Browse SMN2 in pancancer
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF06003 Survival motor neuron protein (SMN) |
||||||||||
| Function |
The SMN complex plays a catalyst role in the assembly of small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Thereby, plays an important role in the splicing of cellular pre-mRNAs. Most spliceosomal snRNPs contain a common set of Sm proteins SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP. In the cytosol, the Sm proteins SNRPD1, SNRPD2, SNRPE, SNRPF and SNRPG are trapped in an inactive 6S pICln-Sm complex by the chaperone CLNS1A that controls the assembly of the core snRNP. Dissociation by the SMN complex of CLNS1A from the trapped Sm proteins and their transfer to an SMN-Sm complex triggers the assembly of core snRNPs and their transport to the nucleus. Ensures the correct splicing of U12 intron-containing genes that may be important for normal motor and proprioceptive neurons development. Also required for resolving RNA-DNA hybrids created by RNA polymerase II, that form R-loop in transcription terminal regions, an important step in proper transcription termination. May also play a role in the metabolism of small nucleolar ribonucleoprotein (snoRNPs). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000245 spliceosomal complex assembly GO:0000375 RNA splicing, via transesterification reactions GO:0000377 RNA splicing, via transesterification reactions with bulged adenosine as nucleophile GO:0000387 spliceosomal snRNP assembly GO:0000398 mRNA splicing, via spliceosome GO:0006353 DNA-templated transcription, termination GO:0006397 mRNA processing GO:0006913 nucleocytoplasmic transport GO:0008380 RNA splicing GO:0022613 ribonucleoprotein complex biogenesis GO:0022618 ribonucleoprotein complex assembly GO:0051169 nuclear transport GO:0051170 nuclear import GO:0071826 ribonucleoprotein complex subunit organization |
| Molecular Function | - |
| Cellular Component |
GO:0015030 Cajal body GO:0016604 nuclear body GO:0030016 myofibril GO:0030017 sarcomere GO:0030018 Z disc GO:0031674 I band GO:0032797 SMN complex GO:0034719 SMN-Sm protein complex GO:0035770 ribonucleoprotein granule GO:0036464 cytoplasmic ribonucleoprotein granule GO:0043025 neuronal cell body GO:0043204 perikaryon GO:0043292 contractile fiber GO:0044297 cell body GO:0044449 contractile fiber part GO:0097504 Gemini of coiled bodies |
| KEGG |
hsa03013 RNA transport |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record for SMN2. |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
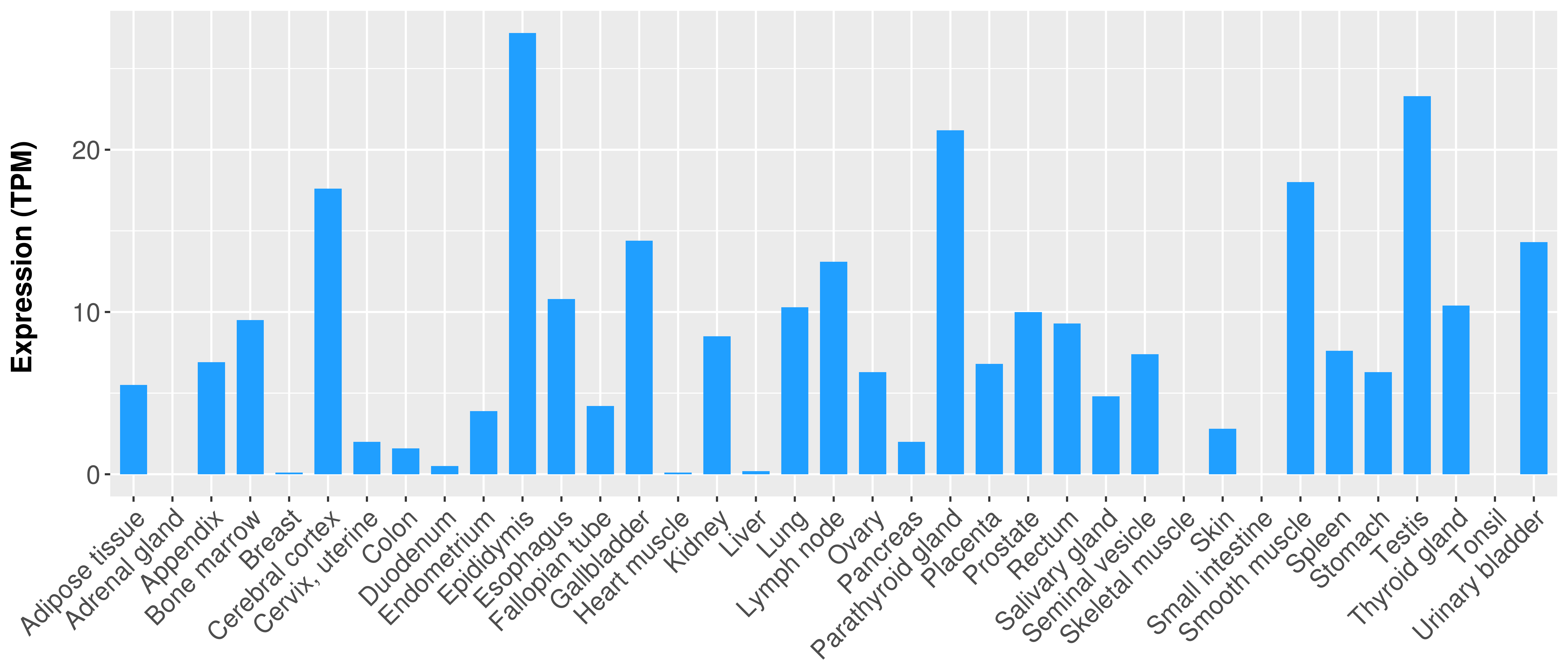
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
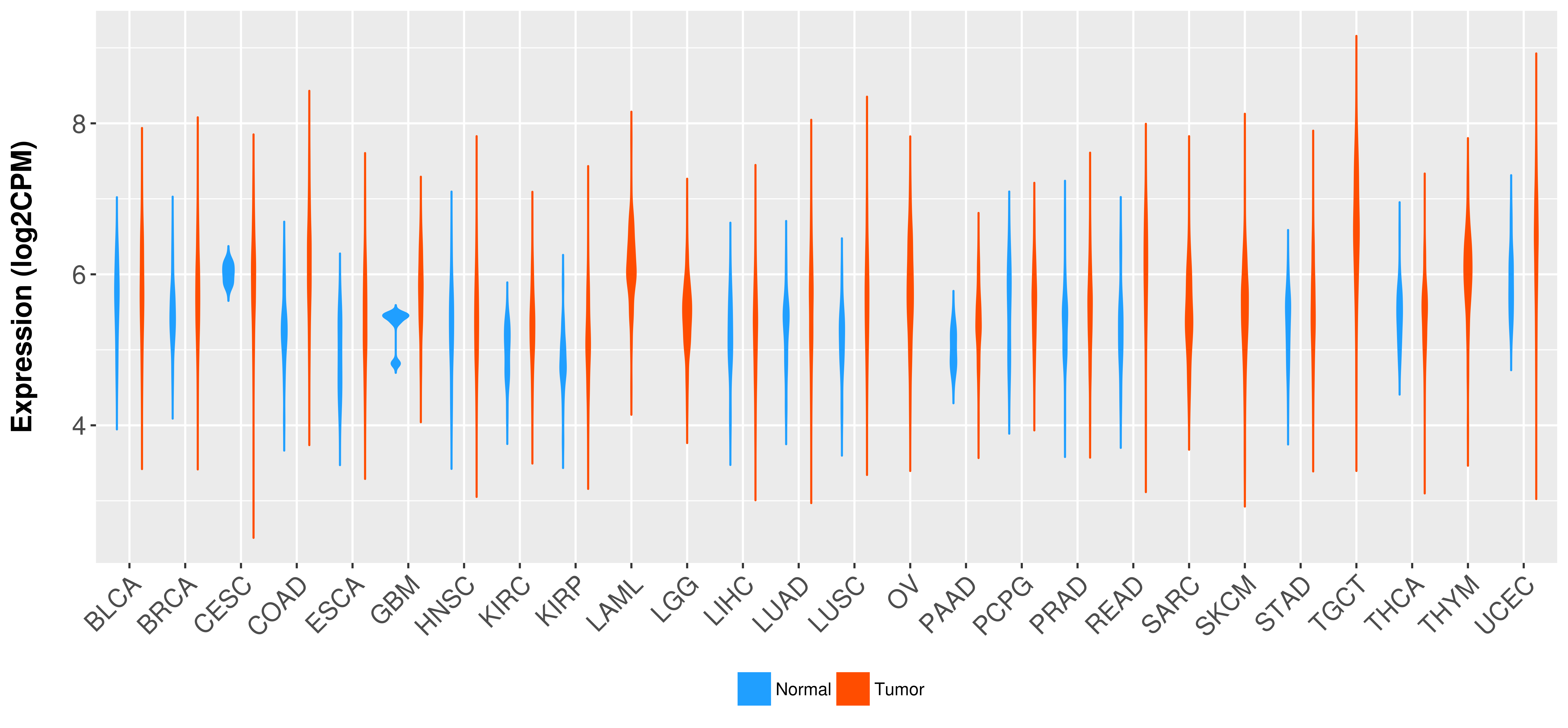
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |

Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
| There is no record. |
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
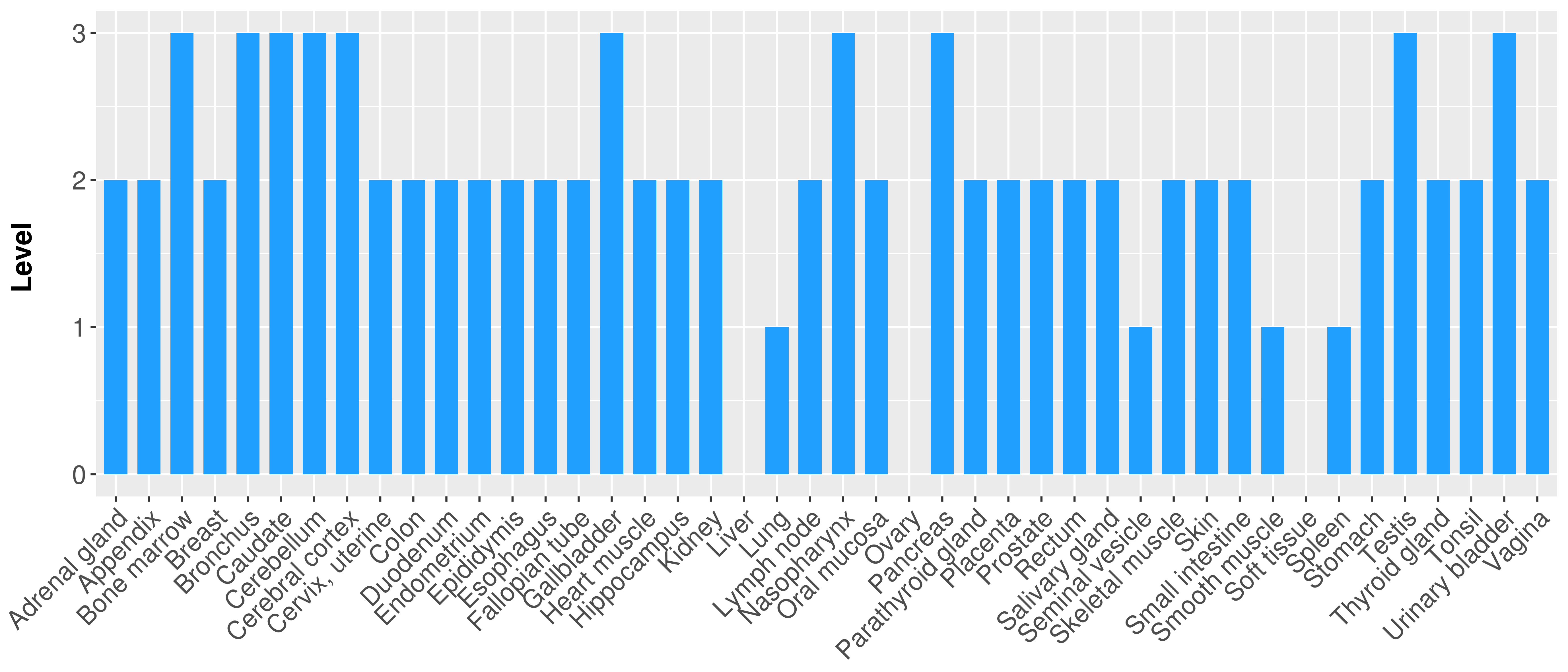
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
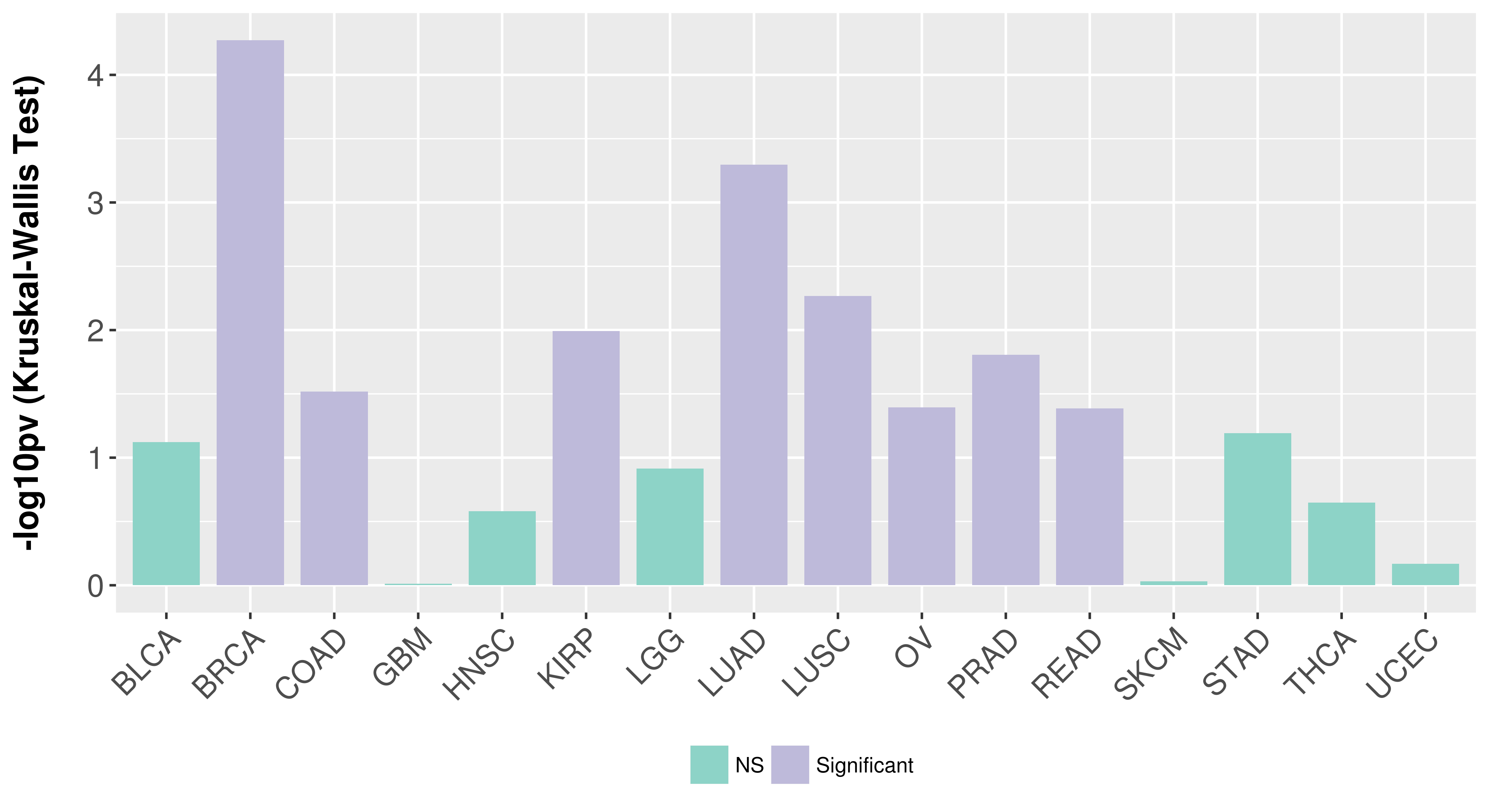
Association between expresson and subtype.
|
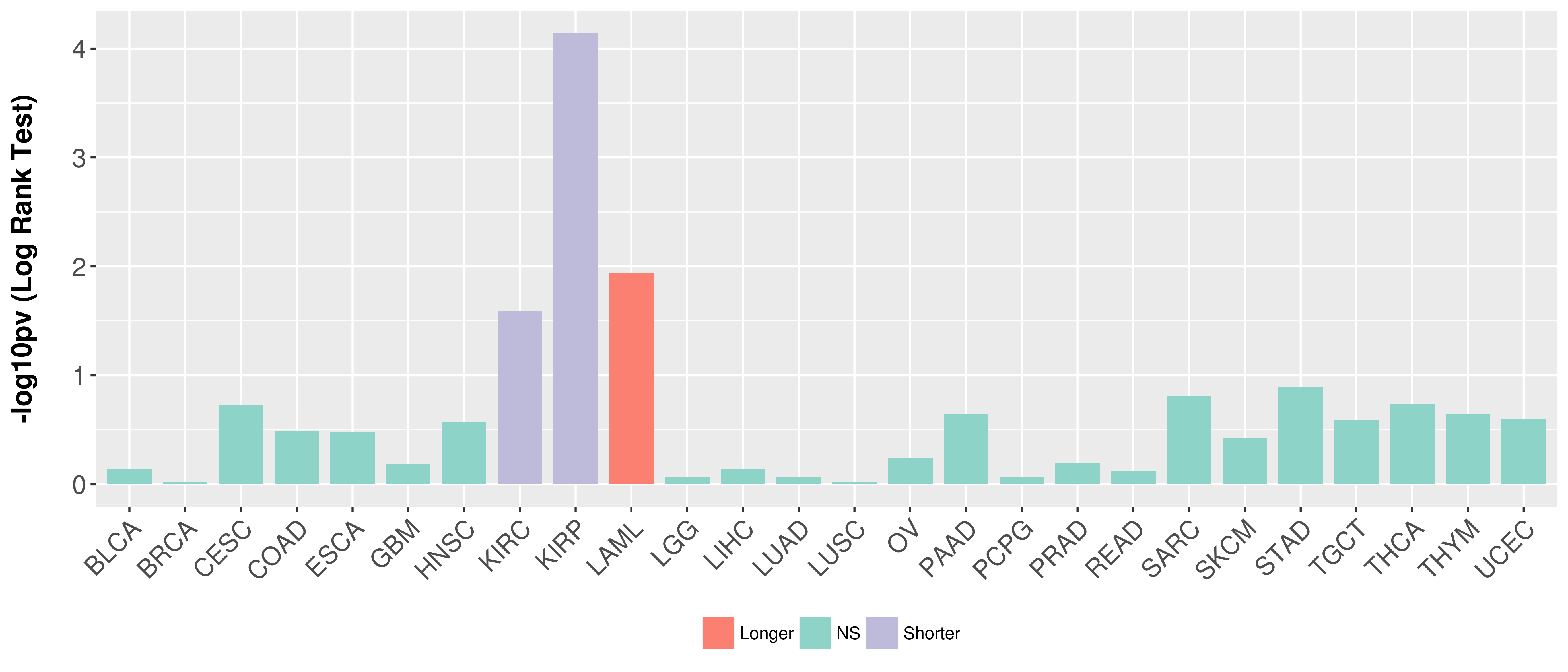
Overall survival analysis based on expression.
|
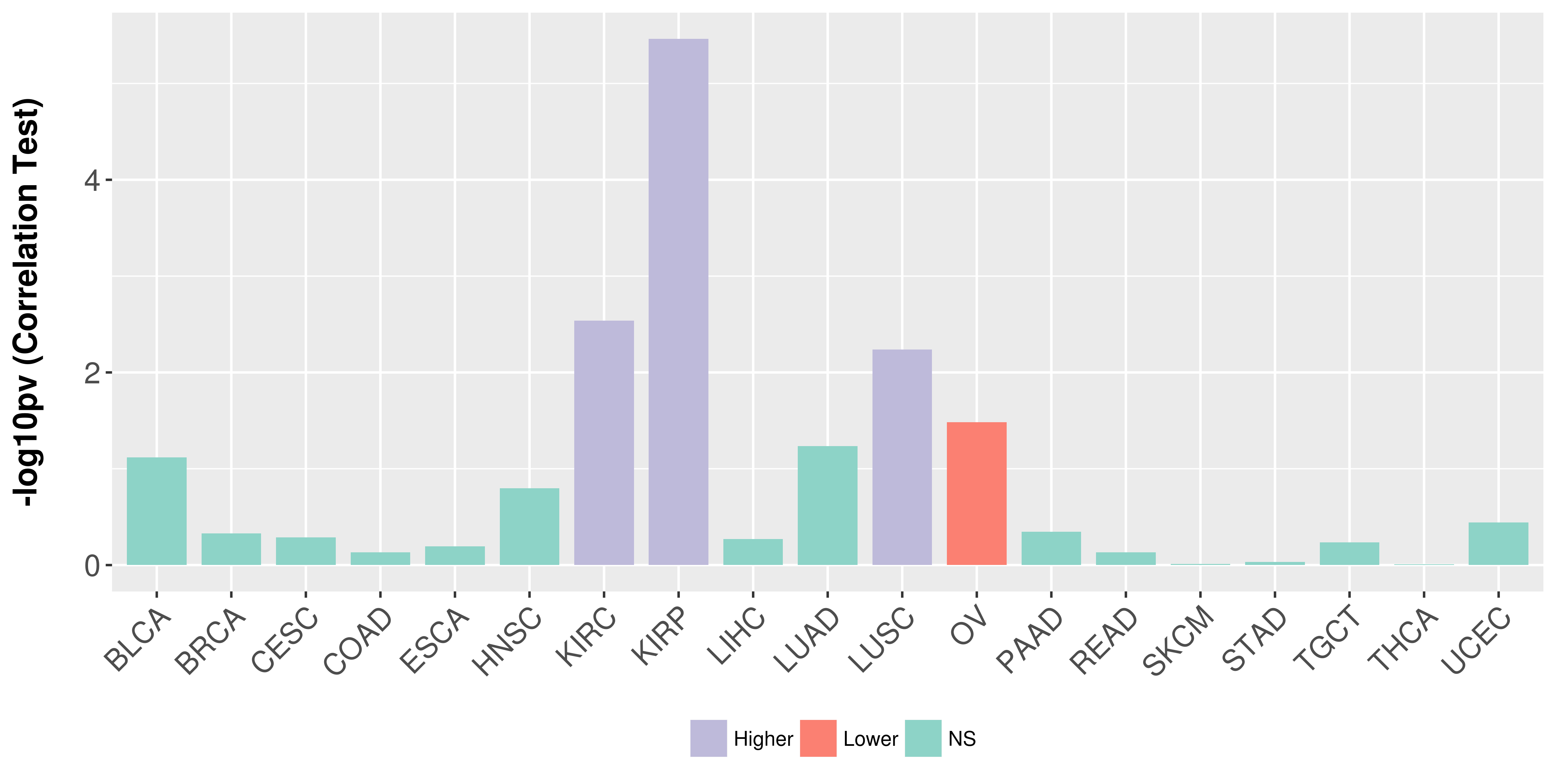
Association between expresson and stage.
|
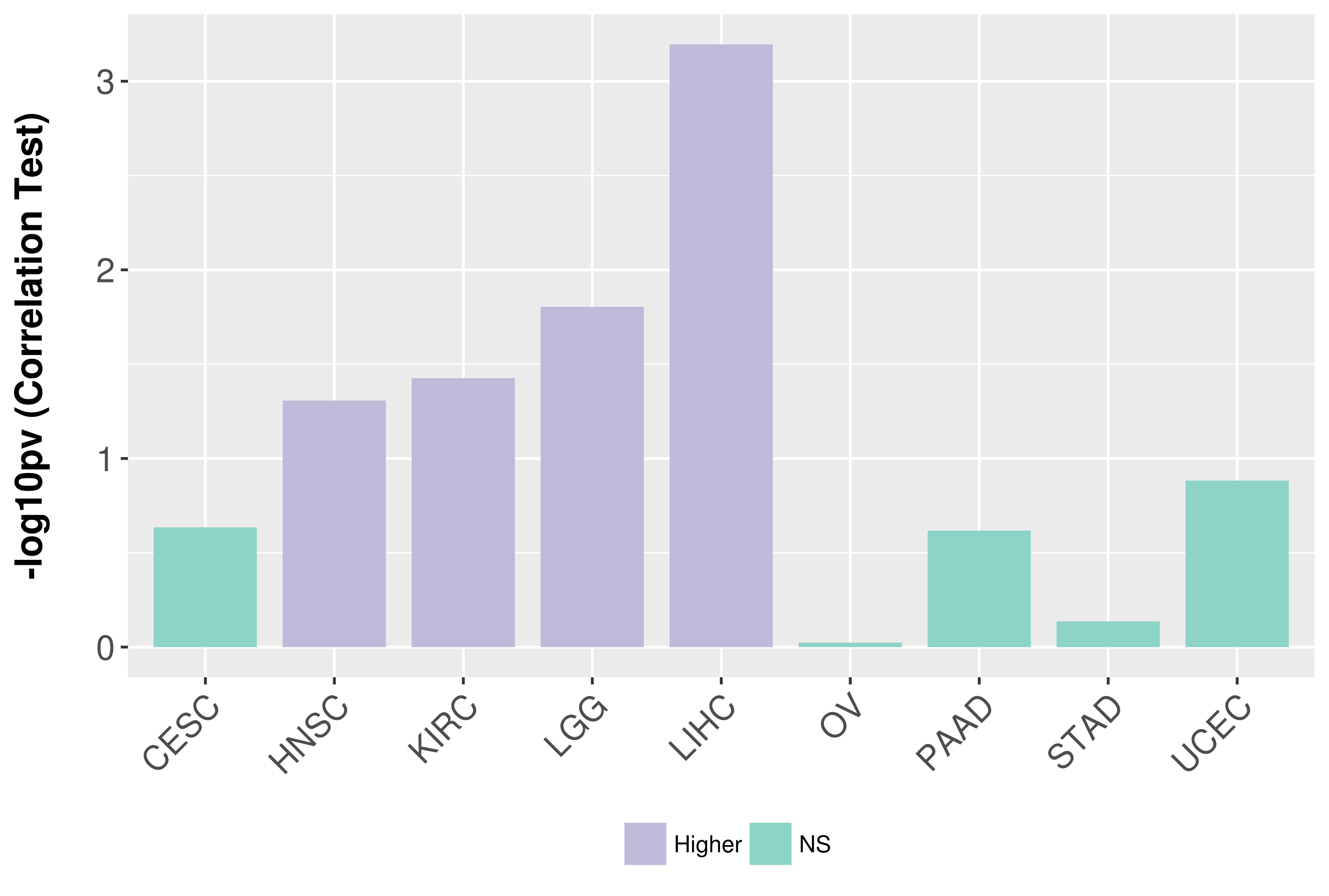
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SMN2. |
| Summary | |
|---|---|
| Symbol | SMN2 |
| Name | survival of motor neuron 2, centromeric |
| Aliases | SMNC; TDRD16B; tudor domain containing 16B; C-BCD541 |
| Location | 5q13.2 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|