative regulation of glycolytic process GO:0045912 negative regulation of carbohydrate metabolic process GO:0045978 negative regulation of nucleoside metabolic process GO:0045980 negative regulation of nucleotide metabolic process GO:0046031 ADP metabolic process GO:0046034 ATP metabolic process GO:0046128 purine ribonucleoside metabolic process GO:0046323 glucose import GO:0046324 regulation of glucose import GO:0046325 negative regulation of glucose import GO:0046496 nicotinamide nucleotide metabolic process GO:0046939 nucleotide phosphorylation GO:0048144 fibroblast proliferation GO:0048145 regulation of fibroblast proliferation GO:0048146 positive regulation of fibroblast proliferation GO:0048588 developmental cell growth GO:0048644 muscle organ morphogenesis GO:0048738 cardiac muscle tissue development GO:0051051 negative regulation of transport GO:0051052 regulation of DNA metabolic process GO:0051186 cofactor metabolic process GO:0051193 regulation of cofactor metabolic process GO:0051195 negative regulation of cofactor metabolic process GO:0051196 regulation of coenzyme metabolic process GO:0051198 negative regulation of coenzyme metabolic process GO:0055008 cardiac muscle tissue morphogenesis GO:0055017 cardiac muscle tissue growth GO:0060415 muscle tissue morphogenesis GO:0060419 heart growth GO:0060537 muscle tissue development GO:0060560 developmental growth involved in morphogenesis GO:0061647 histone H3-K9 modification GO:0070085 glycosylation GO:0070932 histone H3 deacetylation GO:0072089 stem cell proliferation GO:0072091 regulation of stem cell proliferation GO:0072524 pyridine-containing compound metabolic process GO:0098732 macromolecule deacylation GO:1900542 regulation of purine nucleotide metabolic process GO:1900543 negative regulation of purine nucleotide metabolic process GO:1901657 glycosyl compound metabolic process GO:1903578 regulation of ATP metabolic process GO:1903579 negative regulation of ATP metabolic process GO:1990619 histone H3-K9 deacetylation GO:2000648 positive regulation of stem cell proliferation GO:2000779 regulation of double-strand break repair GO:2001020 regulation of response to DNA damage stimulus |
| Molecular Function |
GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0003950 NAD+ ADP-ribosyltransferase activity GO:0003956 NAD(P)+-protein-arginine ADP-ribosyltransferase activity GO:0004407 histone deacetylase activity GO:0016757 transferase activity, transferring glycosyl groups GO:0016763 transferase activity, transferring pentosyl groups GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0032129 histone deacetylase activity (H3-K9 specific) GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0046969 NAD-dependent histone deacetylase activity (H3-K9 specific) GO:0048037 cofactor binding GO:0050662 coenzyme binding GO:0051287 NAD binding GO:0070403 NAD+ binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000792 heterochromatin GO:0005720 nuclear heterochromatin GO:0005724 nuclear telomeric heterochromatin GO:0031933 telomeric heterochromatin GO:0044454 nuclear chromosome part GO:0098687 chromosomal region |
| KEGG | - |
| Reactome |
R-HSA-5693532: DNA Double-Strand Break Repair R-HSA-73894: DNA Repair R-HSA-5693567: HDR through Homologous Recombination (HR) or Single Strand Annealing (SSA) R-HSA-5693538: Homology Directed Repair R-HSA-5693607: Processing of DNA double-strand break ends |
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SIRT6. |
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
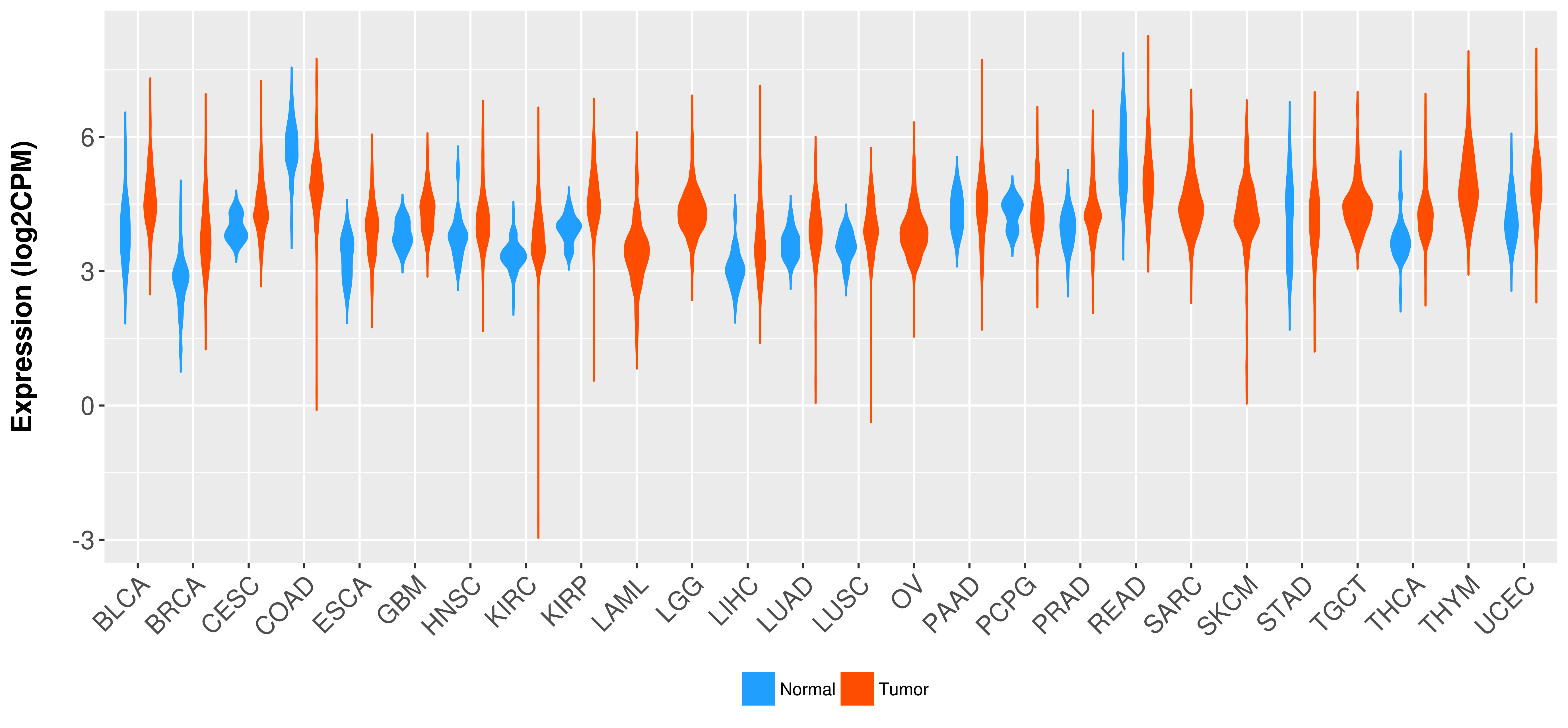
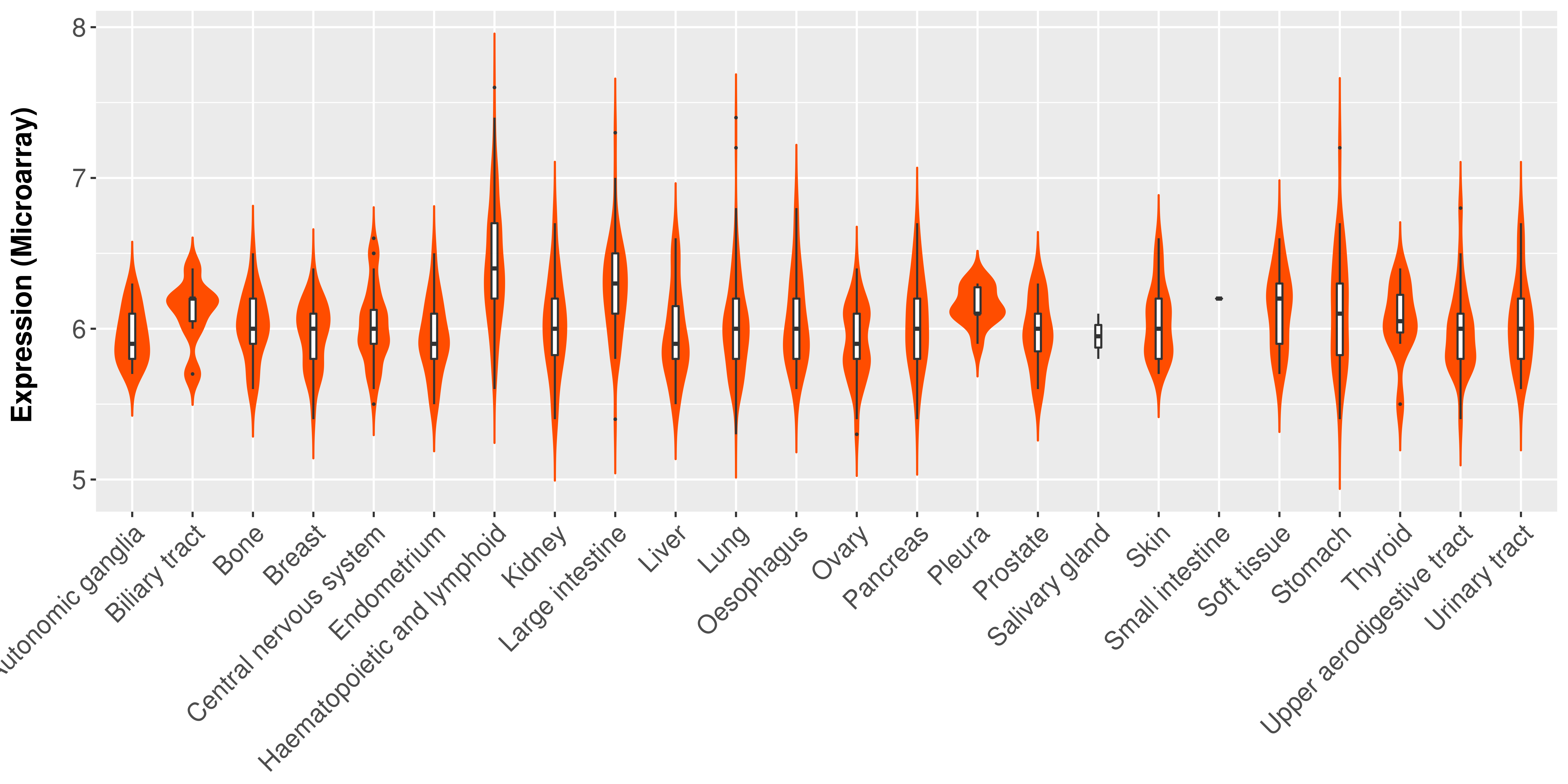
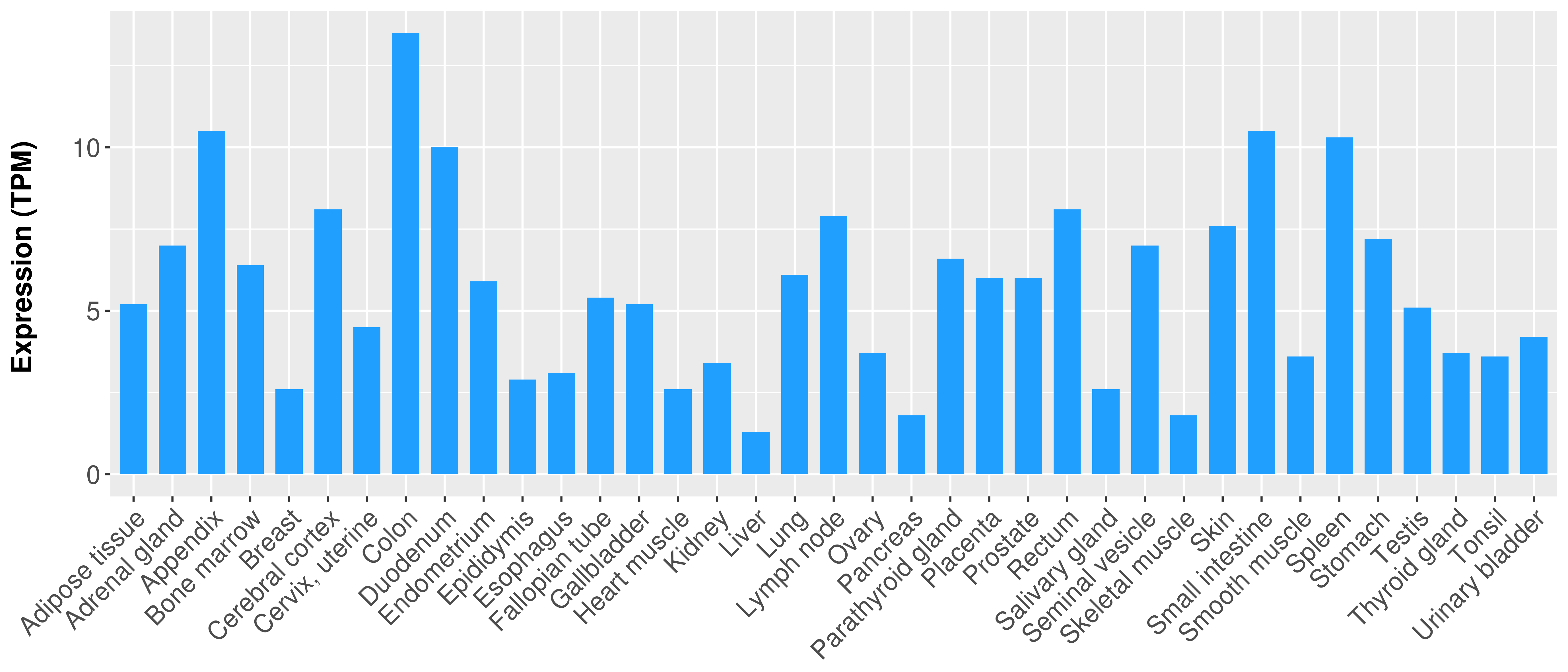
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
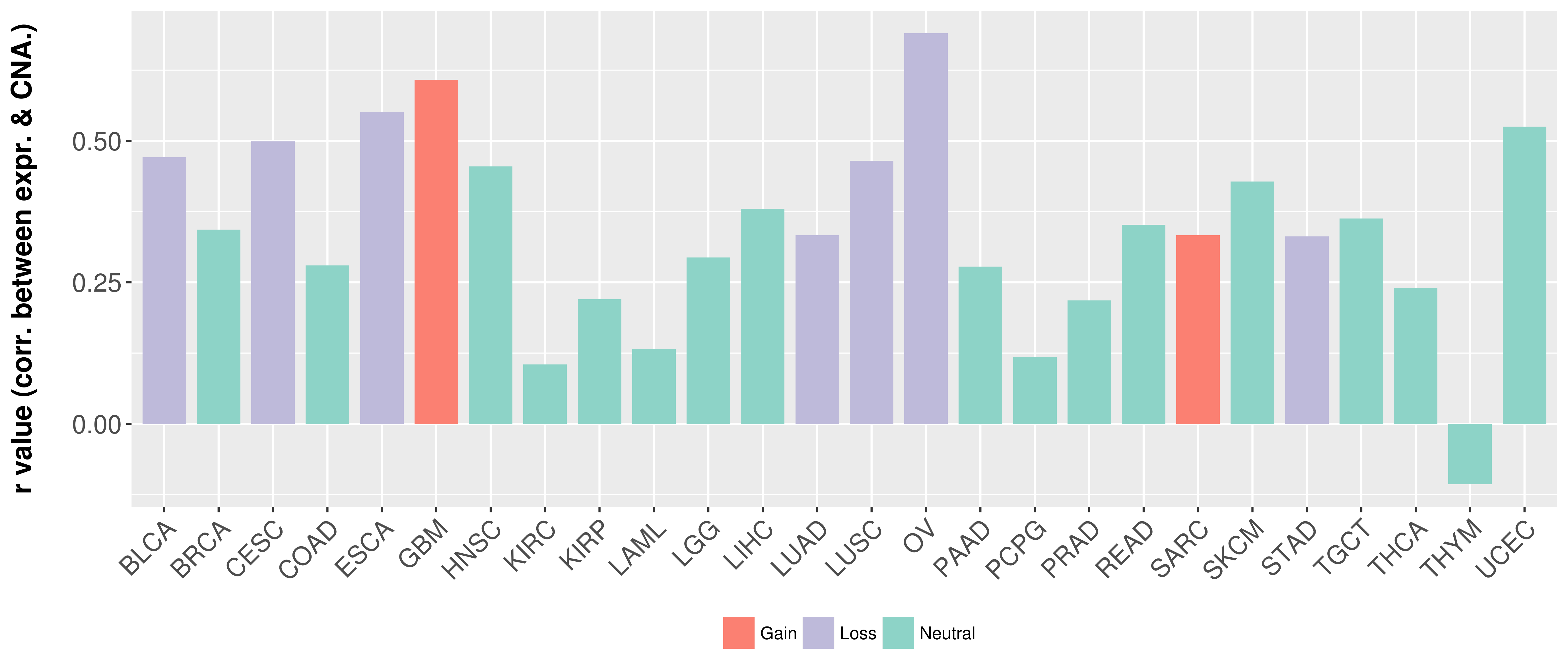
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
| There is no record. |
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
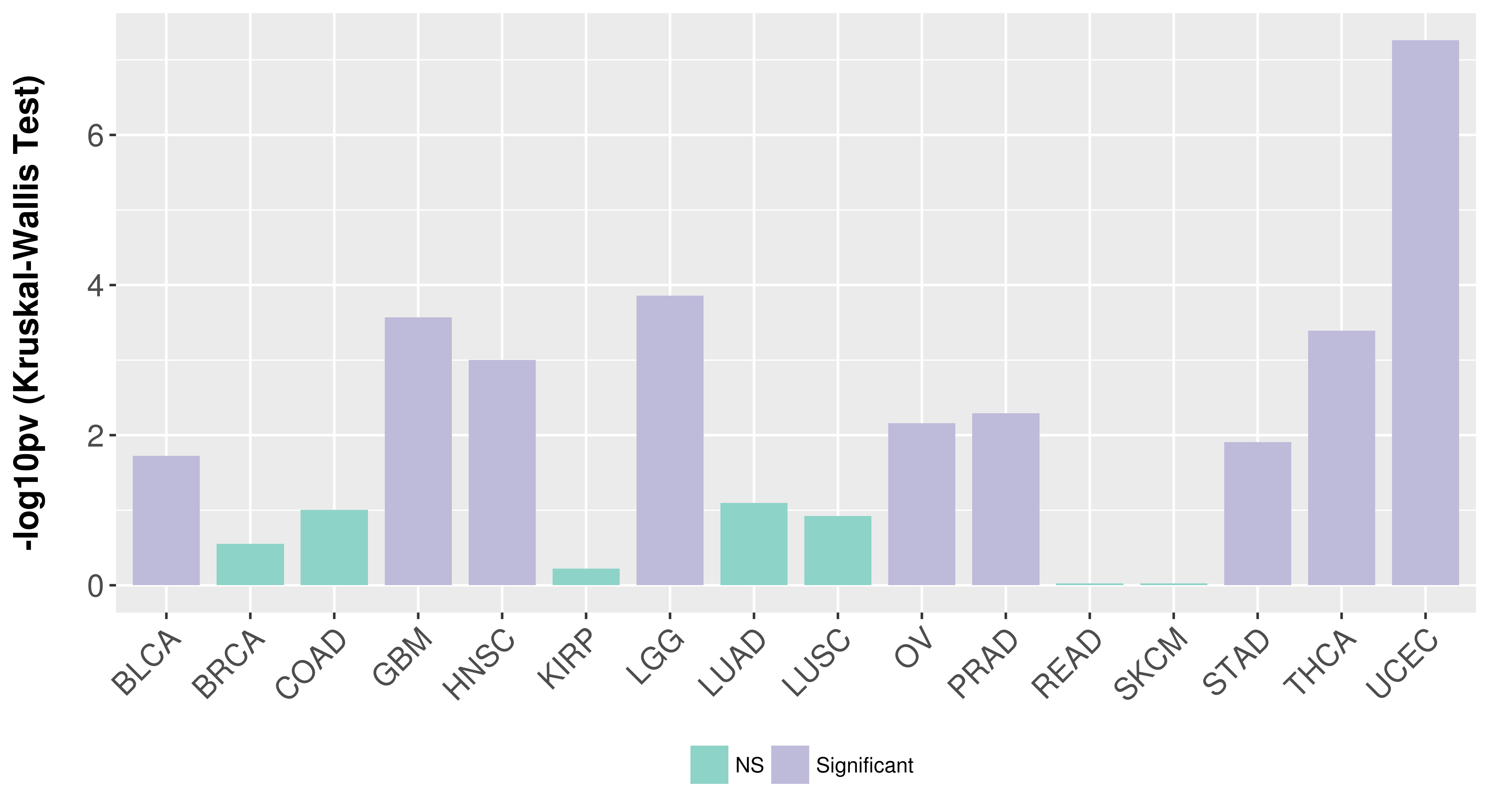
Association between expresson and subtype.
|
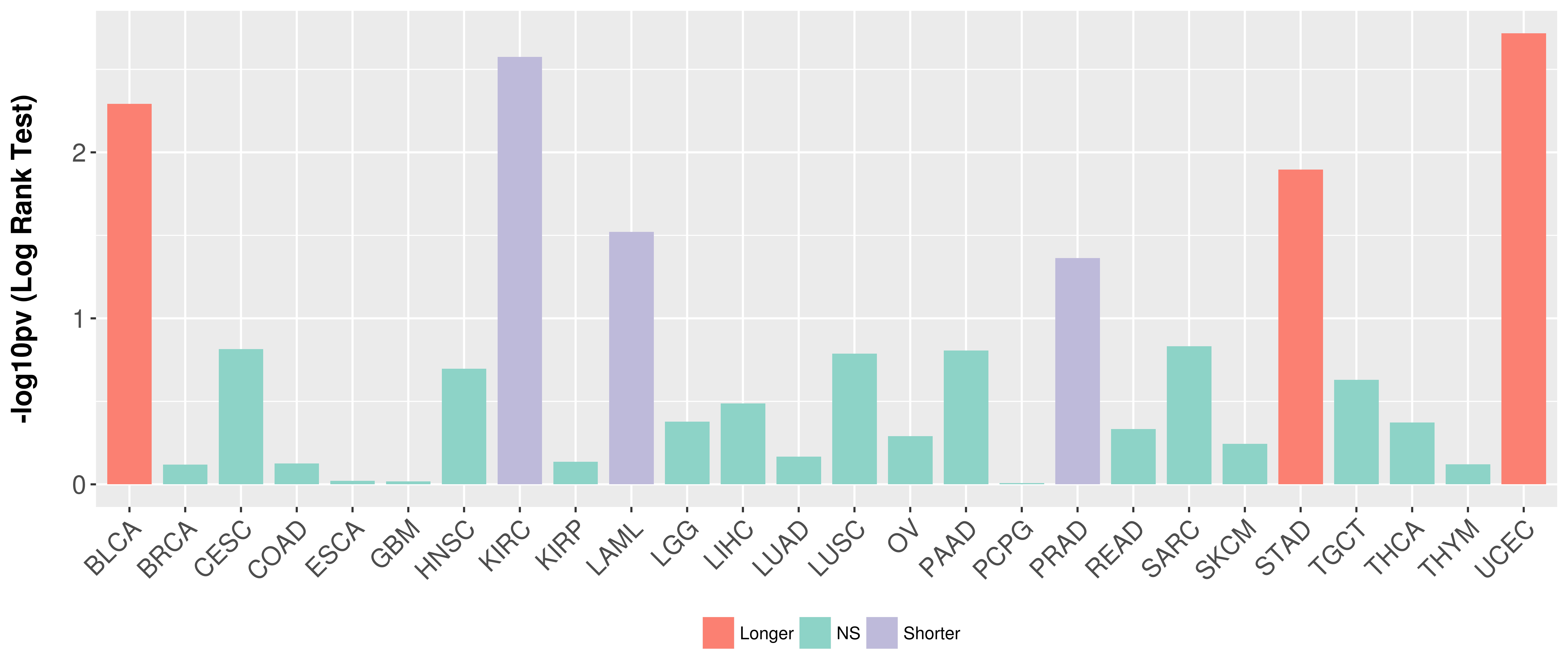
Overall survival analysis based on expression.
|
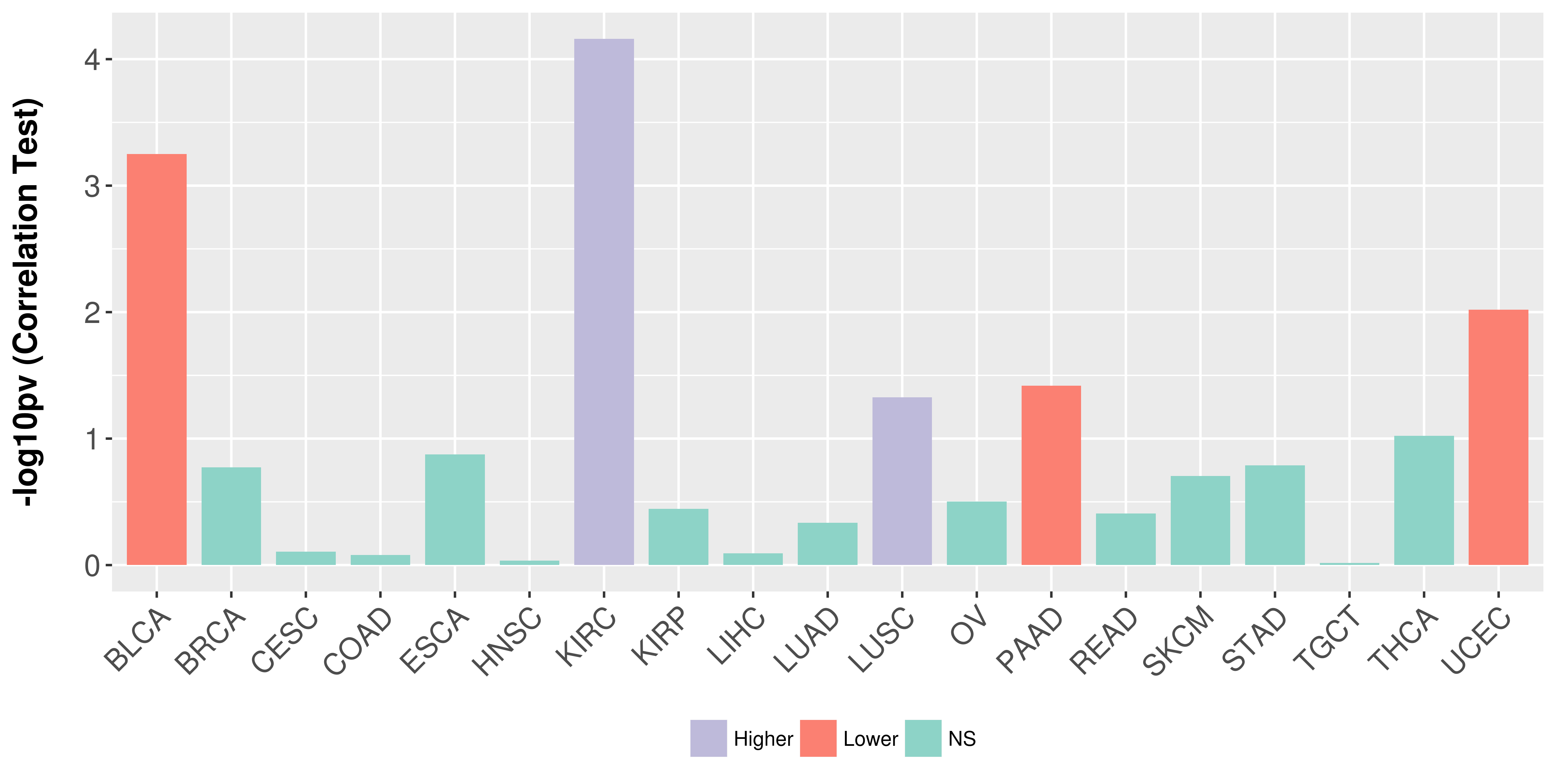
Association between expresson and stage.
|
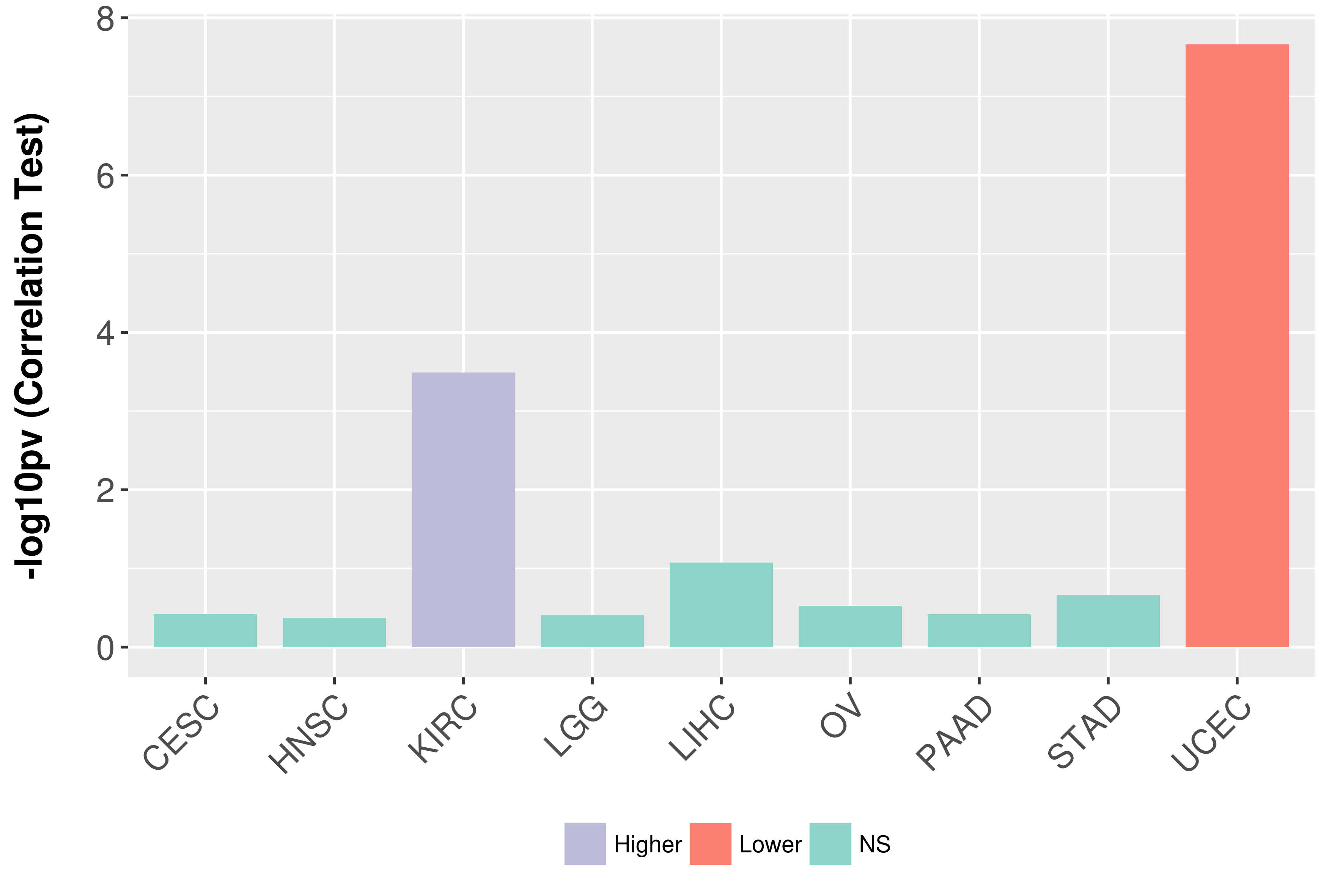
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SIRT6. |
| Summary | |
|---|---|
| Symbol | SIRT6 |
| Name | sirtuin 6 |
| Aliases | sirtuin (silent mating type information regulation 2, S. cerevisiae, homolog) 6; sirtuin (silent mating type ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SIRT6. |
|