Browse SMARCA4 in pancancer
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF07533 BRK domain PF00439 Bromodomain PF00271 Helicase conserved C-terminal domain PF07529 HSA PF08880 QLQ PF14619 Snf2-ATP coupling PF00176 SNF2 family N-terminal domain |
||||||||||
| Function |
Transcriptional coactivator cooperating with nuclear hormone receptors to potentiate transcriptional activation. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a post-mitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to post-mitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth. SMARCA4/BAF190A may promote neural stem cell self-renewal/proliferation by enhancing Notch-dependent proliferative signals, while concurrently making the neural stem cell insensitive to SHH-dependent differentiating cues (By similarity). Acts as a corepressor of ZEB1 to regulate E-cadherin transcription and is required for induction of epithelial-mesenchymal transition (EMT) by ZEB1. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000082 G1/S transition of mitotic cell cycle GO:0001558 regulation of cell growth GO:0001654 eye development GO:0001678 cellular glucose homeostasis GO:0003407 neural retina development GO:0006333 chromatin assembly or disassembly GO:0006337 nucleosome disassembly GO:0006338 chromatin remodeling GO:0006356 regulation of transcription from RNA polymerase I promoter GO:0006360 transcription from RNA polymerase I promoter GO:0007067 mitotic nuclear division GO:0007068 negative regulation of transcription during mitosis GO:0007070 negative regulation of transcription from RNA polymerase II promoter during mitosis GO:0007346 regulation of mitotic cell cycle GO:0007423 sensory organ development GO:0009303 rRNA transcription GO:0009743 response to carbohydrate GO:0009746 response to hexose GO:0009749 response to glucose GO:0009755 hormone-mediated signaling pathway GO:0009756 carbohydrate mediated signaling GO:0009757 hexose mediated signaling GO:0010182 sugar mediated signaling pathway GO:0010255 glucose mediated signaling pathway GO:0010948 negative regulation of cell cycle process GO:0016049 cell growth GO:0016055 Wnt signaling pathway GO:0019058 viral life cycle GO:0019080 viral gene expression GO:0019083 viral transcription GO:0030111 regulation of Wnt signaling pathway GO:0030177 positive regulation of Wnt signaling pathway GO:0030308 negative regulation of cell growth GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0031498 chromatin disassembly GO:0032844 regulation of homeostatic process GO:0032846 positive regulation of homeostatic process GO:0032984 macromolecular complex disassembly GO:0032986 protein-DNA complex disassembly GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0033144 negative regulation of intracellular steroid hormone receptor signaling pathway GO:0033500 carbohydrate homeostasis GO:0034284 response to monosaccharide GO:0034728 nucleosome organization GO:0035821 modification of morphology or physiology of other organism GO:0042593 glucose homeostasis GO:0042790 transcription of nuclear large rRNA transcript from RNA polymerase I promoter GO:0043010 camera-type eye development GO:0043044 ATP-dependent chromatin remodeling GO:0043241 protein complex disassembly GO:0043401 steroid hormone mediated signaling pathway GO:0043900 regulation of multi-organism process GO:0043902 positive regulation of multi-organism process GO:0043903 regulation of symbiosis, encompassing mutualism through parasitism GO:0043921 modulation by host of viral transcription GO:0043923 positive regulation by host of viral transcription GO:0044033 multi-organism metabolic process GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044843 cell cycle G1/S phase transition GO:0045786 negative regulation of cell cycle GO:0045896 regulation of transcription during mitosis GO:0045926 negative regulation of growth GO:0045930 negative regulation of mitotic cell cycle GO:0045943 positive regulation of transcription from RNA polymerase I promoter GO:0046021 regulation of transcription from RNA polymerase II promoter, mitotic GO:0046782 regulation of viral transcription GO:0048524 positive regulation of viral process GO:0048545 response to steroid hormone GO:0050434 positive regulation of viral transcription GO:0050792 regulation of viral process GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051702 interaction with symbiont GO:0051817 modification of morphology or physiology of other organism involved in symbiotic interaction GO:0051851 modification by host of symbiont morphology or physiology GO:0052312 modulation of transcription in other organism involved in symbiotic interaction GO:0052472 modulation by host of symbiont transcription GO:0060041 retina development in camera-type eye GO:0060765 regulation of androgen receptor signaling pathway GO:0060766 negative regulation of androgen receptor signaling pathway GO:0061614 pri-miRNA transcription from RNA polymerase II promoter GO:0071322 cellular response to carbohydrate stimulus GO:0071326 cellular response to monosaccharide stimulus GO:0071331 cellular response to hexose stimulus GO:0071333 cellular response to glucose stimulus GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071824 protein-DNA complex subunit organization GO:0098781 ncRNA transcription GO:0198738 cell-cell signaling by wnt GO:1901836 regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter GO:1901838 positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901991 negative regulation of mitotic cell cycle phase transition GO:1902659 regulation of glucose mediated signaling pathway GO:1902661 positive regulation of glucose mediated signaling pathway GO:1902806 regulation of cell cycle G1/S phase transition GO:1902807 negative regulation of cell cycle G1/S phase transition GO:1902893 regulation of pri-miRNA transcription from RNA polymerase II promoter GO:1902895 positive regulation of pri-miRNA transcription from RNA polymerase II promoter GO:1903900 regulation of viral life cycle GO:1903902 positive regulation of viral life cycle GO:1904837 beta-catenin-TCF complex assembly GO:2000045 regulation of G1/S transition of mitotic cell cycle GO:2000134 negative regulation of G1/S transition of mitotic cell cycle |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001013 RNA polymerase I regulatory region DNA binding GO:0001046 core promoter sequence-specific DNA binding GO:0001047 core promoter binding GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001104 RNA polymerase II transcription cofactor activity GO:0001105 RNA polymerase II transcription coactivator activity GO:0001158 enhancer sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0001163 RNA polymerase I regulatory region sequence-specific DNA binding GO:0001164 RNA polymerase I CORE element sequence-specific DNA binding GO:0001190 transcriptional activator activity, RNA polymerase II transcription factor binding GO:0001191 transcriptional repressor activity, RNA polymerase II transcription factor binding GO:0002039 p53 binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0003714 transcription corepressor activity GO:0004386 helicase activity GO:0008094 DNA-dependent ATPase activity GO:0008134 transcription factor binding GO:0016887 ATPase activity GO:0030957 Tat protein binding GO:0031490 chromatin DNA binding GO:0031491 nucleosome binding GO:0031492 nucleosomal DNA binding GO:0035257 nuclear hormone receptor binding GO:0035258 steroid hormone receptor binding GO:0035326 enhancer binding GO:0042393 histone binding GO:0042623 ATPase activity, coupled GO:0043566 structure-specific DNA binding GO:0047485 protein N-terminus binding GO:0050681 androgen receptor binding GO:0051427 hormone receptor binding GO:0070182 DNA polymerase binding GO:0070577 lysine-acetylated histone binding GO:0098811 transcriptional repressor activity, RNA polymerase II activating transcription factor binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016514 SWI/SNF complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0071564 npBAF complex GO:0071565 nBAF complex GO:0090544 BAF-type complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-1280215: Cytokine Signaling in Immune system R-HSA-201722: Formation of the beta-catenin R-HSA-168256: Immune System R-HSA-1266695: Interleukin-7 signaling R-HSA-3214858: RMTs methylate histone arginines R-HSA-162582: Signal Transduction R-HSA-449147: Signaling by Interleukins R-HSA-195721: Signaling by Wnt R-HSA-201681: TCF dependent signaling in response to WNT |
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
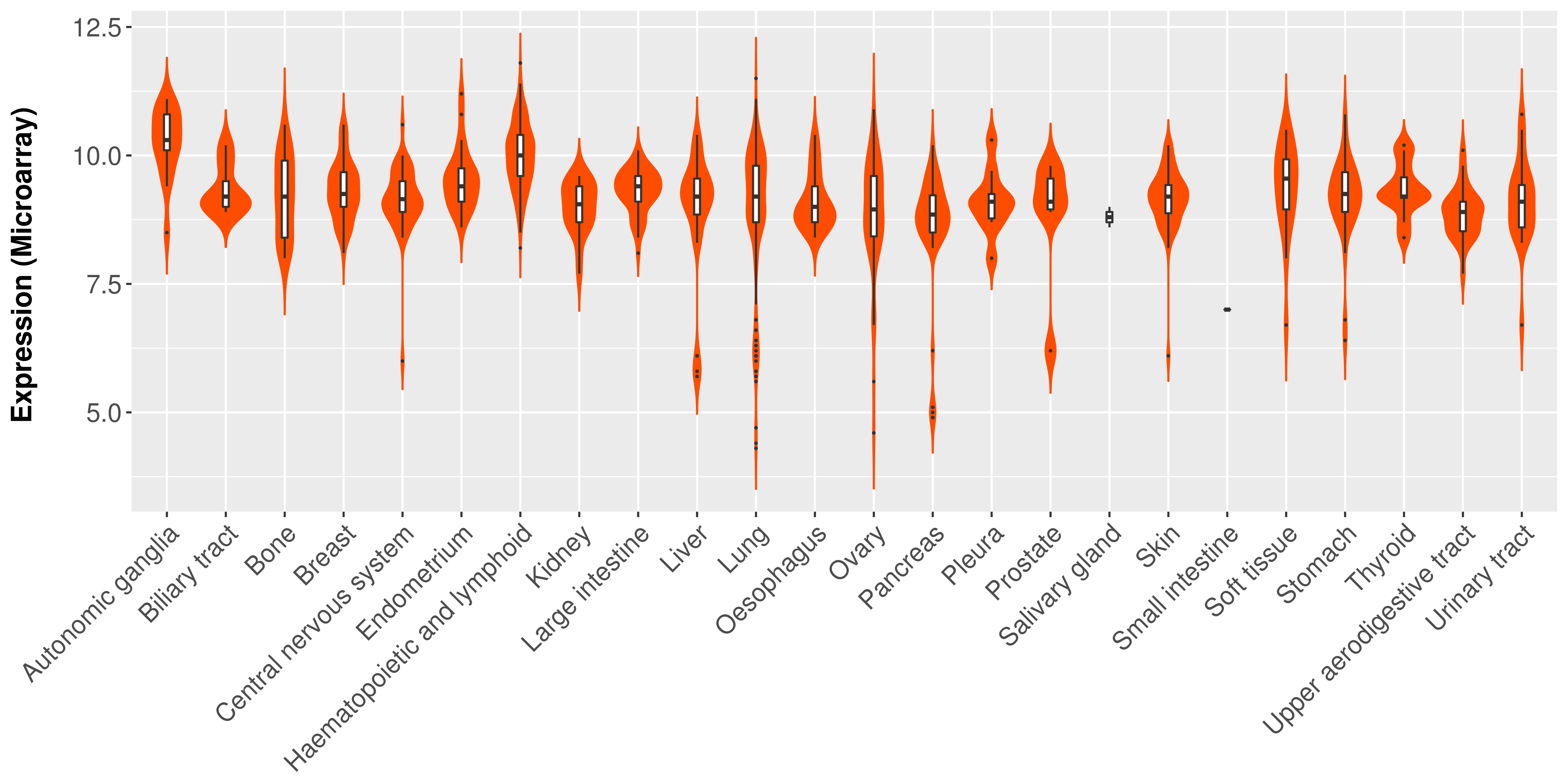
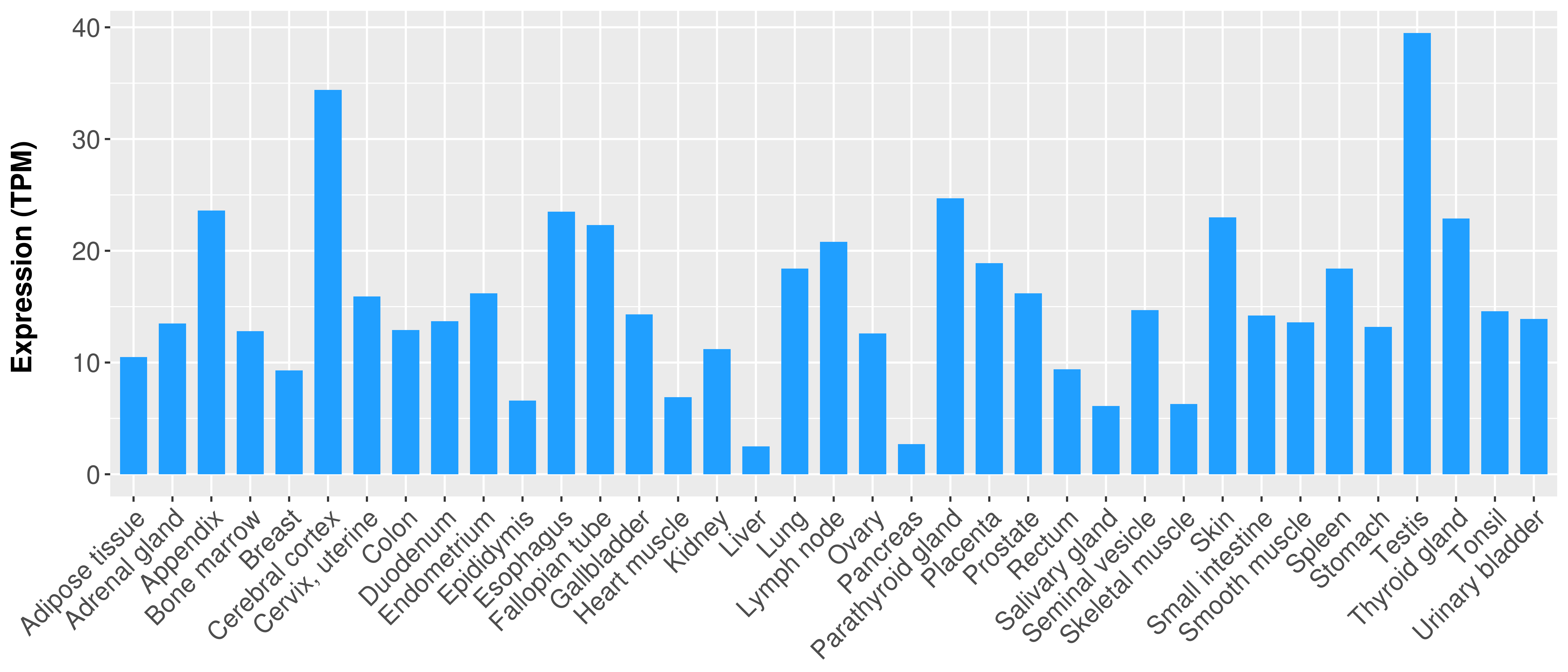
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
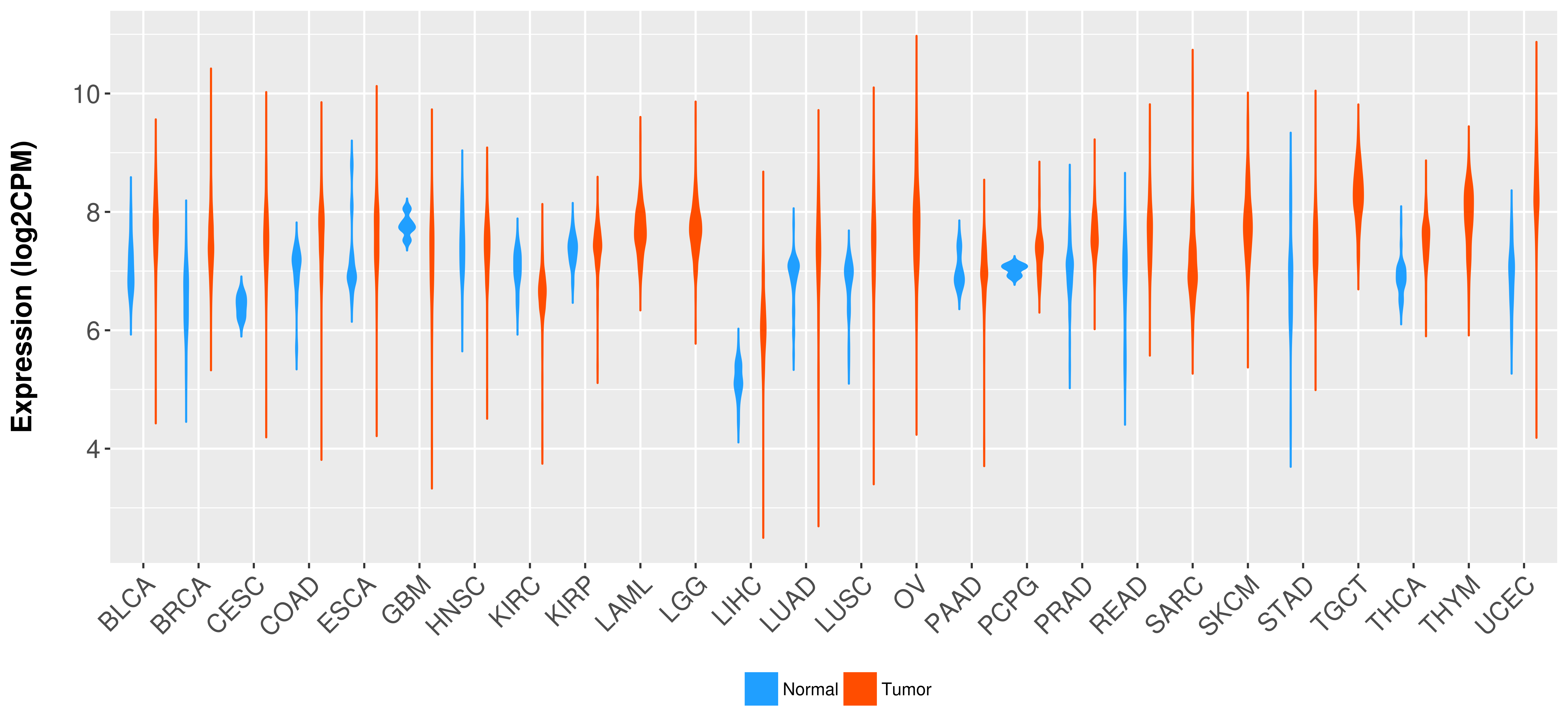
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
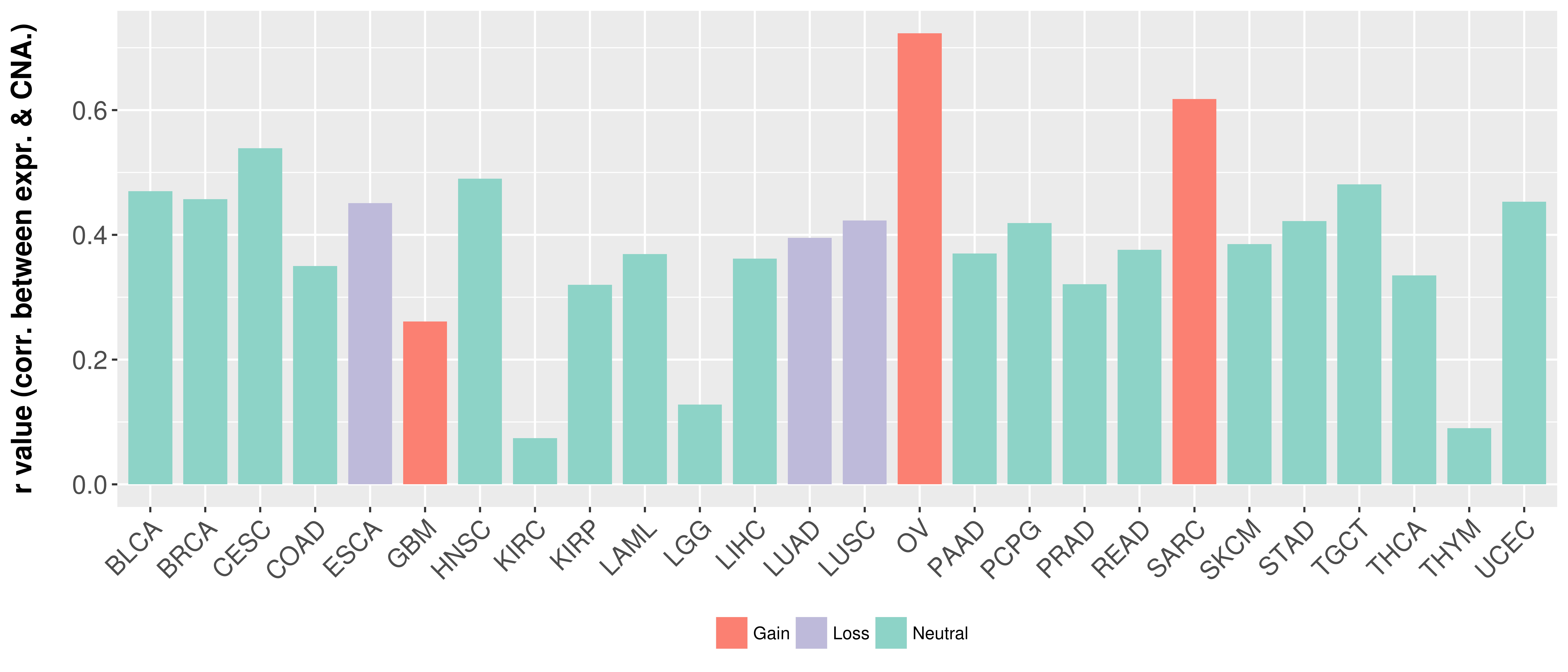
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
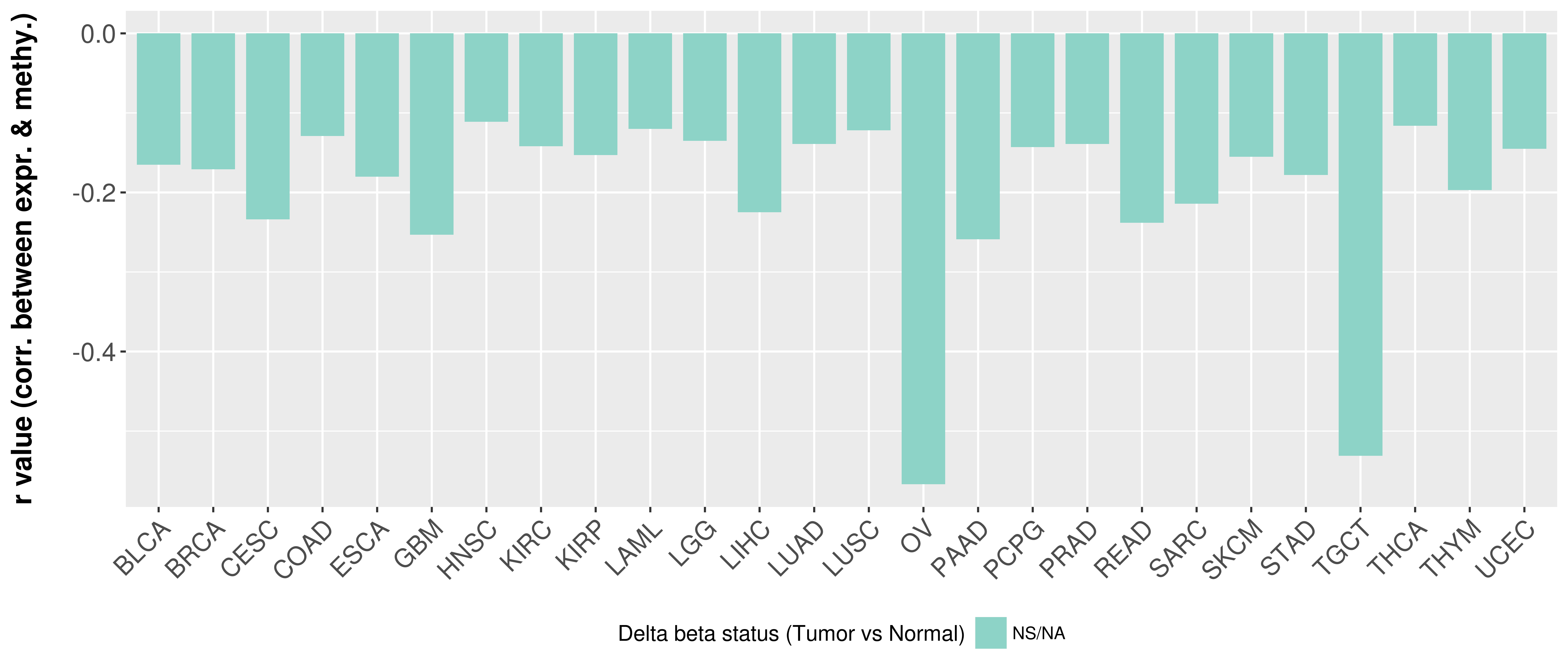
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
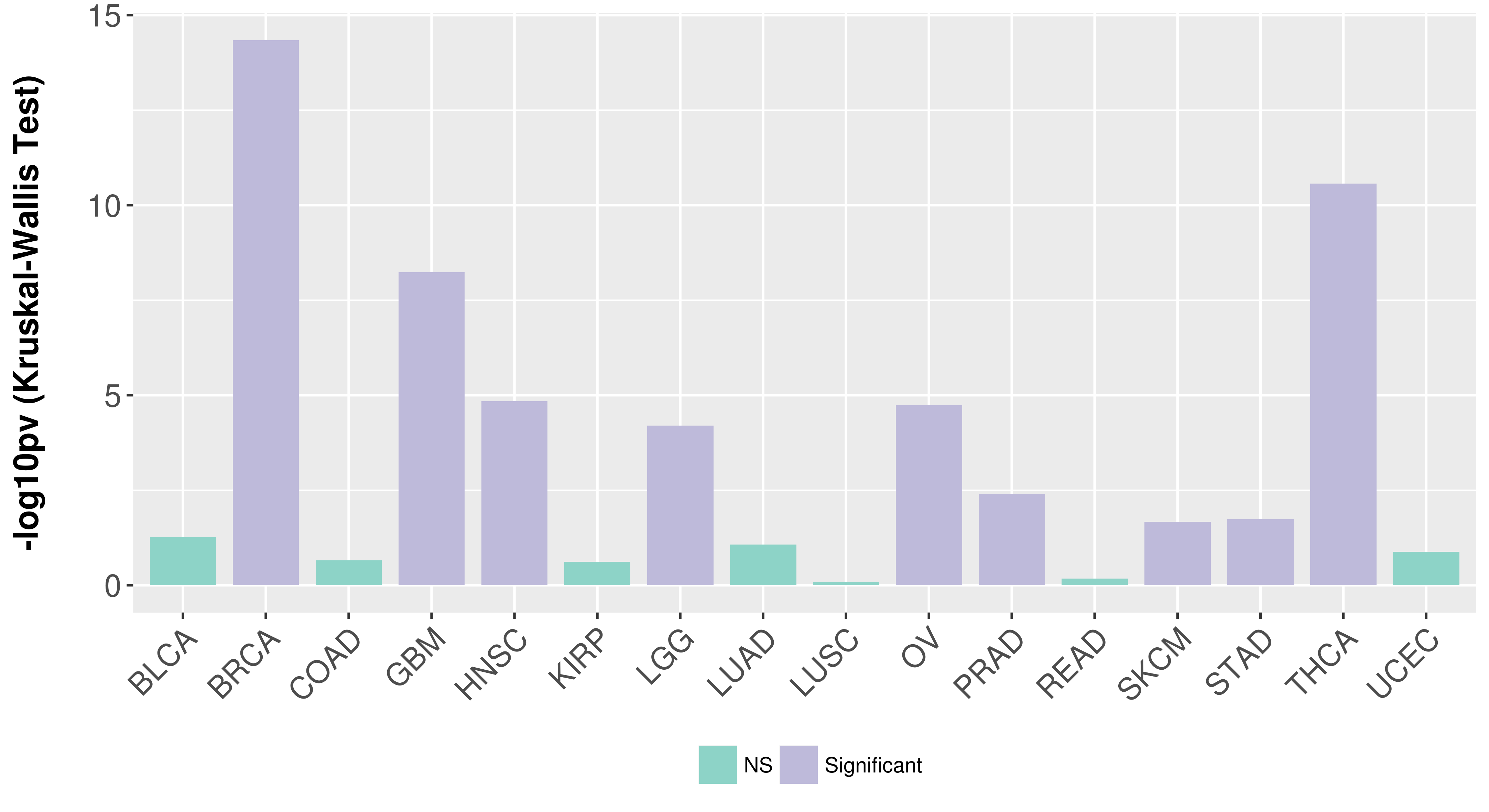
Association between expresson and subtype.
|
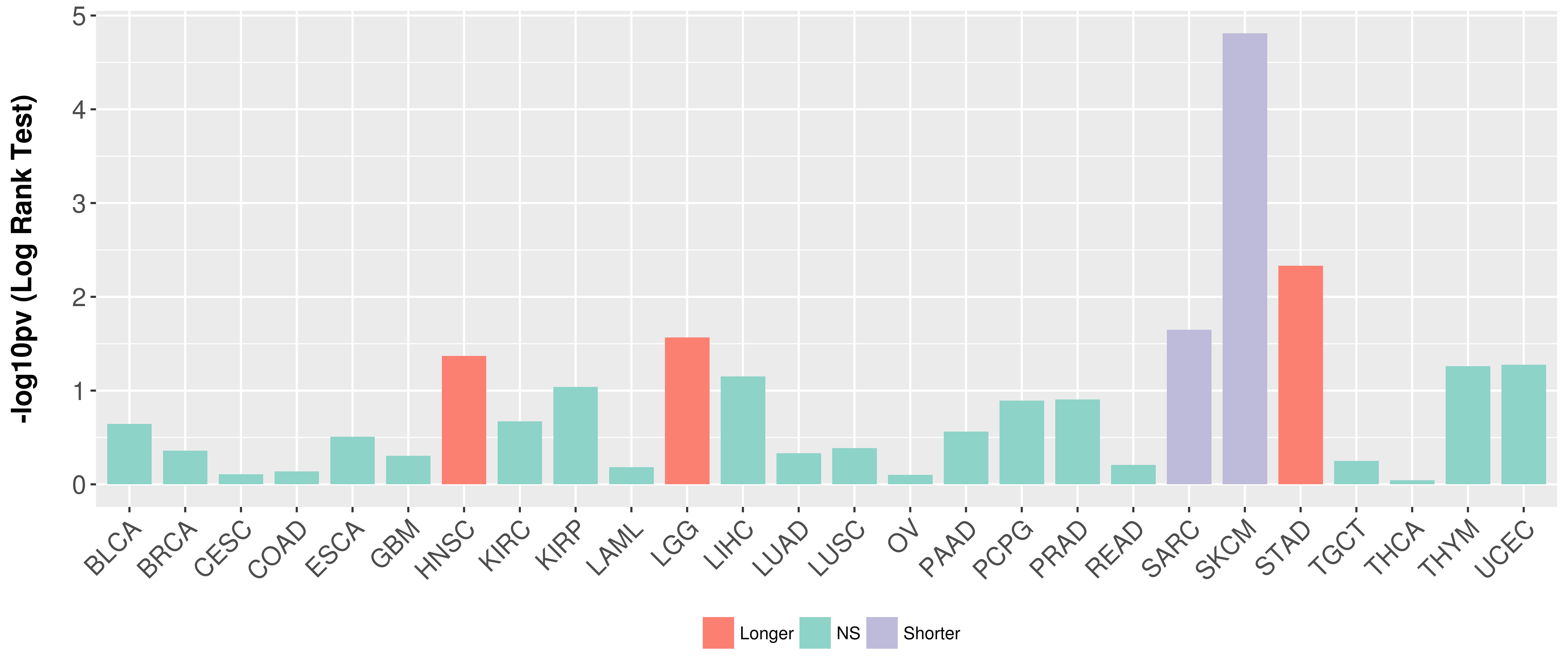
Overall survival analysis based on expression.
|
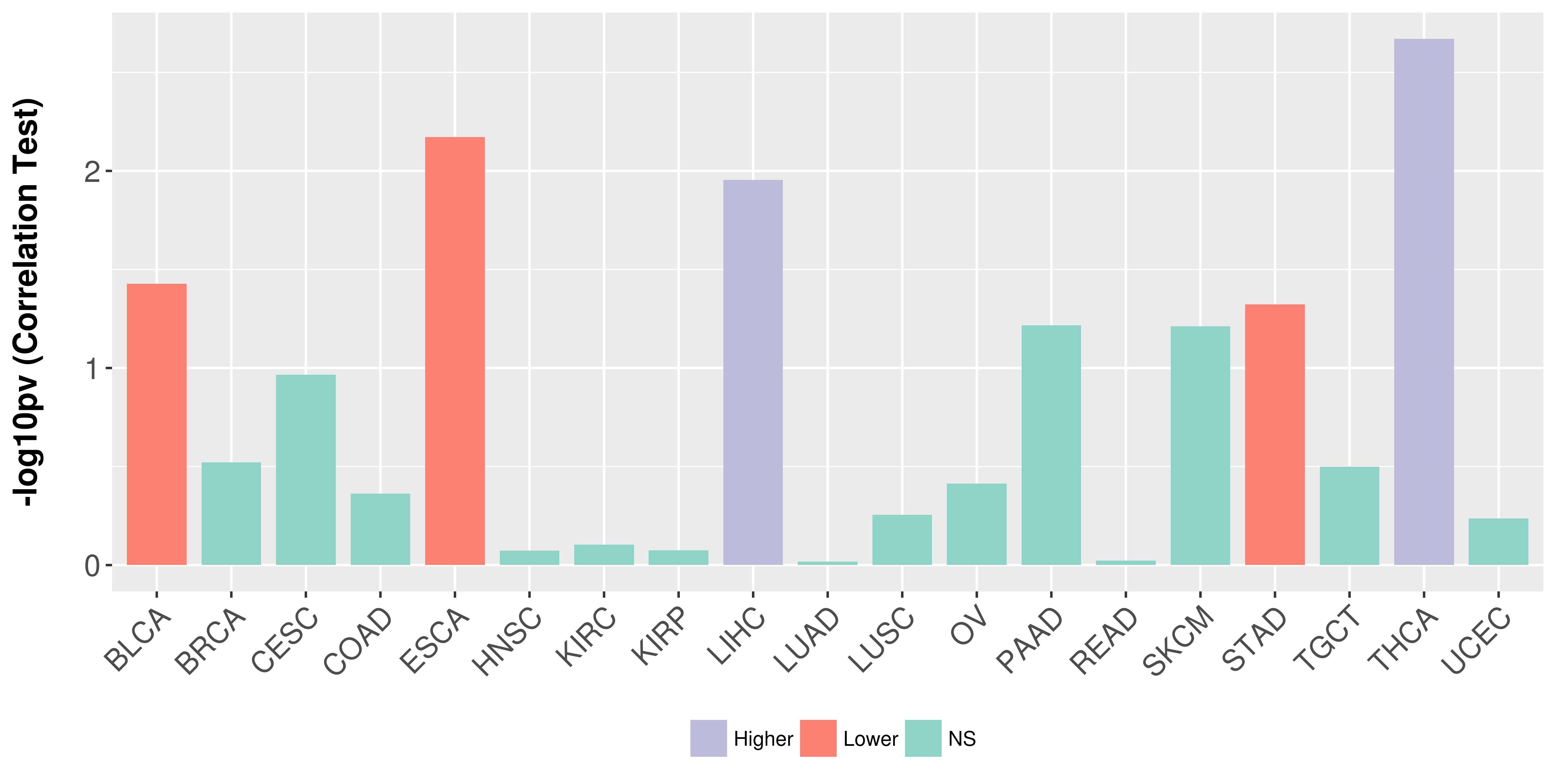
Association between expresson and stage.
|
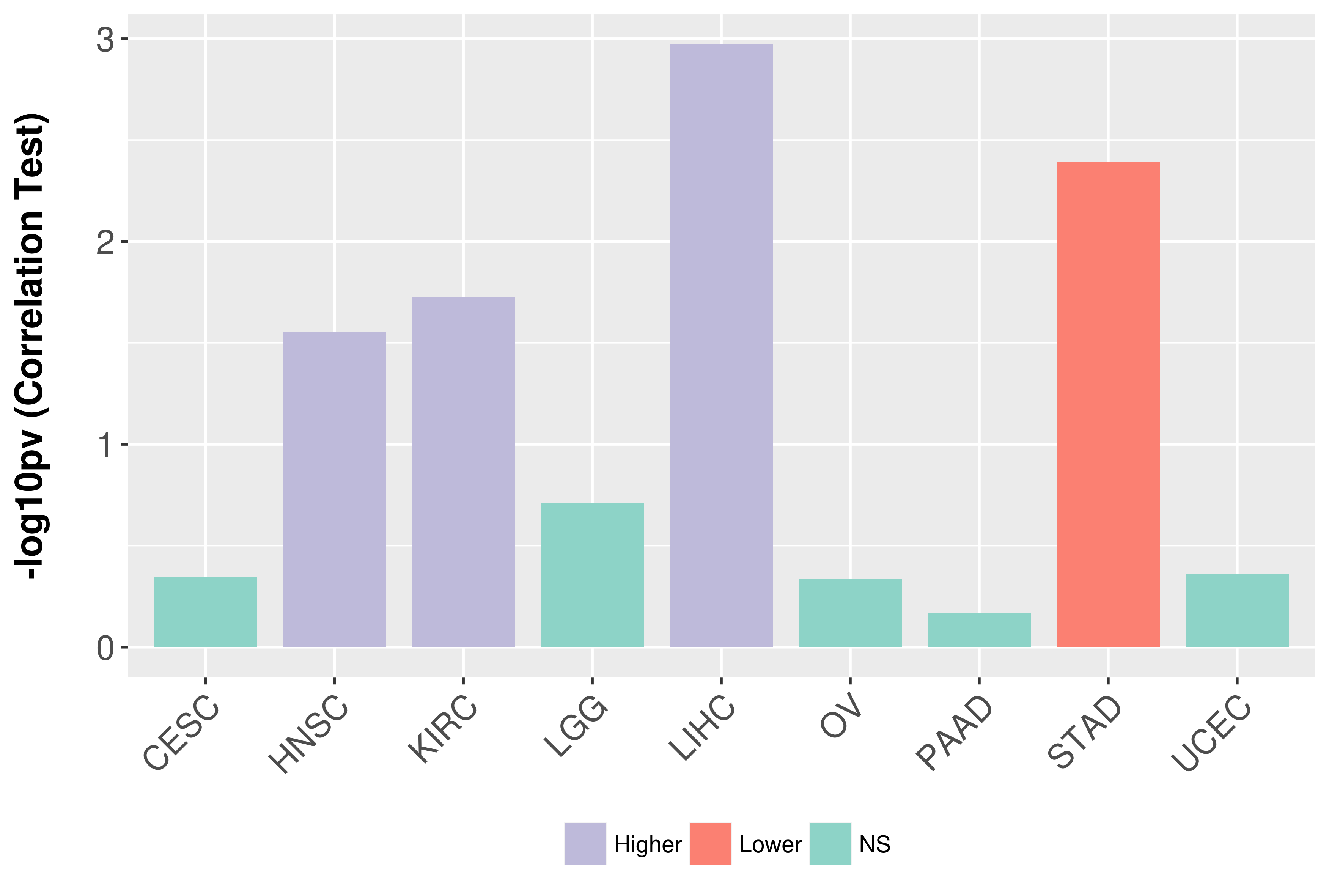
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SMARCA4. |
| Summary | |
|---|---|
| Symbol | SMARCA4 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Aliases | hSNF2b; BRG1; SNF2-BETA; FLJ39786; SNF2-like 4; sucrose nonfermenting-like 4; mitotic growth and transcripti ...... |
| Location | 19p13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|