Browse SMARCD3 in pancancer
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF02201 SWIB/MDM2 domain |
||||||||||
| Function |
Plays a role in ATP dependent nucleosome remodeling by SMARCA4 containing complexes. Stimulates nuclear receptor mediated transcription. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a post-mitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to post-mitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000086 G2/M transition of mitotic cell cycle GO:0001654 eye development GO:0002052 positive regulation of neuroblast proliferation GO:0003002 regionalization GO:0003007 heart morphogenesis GO:0003128 heart field specification GO:0003139 secondary heart field specification GO:0003205 cardiac chamber development GO:0003206 cardiac chamber morphogenesis GO:0003207 cardiac chamber formation GO:0003208 cardiac ventricle morphogenesis GO:0003211 cardiac ventricle formation GO:0003215 cardiac right ventricle morphogenesis GO:0003219 cardiac right ventricle formation GO:0003231 cardiac ventricle development GO:0003407 neural retina development GO:0006333 chromatin assembly or disassembly GO:0006337 nucleosome disassembly GO:0006338 chromatin remodeling GO:0007346 regulation of mitotic cell cycle GO:0007389 pattern specification process GO:0007405 neuroblast proliferation GO:0007423 sensory organ development GO:0007507 heart development GO:0010092 specification of animal organ identity GO:0010389 regulation of G2/M transition of mitotic cell cycle GO:0010720 positive regulation of cell development GO:0010971 positive regulation of G2/M transition of mitotic cell cycle GO:0031498 chromatin disassembly GO:0032984 macromolecular complex disassembly GO:0032986 protein-DNA complex disassembly GO:0034728 nucleosome organization GO:0042692 muscle cell differentiation GO:0043010 camera-type eye development GO:0043241 protein complex disassembly GO:0043393 regulation of protein binding GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044839 cell cycle G2/M phase transition GO:0045787 positive regulation of cell cycle GO:0045931 positive regulation of mitotic cell cycle GO:0048645 animal organ formation GO:0050769 positive regulation of neurogenesis GO:0051098 regulation of binding GO:0051145 smooth muscle cell differentiation GO:0051147 regulation of muscle cell differentiation GO:0051149 positive regulation of muscle cell differentiation GO:0051150 regulation of smooth muscle cell differentiation GO:0051152 positive regulation of smooth muscle cell differentiation GO:0051962 positive regulation of nervous system development GO:0060041 retina development in camera-type eye GO:0060914 heart formation GO:0061351 neural precursor cell proliferation GO:0071824 protein-DNA complex subunit organization GO:0072089 stem cell proliferation GO:0072091 regulation of stem cell proliferation GO:0090068 positive regulation of cell cycle process GO:1901987 regulation of cell cycle phase transition GO:1901989 positive regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901992 positive regulation of mitotic cell cycle phase transition GO:1902692 regulation of neuroblast proliferation GO:1902749 regulation of cell cycle G2/M phase transition GO:1902751 positive regulation of cell cycle G2/M phase transition GO:2000177 regulation of neural precursor cell proliferation GO:2000179 positive regulation of neural precursor cell proliferation GO:2000648 positive regulation of stem cell proliferation |
| Molecular Function |
GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0008134 transcription factor binding GO:0035257 nuclear hormone receptor binding GO:0051427 hormone receptor binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016514 SWI/SNF complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0071564 npBAF complex GO:0071565 nBAF complex GO:0090544 BAF-type complex |
| KEGG | - |
| Reactome |
R-HSA-2426168: Activation of gene expression by SREBF (SREBP) R-HSA-1368108: BMAL1 R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-1266738: Developmental Biology R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-1592230: Mitochondrial biogenesis R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-1989781: PPARA activates gene expression R-HSA-3214858: RMTs methylate histone arginines R-HSA-1368082: RORA activates gene expression R-HSA-1655829: Regulation of cholesterol biosynthesis by SREBP (SREBF) R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis R-HSA-381340: Transcriptional regulation of white adipocyte differentiation R-HSA-2032785: YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SMARCD3. |
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
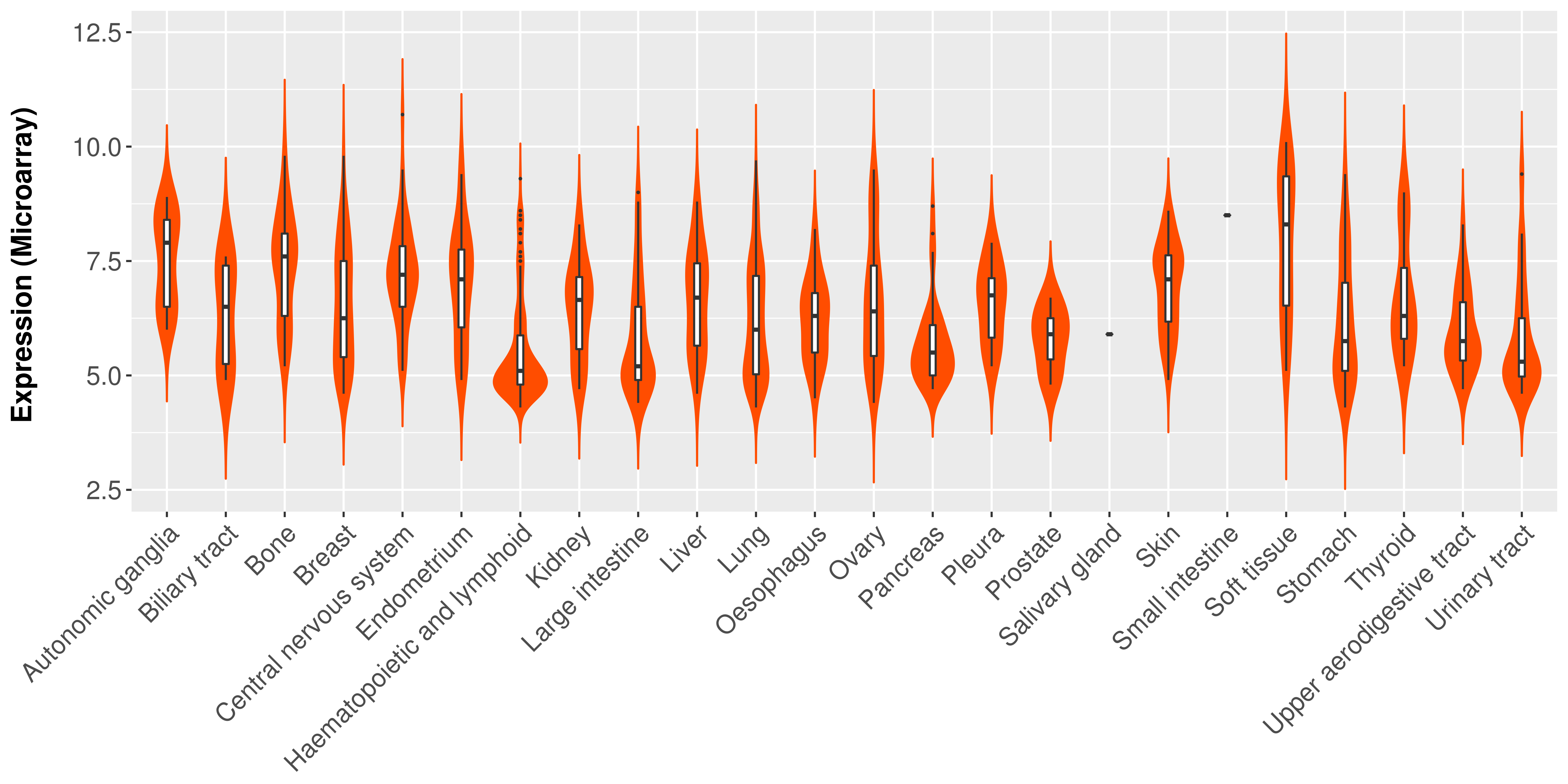
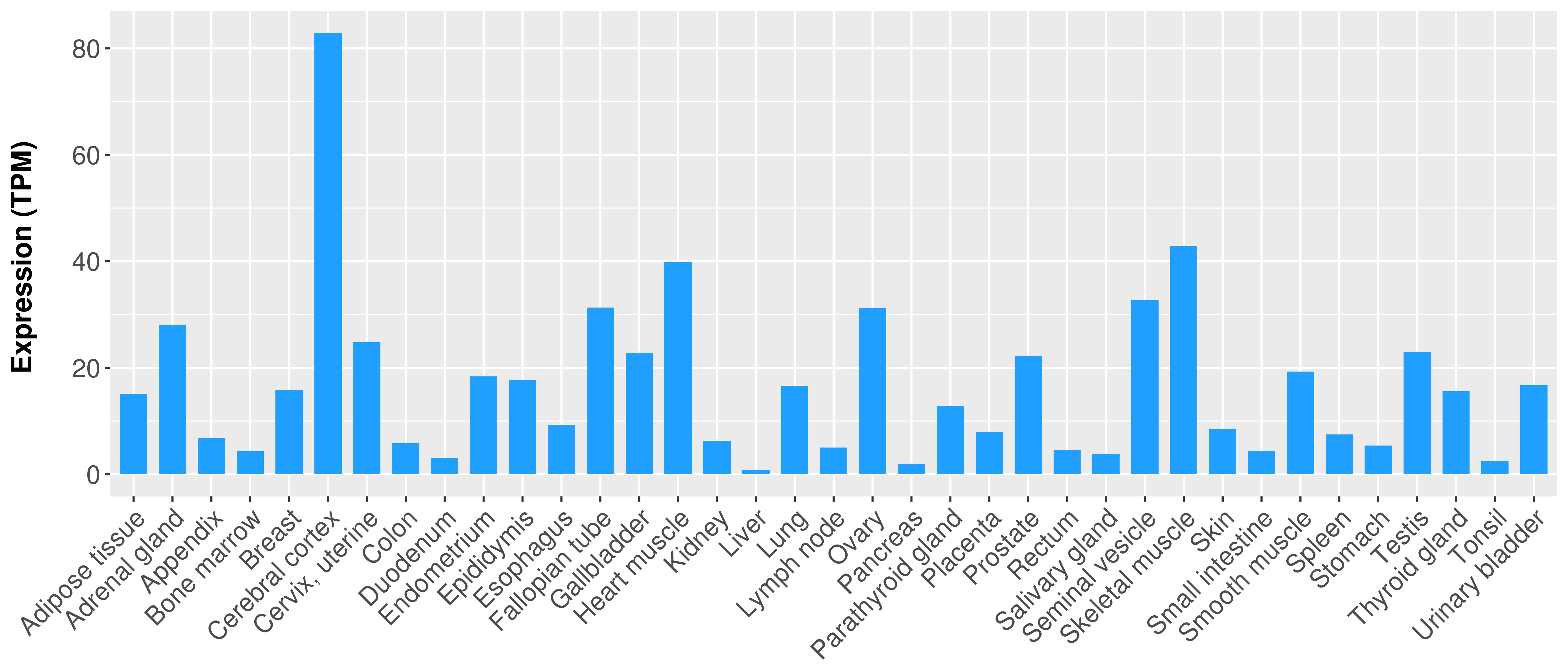
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
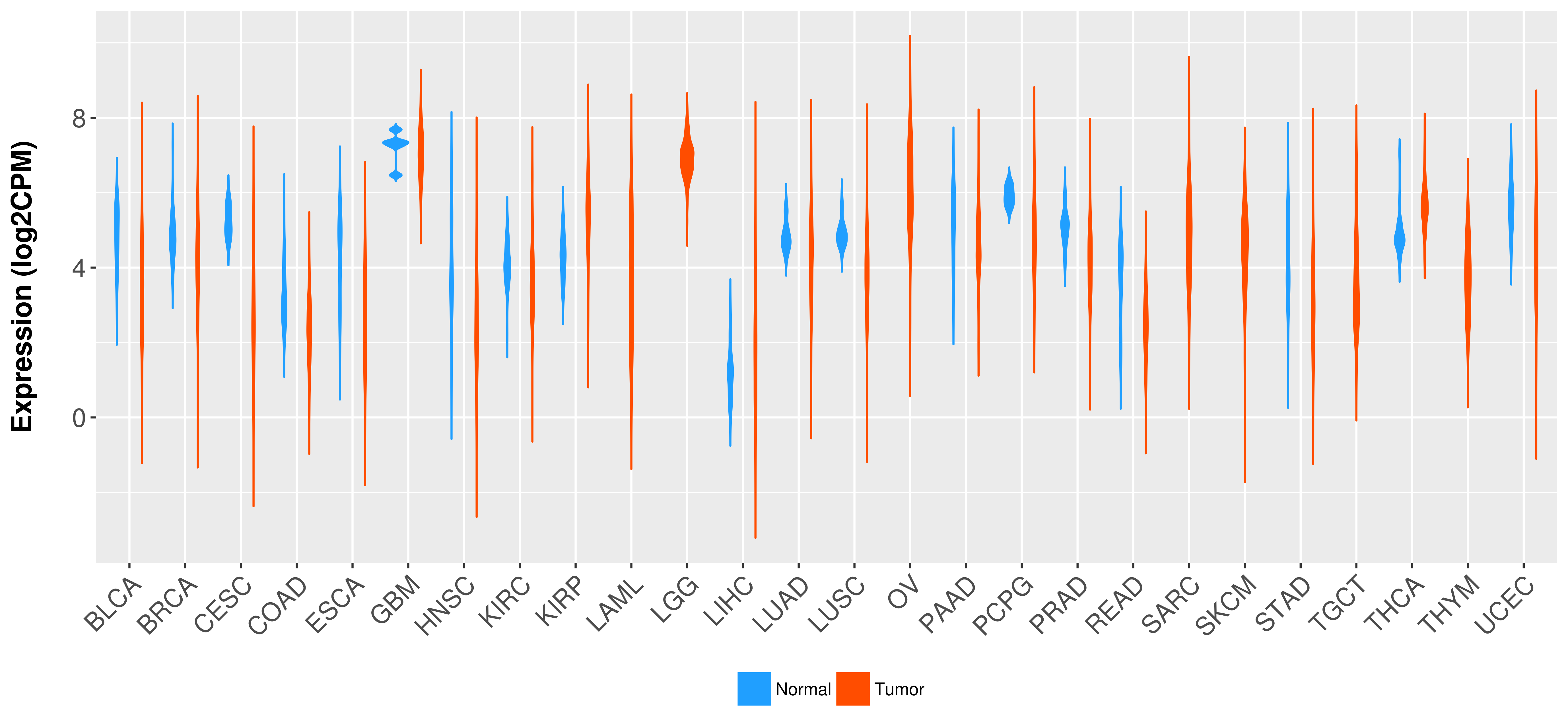
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |

Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
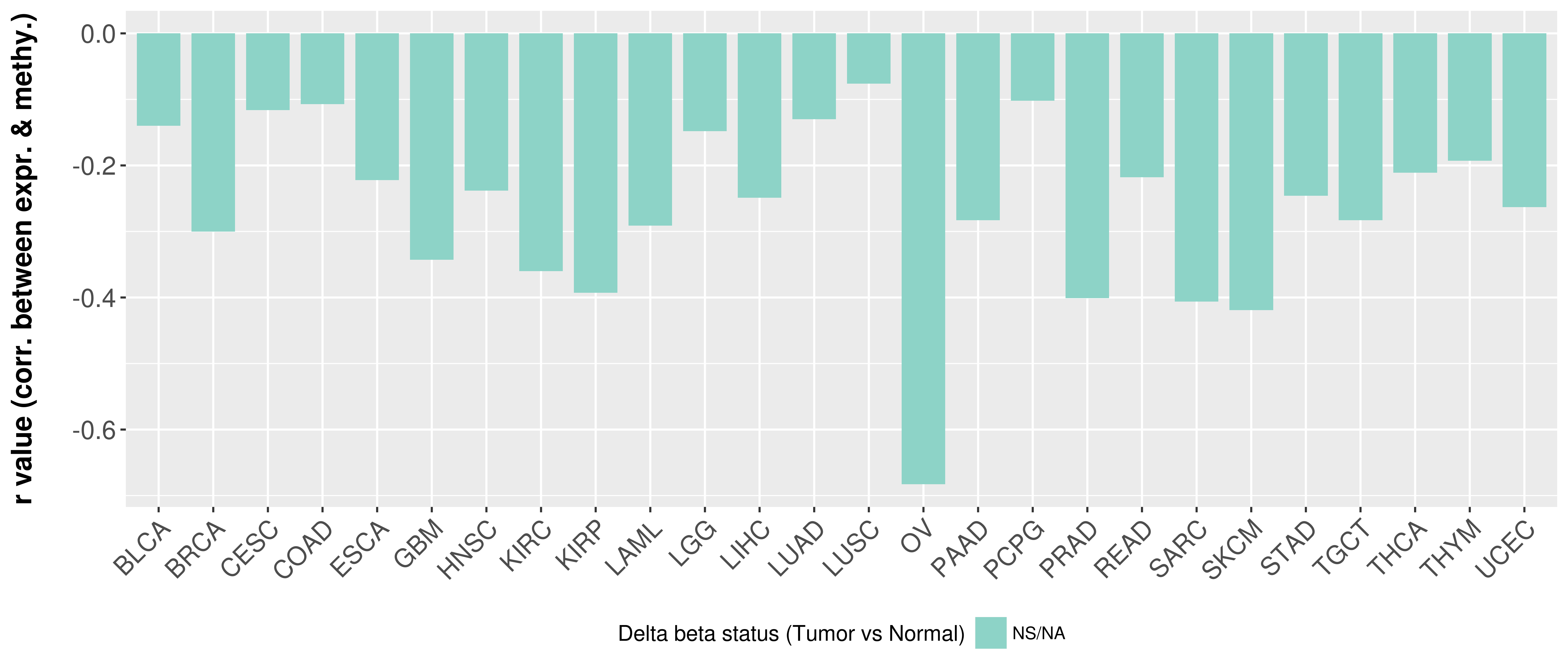
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |

|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |

Association between expresson and subtype.
|
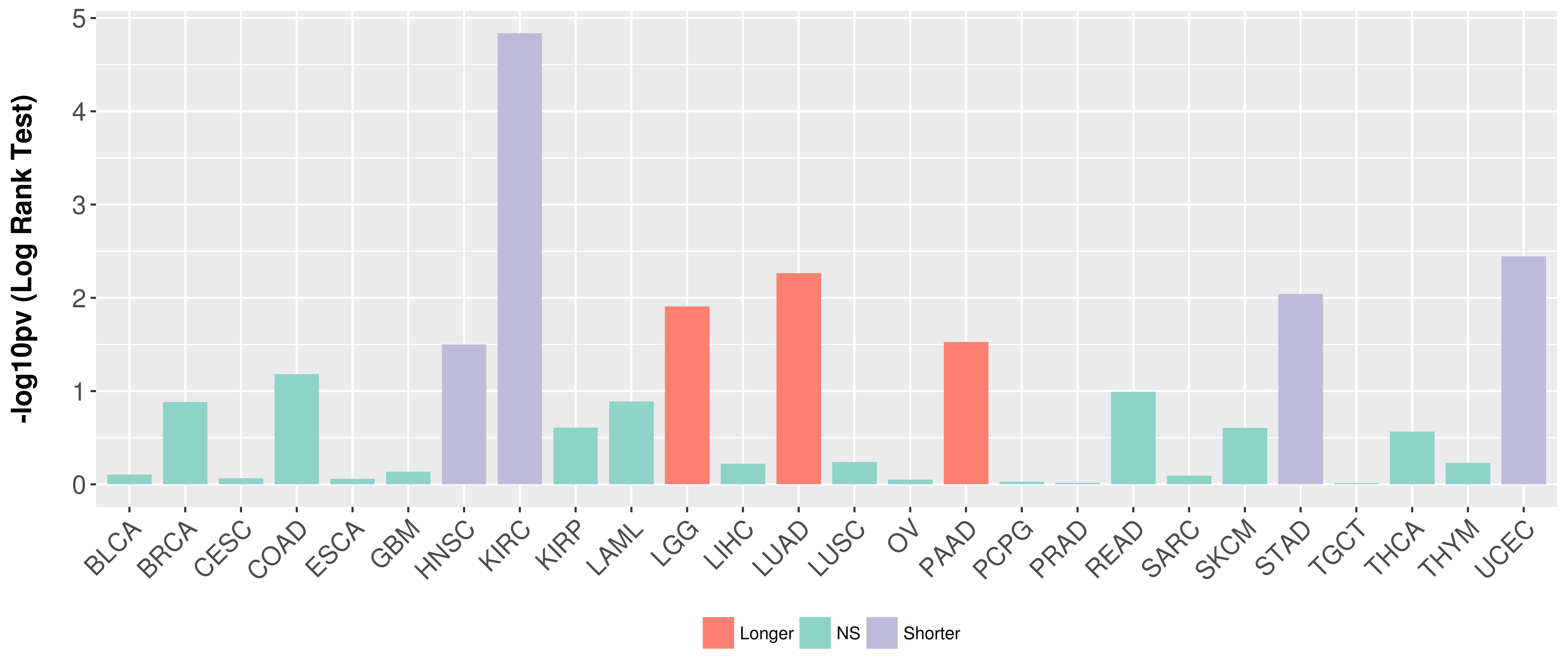
Overall survival analysis based on expression.
|
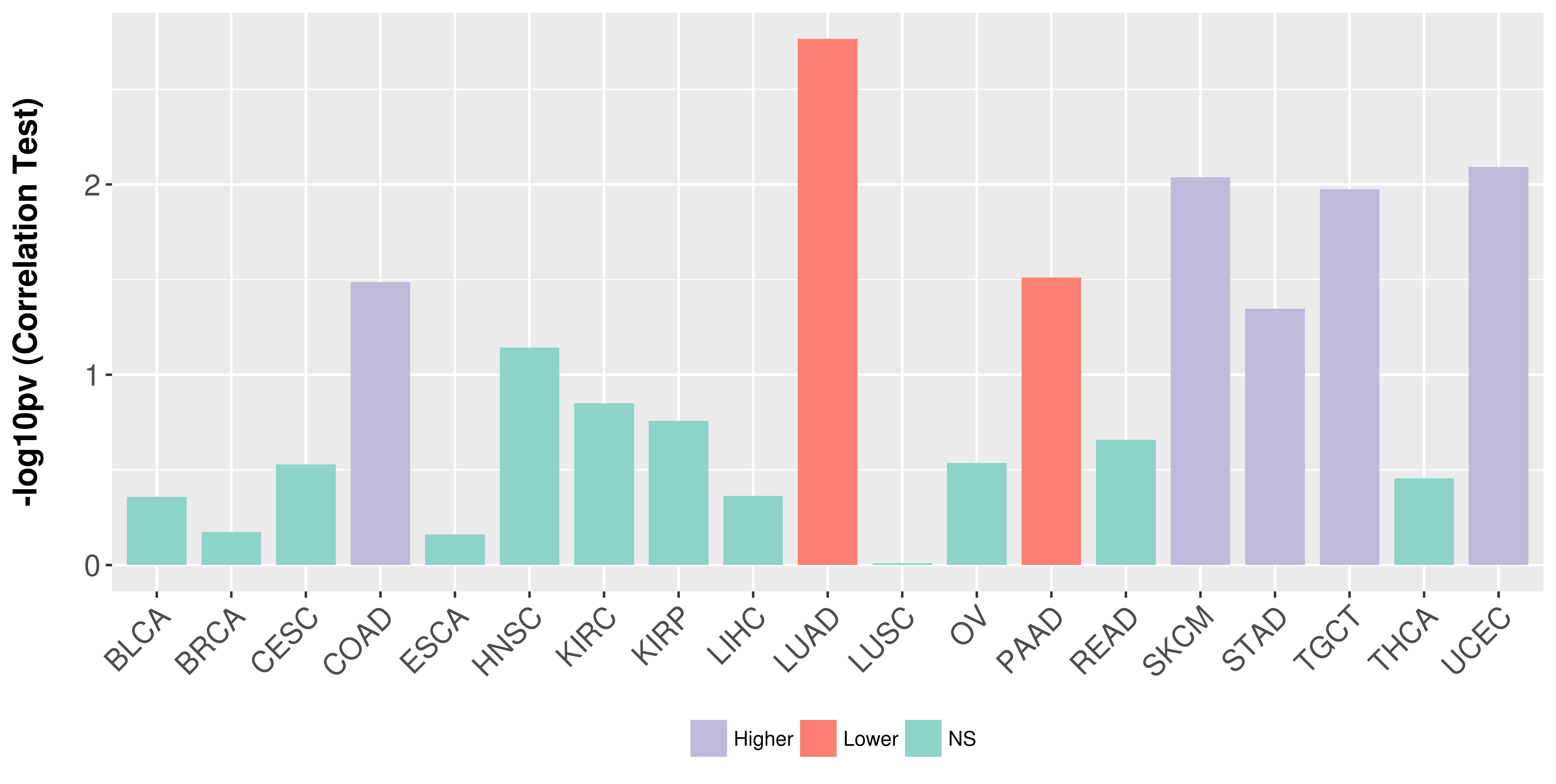
Association between expresson and stage.
|
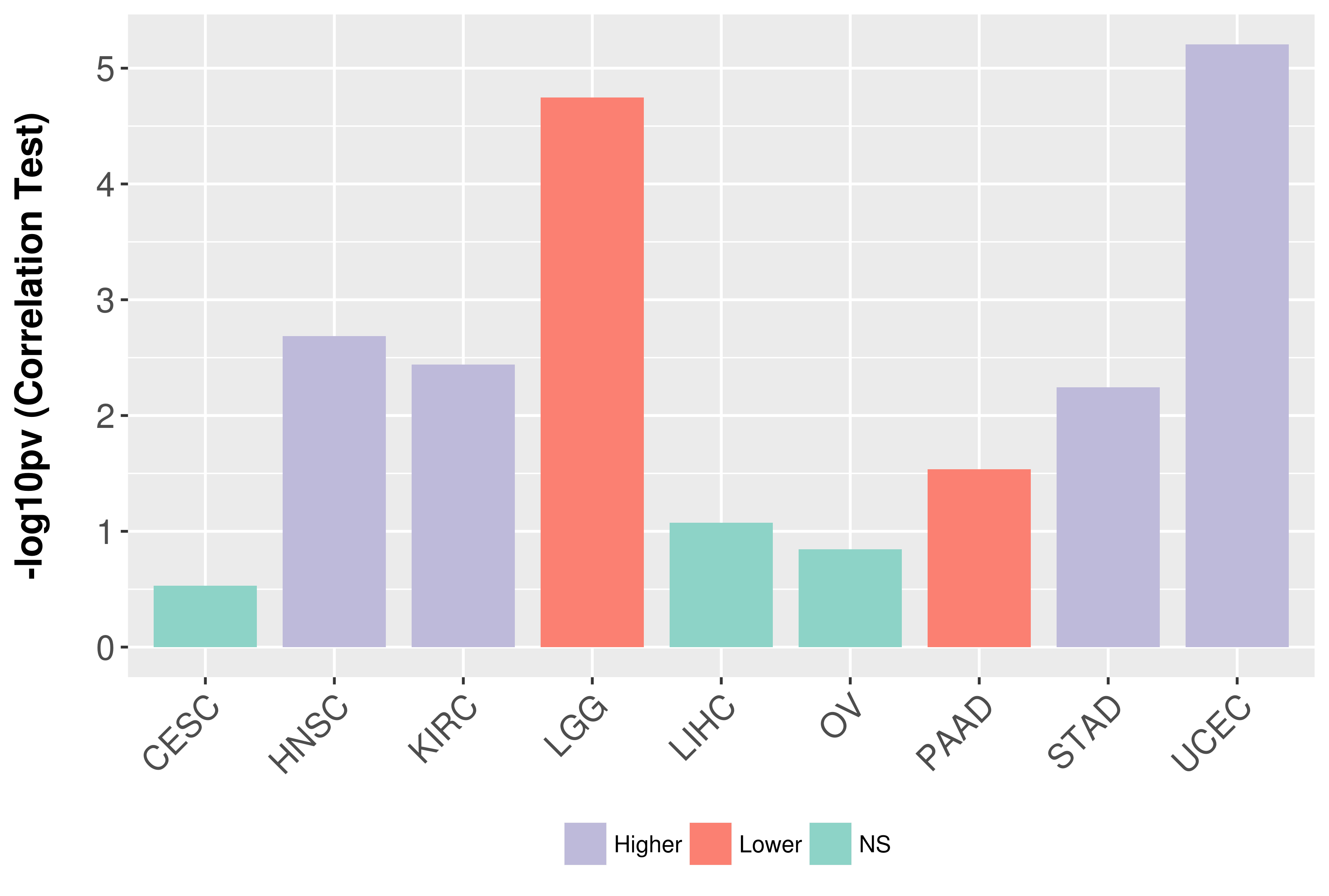
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SMARCD3. |
| Summary | |
|---|---|
| Symbol | SMARCD3 |
| Name | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| Aliases | BAF60C; CRACD3; mammalian chromatin remodeling complex BRG1-associated factor 60C; SWI/SNF complex 60 kDa su ...... |
| Location | 7q36.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SMARCD3. |
| There is no record for SMARCD3. |