Browse SMC1A in pancancer
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF06470 SMC proteins Flexible Hinge Domain PF02463 RecF/RecN/SMC N terminal domain |
||||||||||
| Function |
Involved in chromosome cohesion during cell cycle and in DNA repair. Central component of cohesin complex. The cohesin complex is required for the cohesion of sister chromatids after DNA replication. The cohesin complex apparently forms a large proteinaceous ring within which sister chromatids can be trapped. At anaphase, the complex is cleaved and dissociates from chromatin, allowing sister chromatids to segregate. The cohesin complex may also play a role in spindle pole assembly during mitosis. Involved in DNA repair via its interaction with BRCA1 and its related phosphorylation by ATM, or via its phosphorylation by ATR. Works as a downstream effector both in the ATM/NBS1 branch and in the ATR/MSH2 branch of S-phase checkpoint. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000070 mitotic sister chromatid segregation GO:0000075 cell cycle checkpoint GO:0000819 sister chromatid segregation GO:0006260 DNA replication GO:0006261 DNA-dependent DNA replication GO:0006275 regulation of DNA replication GO:0007052 mitotic spindle organization GO:0007059 chromosome segregation GO:0007062 sister chromatid cohesion GO:0007064 mitotic sister chromatid cohesion GO:0007067 mitotic nuclear division GO:0007093 mitotic cell cycle checkpoint GO:0007126 meiotic nuclear division GO:0007346 regulation of mitotic cell cycle GO:0008156 negative regulation of DNA replication GO:0009314 response to radiation GO:0010948 negative regulation of cell cycle process GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0019827 stem cell population maintenance GO:0032875 regulation of DNA endoreduplication GO:0032876 negative regulation of DNA endoreduplication GO:0042023 DNA endoreduplication GO:0042770 signal transduction in response to DNA damage GO:0044786 cell cycle DNA replication GO:0045786 negative regulation of cell cycle GO:0045930 negative regulation of mitotic cell cycle GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051321 meiotic cell cycle GO:0090329 regulation of DNA-dependent DNA replication GO:0098727 maintenance of cell number GO:0098813 nuclear chromosome segregation GO:1903046 meiotic cell cycle process GO:2000104 negative regulation of DNA-dependent DNA replication |
| Molecular Function |
GO:0003682 chromatin binding GO:0036033 mediator complex binding GO:0046982 protein heterodimerization activity |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0000776 kinetochore GO:0000777 condensed chromosome kinetochore GO:0000779 condensed chromosome, centromeric region GO:0000793 condensed chromosome GO:0000794 condensed nuclear chromosome GO:0008278 cohesin complex GO:0008280 cohesin core heterodimer GO:0030893 meiotic cohesin complex GO:0098687 chromosomal region |
| KEGG |
hsa04110 Cell cycle hsa04114 Oocyte meiosis |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-2470946: Cohesin Loading onto Chromatin R-HSA-2468052: Establishment of Sister Chromatid Cohesion R-HSA-68886: M Phase R-HSA-1500620: Meiosis R-HSA-1221632: Meiotic synapsis R-HSA-392499: Metabolism of proteins R-HSA-68882: Mitotic Anaphase R-HSA-2555396: Mitotic Metaphase and Anaphase R-HSA-68877: Mitotic Prometaphase R-HSA-68884: Mitotic Telophase/Cytokinesis R-HSA-597592: Post-translational protein modification R-HSA-2500257: Resolution of Sister Chromatid Cohesion R-HSA-69242: S Phase R-HSA-3108232: SUMO E3 ligases SUMOylate target proteins R-HSA-2990846: SUMOylation R-HSA-3108214: SUMOylation of DNA damage response and repair proteins R-HSA-2467813: Separation of Sister Chromatids |
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
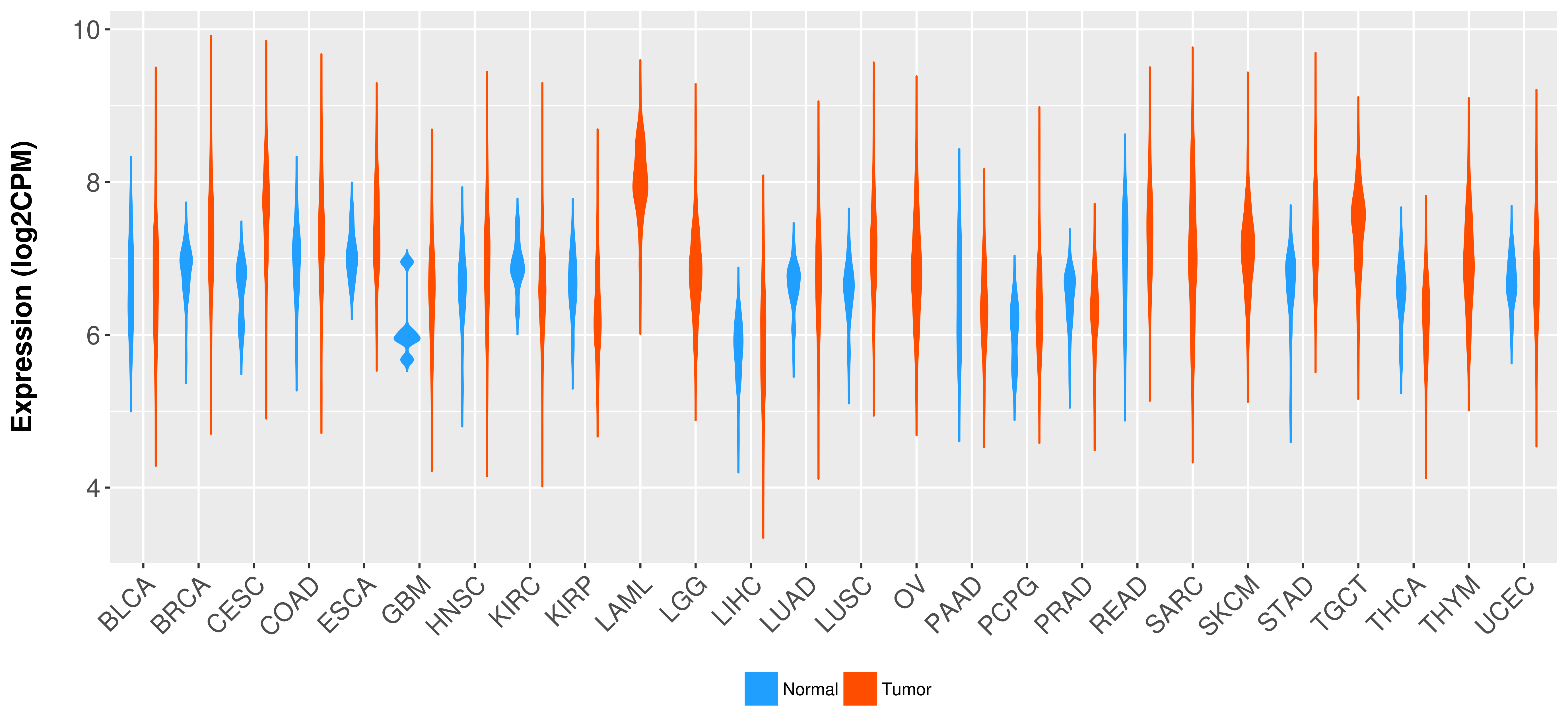
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
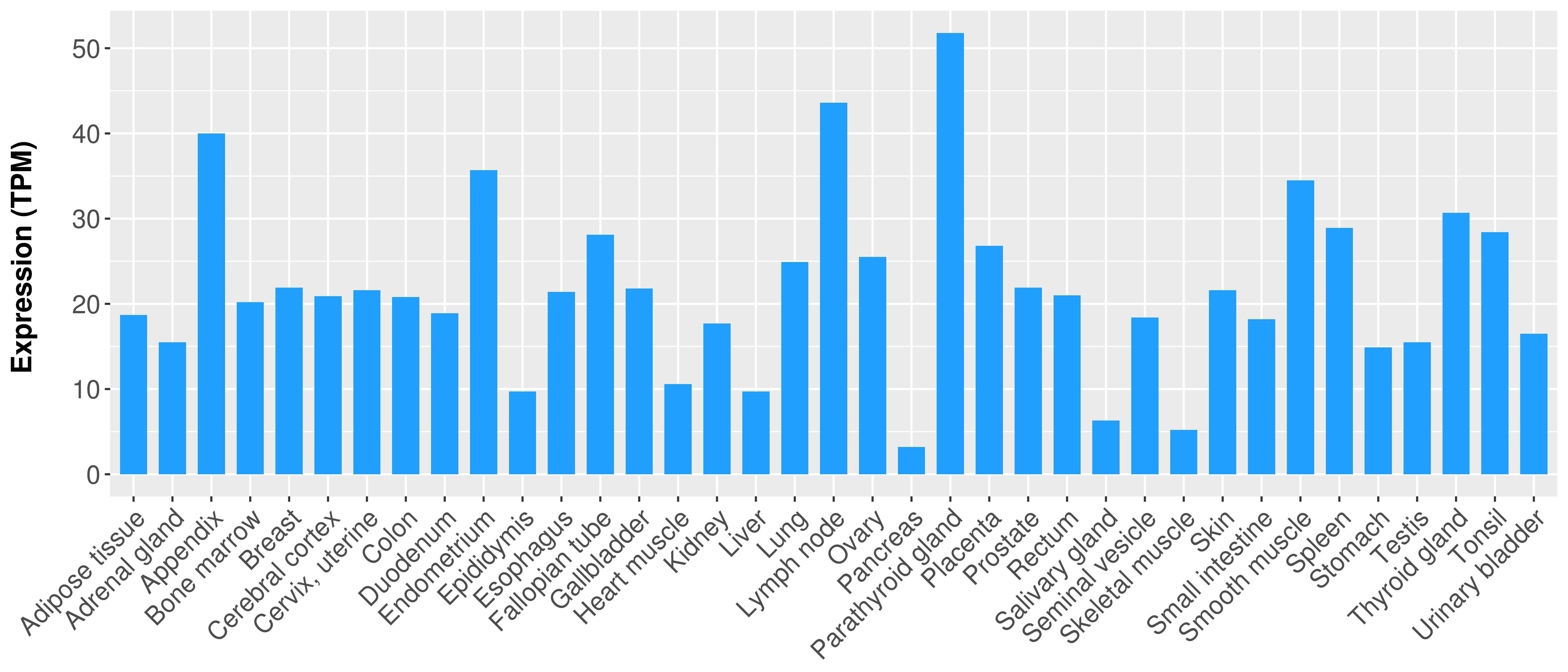
|
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
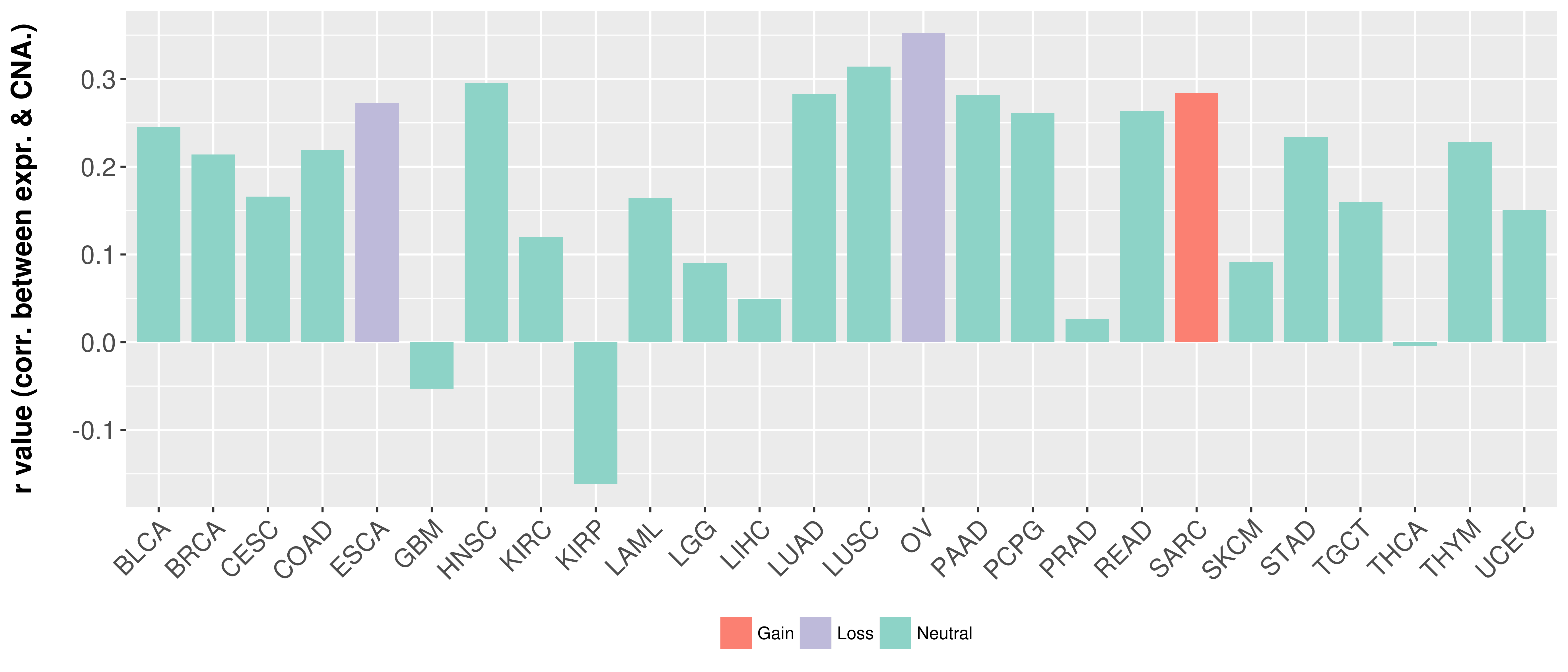
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
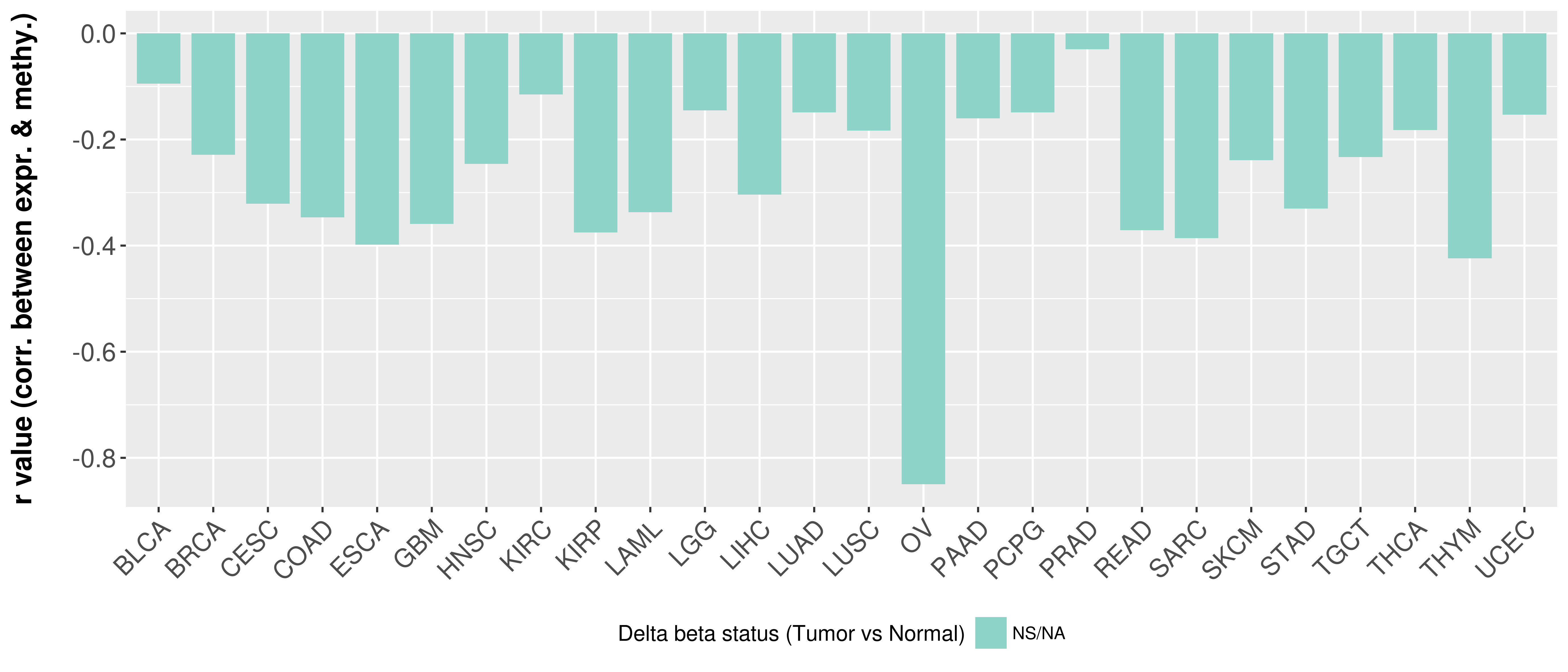
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
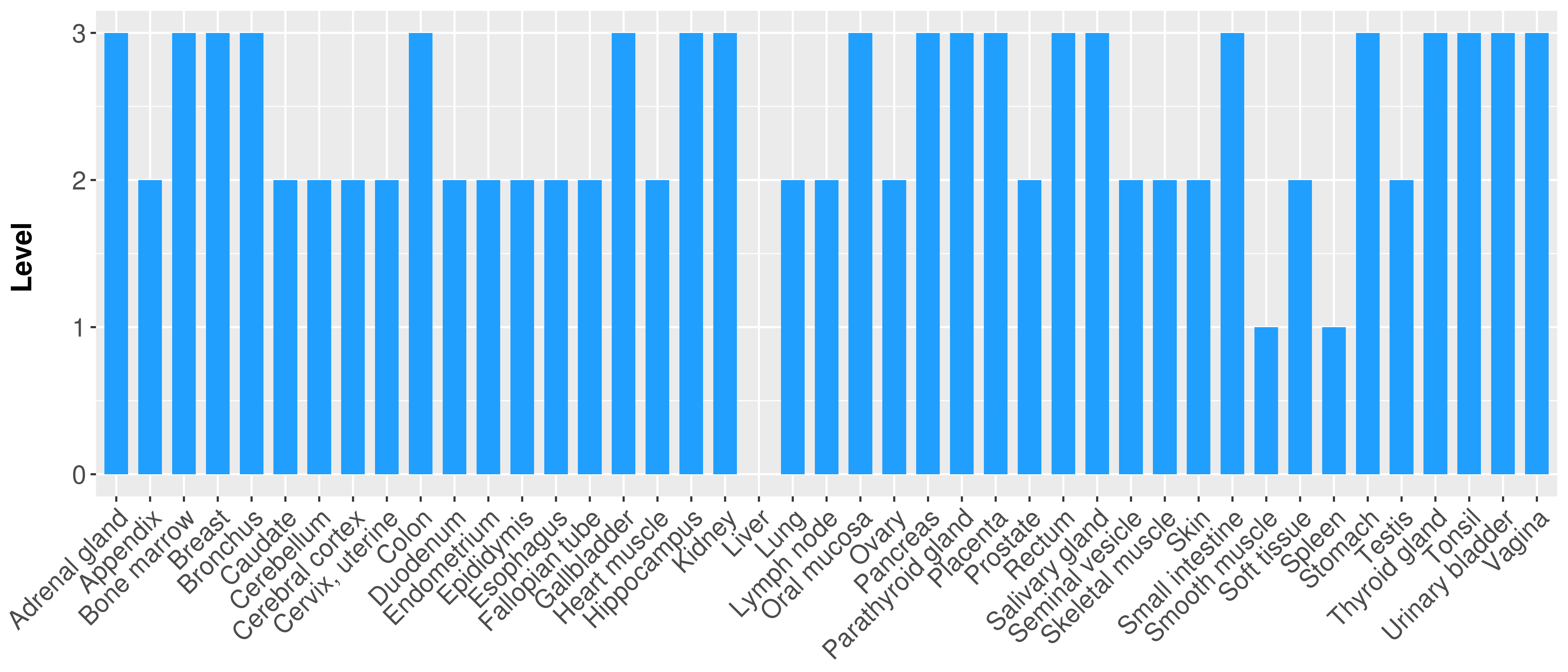
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
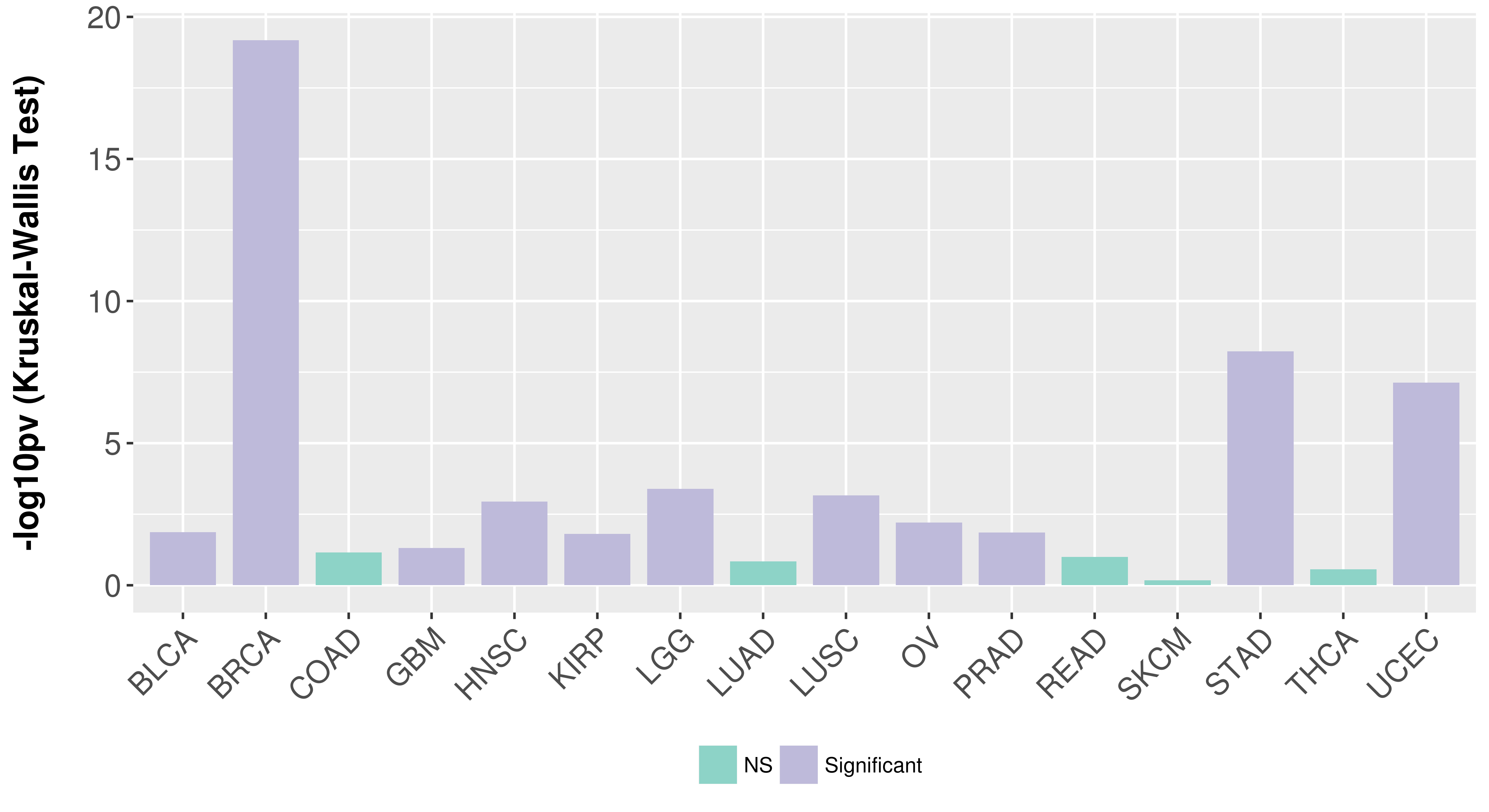
Association between expresson and subtype.
|
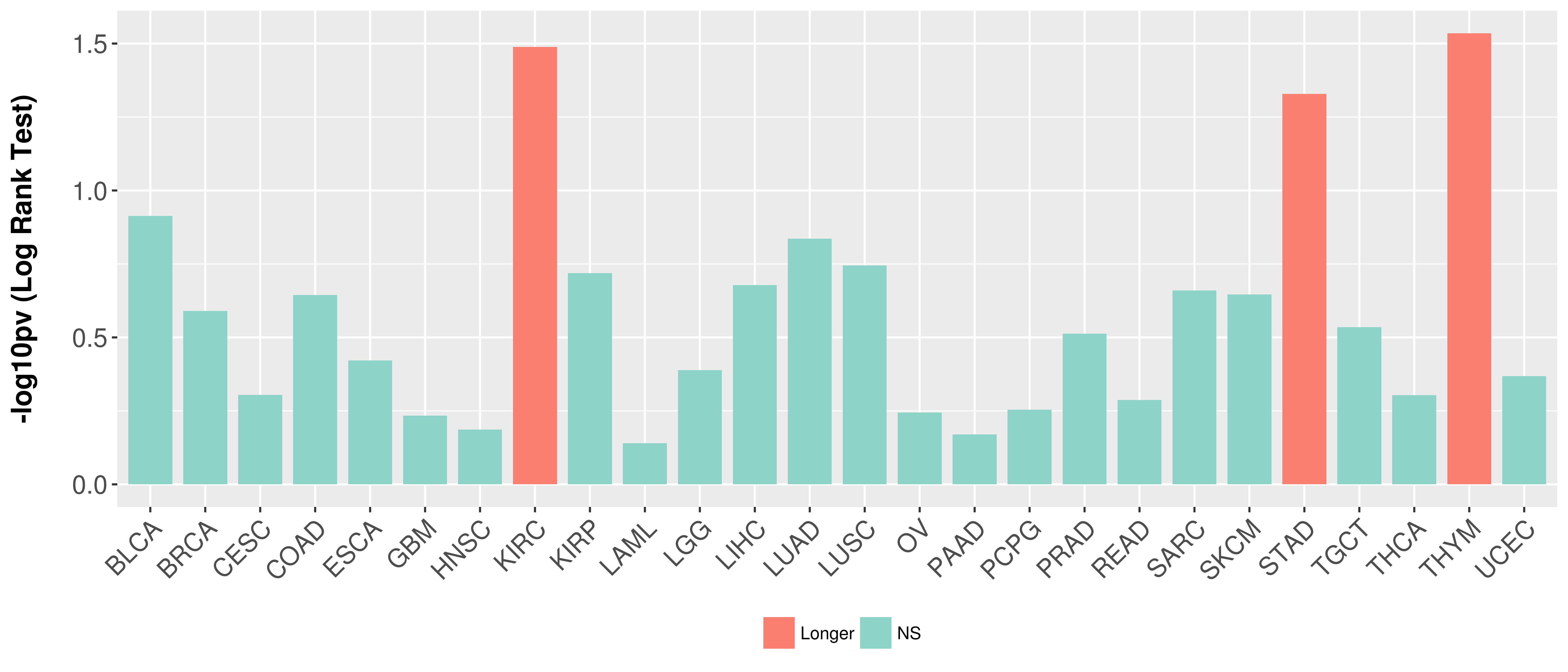
Overall survival analysis based on expression.
|

Association between expresson and stage.
|
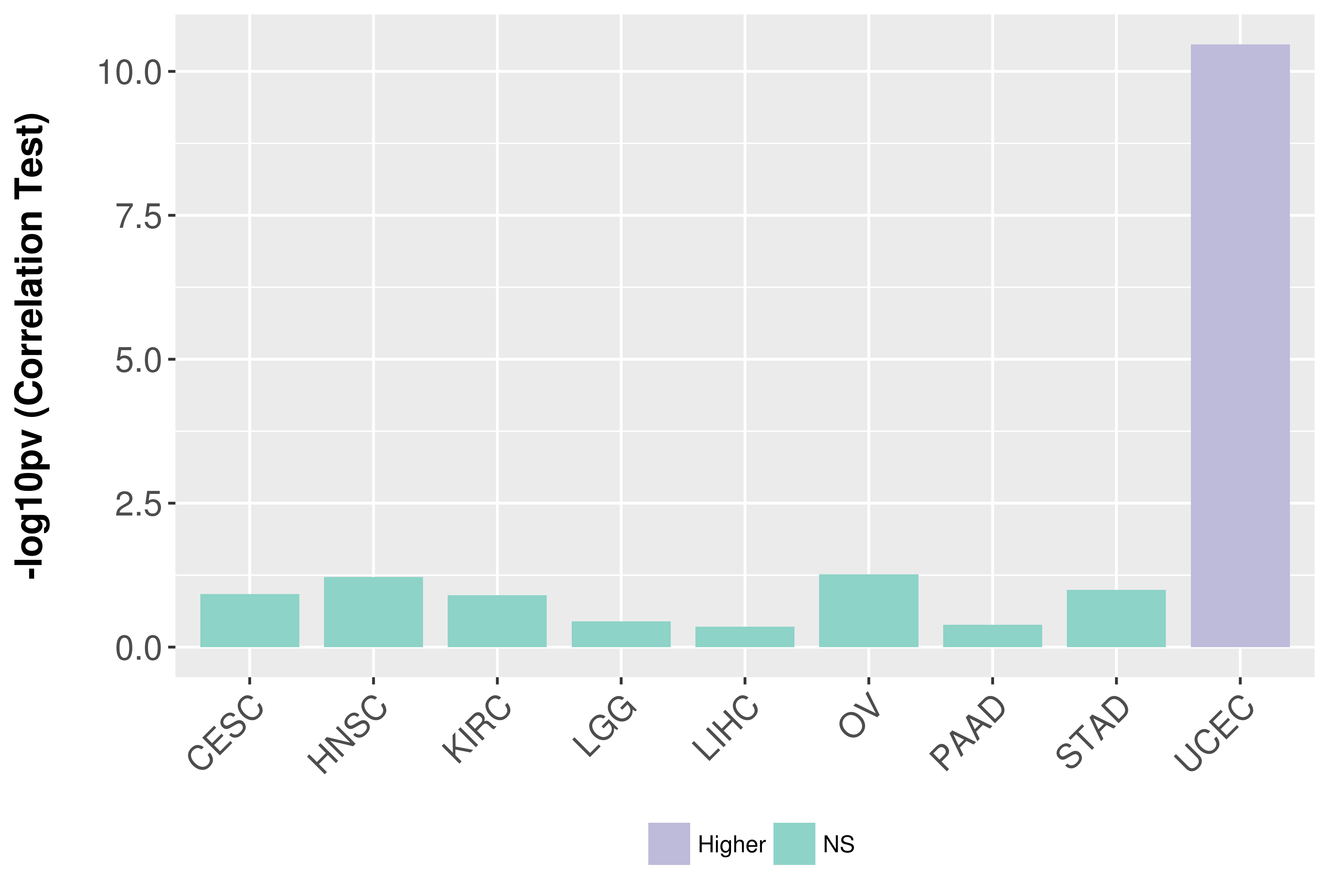
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SMC1A. |
| Summary | |
|---|---|
| Symbol | SMC1A |
| Name | structural maintenance of chromosomes 1A |
| Aliases | DXS423E; KIAA0178; SB1.8; Smcb; SMC1L1; SMC1 structural maintenance of chromosomes 1-like 1 (yeast); CDLS2; ...... |
| Location | Xp11.22 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|