Browse SP100 in pancancer
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00505 HMG (high mobility group) box PF09011 HMG-box domain PF03172 HSR domain PF01342 SAND domain |
||||||||||
| Function |
Together with PML, this tumor suppressor is a major constituent of the PML bodies, a subnuclear organelle involved in a large number of physiological processes including cell growth, differentiation and apoptosis. Functions as a transcriptional coactivator of ETS1 and ETS2 according to PubMed:11909962. Under certain conditions, it may also act as a corepressor of ETS1 preventing its binding to DNA according to PubMed:15247905. Through the regulation of ETS1 it may play a role in angiogenesis, controlling endothelial cell motility and invasion. Through interaction with the MRN complex it may be involved in the regulation of telomeres lengthening. May also regulate TP53-mediated transcription and through CASP8AP2, regulate FAS-mediated apoptosis. Also plays a role in infection by viruses, including human cytomegalovirus and Epstein-Barr virus, through mechanisms that may involve chromatin and/or transcriptional regulation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000723 telomere maintenance GO:0001525 angiogenesis GO:0001667 ameboidal-type cell migration GO:0006611 protein export from nucleus GO:0006913 nucleocytoplasmic transport GO:0006978 DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator GO:0008625 extrinsic apoptotic signaling pathway via death domain receptors GO:0010594 regulation of endothelial cell migration GO:0010596 negative regulation of endothelial cell migration GO:0010631 epithelial cell migration GO:0010632 regulation of epithelial cell migration GO:0010633 negative regulation of epithelial cell migration GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0030330 DNA damage response, signal transduction by p53 class mediator GO:0030336 negative regulation of cell migration GO:0032200 telomere organization GO:0032386 regulation of intracellular transport GO:0032387 negative regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0034340 response to type I interferon GO:0034341 response to interferon-gamma GO:0036337 Fas signaling pathway GO:0040013 negative regulation of locomotion GO:0042770 signal transduction in response to DNA damage GO:0042772 DNA damage response, signal transduction resulting in transcription GO:0043392 negative regulation of DNA binding GO:0043433 negative regulation of sequence-specific DNA binding transcription factor activity GO:0043542 endothelial cell migration GO:0045765 regulation of angiogenesis GO:0046822 regulation of nucleocytoplasmic transport GO:0046823 negative regulation of nucleocytoplasmic transport GO:0046825 regulation of protein export from nucleus GO:0046826 negative regulation of protein export from nucleus GO:0048514 blood vessel morphogenesis GO:0051051 negative regulation of transport GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051098 regulation of binding GO:0051100 negative regulation of binding GO:0051101 regulation of DNA binding GO:0051168 nuclear export GO:0051169 nuclear transport GO:0051224 negative regulation of protein transport GO:0051271 negative regulation of cellular component movement GO:0060249 anatomical structure homeostasis GO:0060333 interferon-gamma-mediated signaling pathway GO:0060337 type I interferon signaling pathway GO:0071346 cellular response to interferon-gamma GO:0071357 cellular response to type I interferon GO:0072331 signal transduction by p53 class mediator GO:0090130 tissue migration GO:0090132 epithelium migration GO:0090317 negative regulation of intracellular protein transport GO:0097191 extrinsic apoptotic signaling pathway GO:1901342 regulation of vasculature development GO:1902041 regulation of extrinsic apoptotic signaling pathway via death domain receptors GO:1902044 regulation of Fas signaling pathway GO:1903828 negative regulation of cellular protein localization GO:1904950 negative regulation of establishment of protein localization GO:2000146 negative regulation of cell motility GO:2001233 regulation of apoptotic signaling pathway GO:2001236 regulation of extrinsic apoptotic signaling pathway |
| Molecular Function |
GO:0003713 transcription coactivator activity GO:0003714 transcription corepressor activity GO:0005125 cytokine activity GO:0070087 chromo shadow domain binding |
| Cellular Component |
GO:0000781 chromosome, telomeric region GO:0000784 nuclear chromosome, telomeric region GO:0016604 nuclear body GO:0016605 PML body GO:0030870 Mre11 complex GO:0044454 nuclear chromosome part GO:0098687 chromosomal region |
| KEGG | - |
| Reactome |
R-HSA-1280215: Cytokine Signaling in Immune system R-HSA-168256: Immune System R-HSA-913531: Interferon Signaling R-HSA-877300: Interferon gamma signaling R-HSA-392499: Metabolism of proteins R-HSA-597592: Post-translational protein modification R-HSA-3108232: SUMO E3 ligases SUMOylate target proteins R-HSA-2990846: SUMOylation R-HSA-3108214: SUMOylation of DNA damage response and repair proteins |
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for SP100. |
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
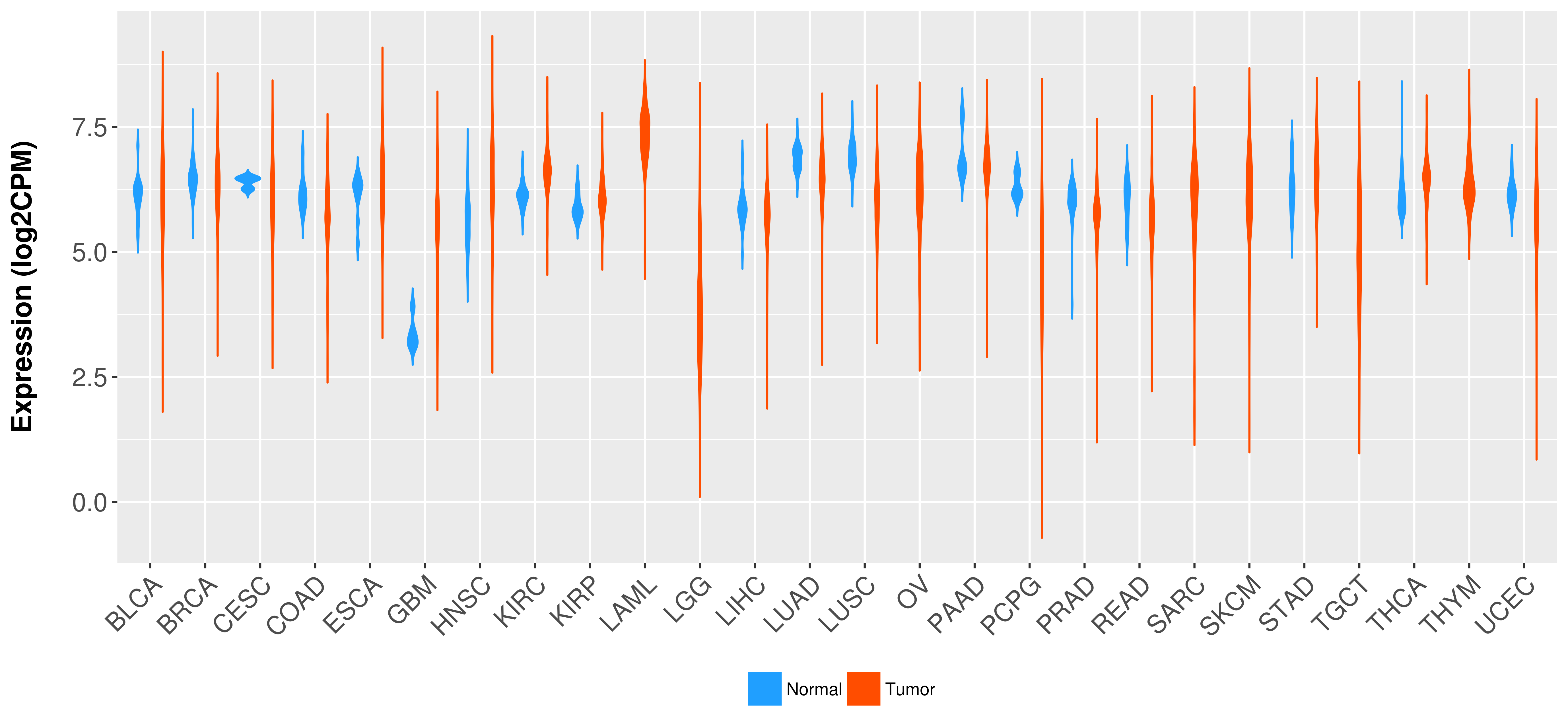
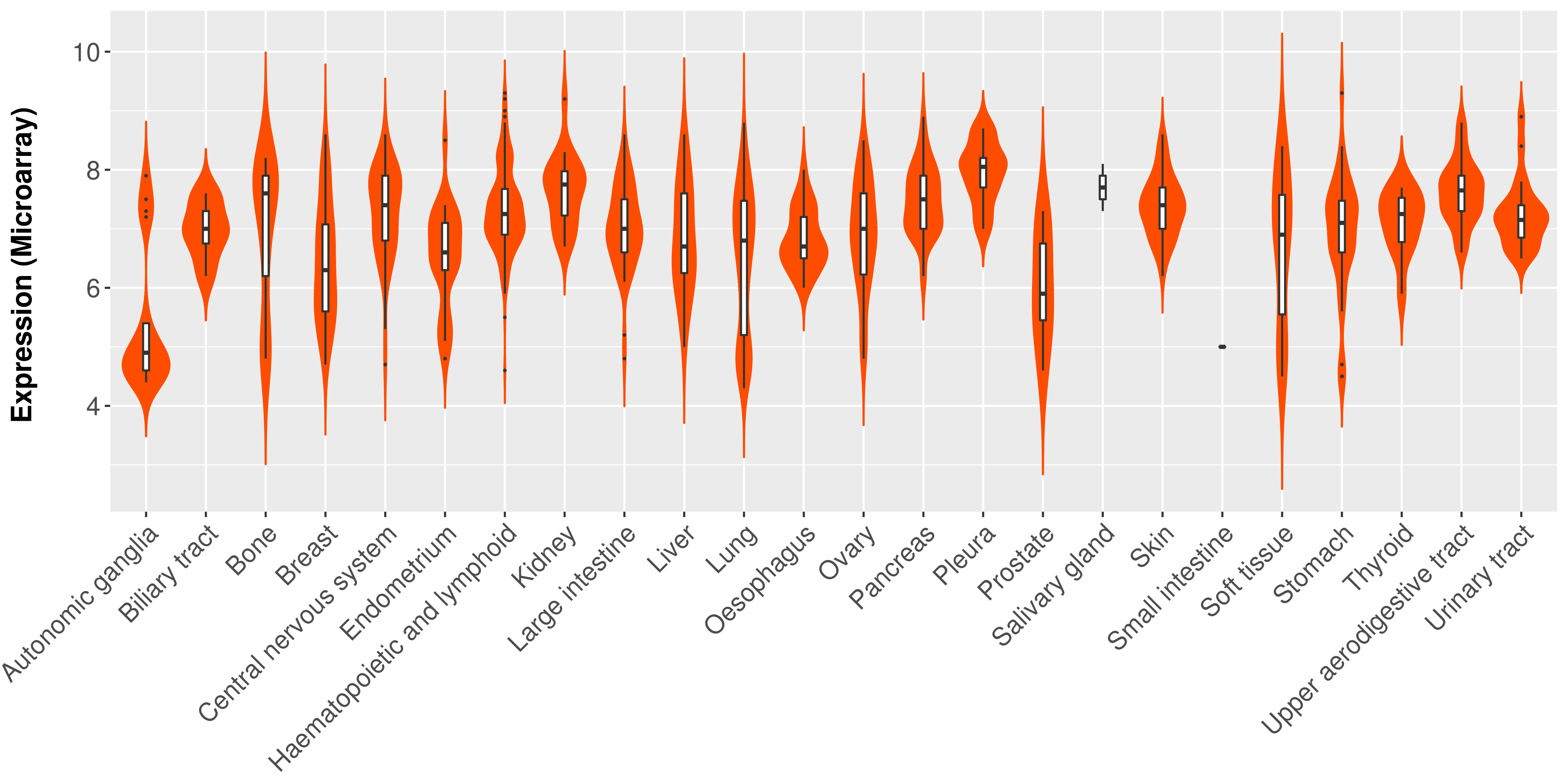
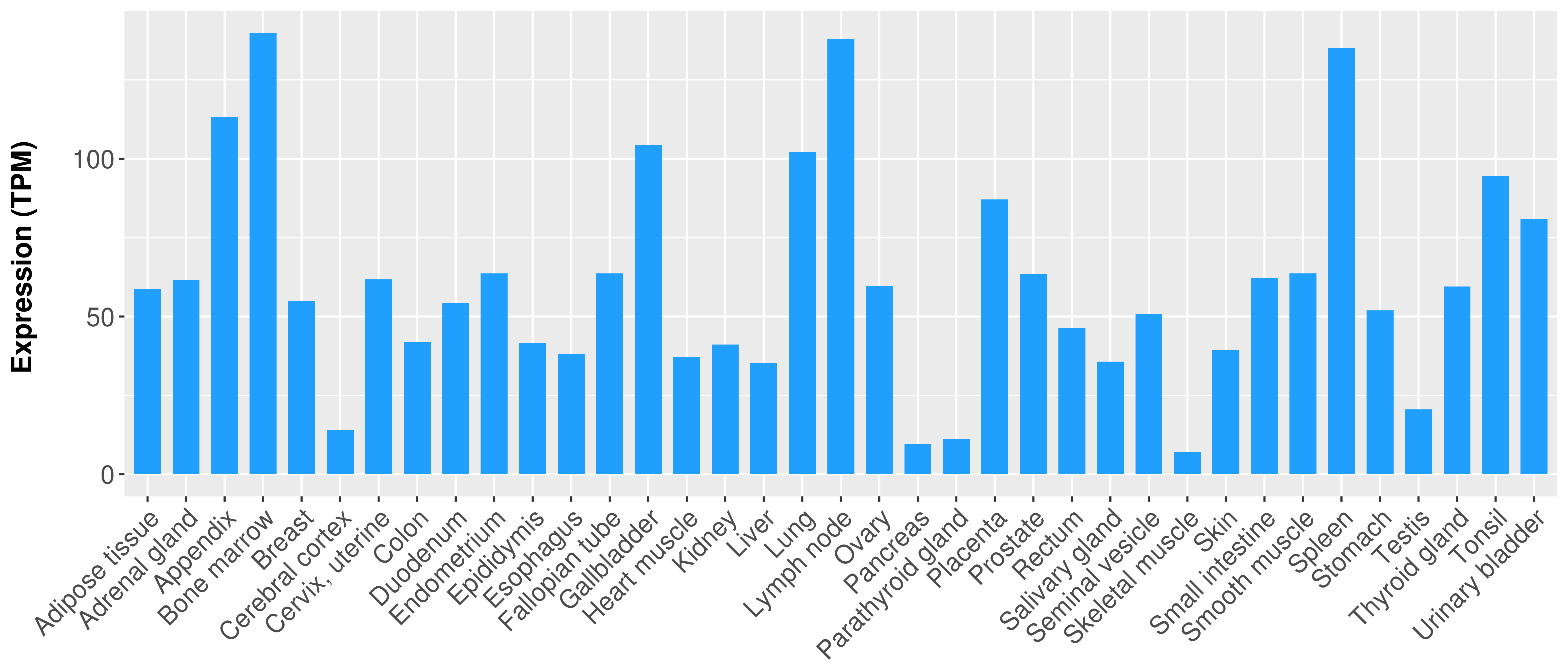
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
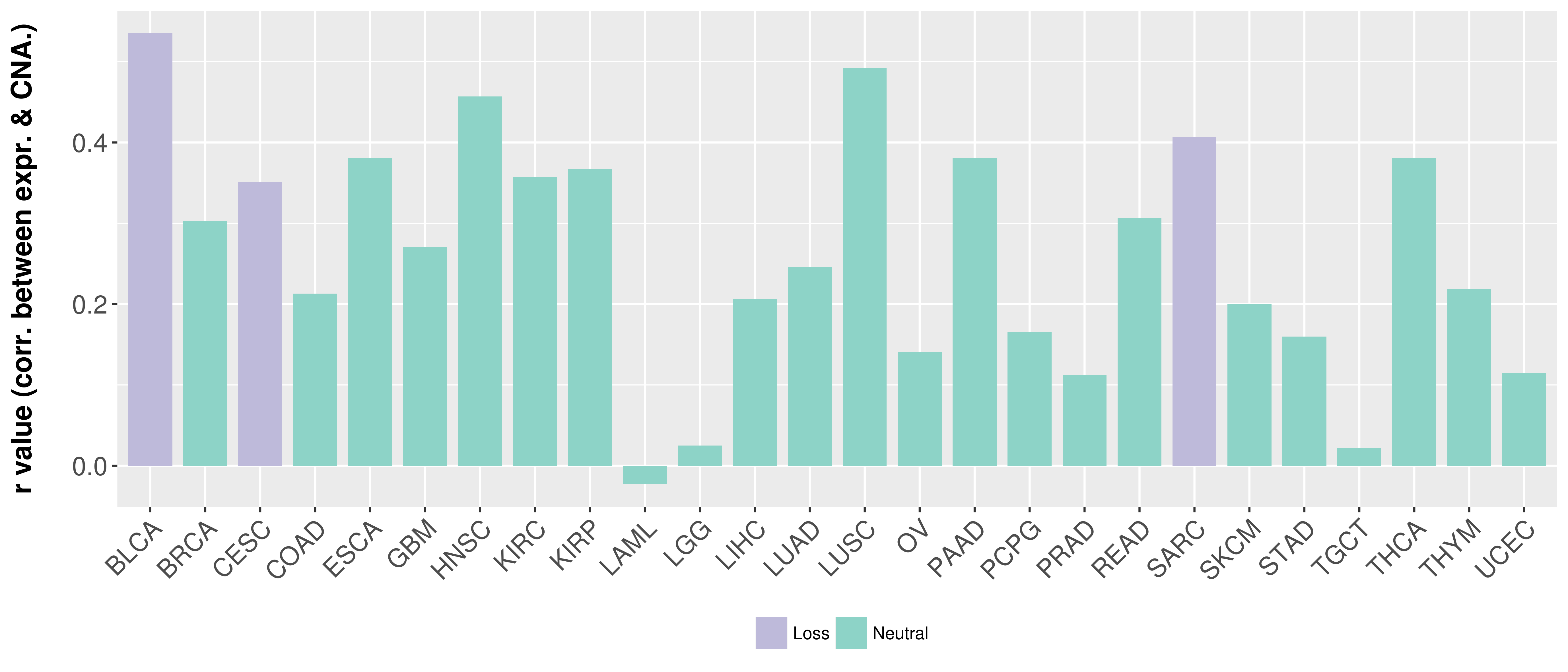
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
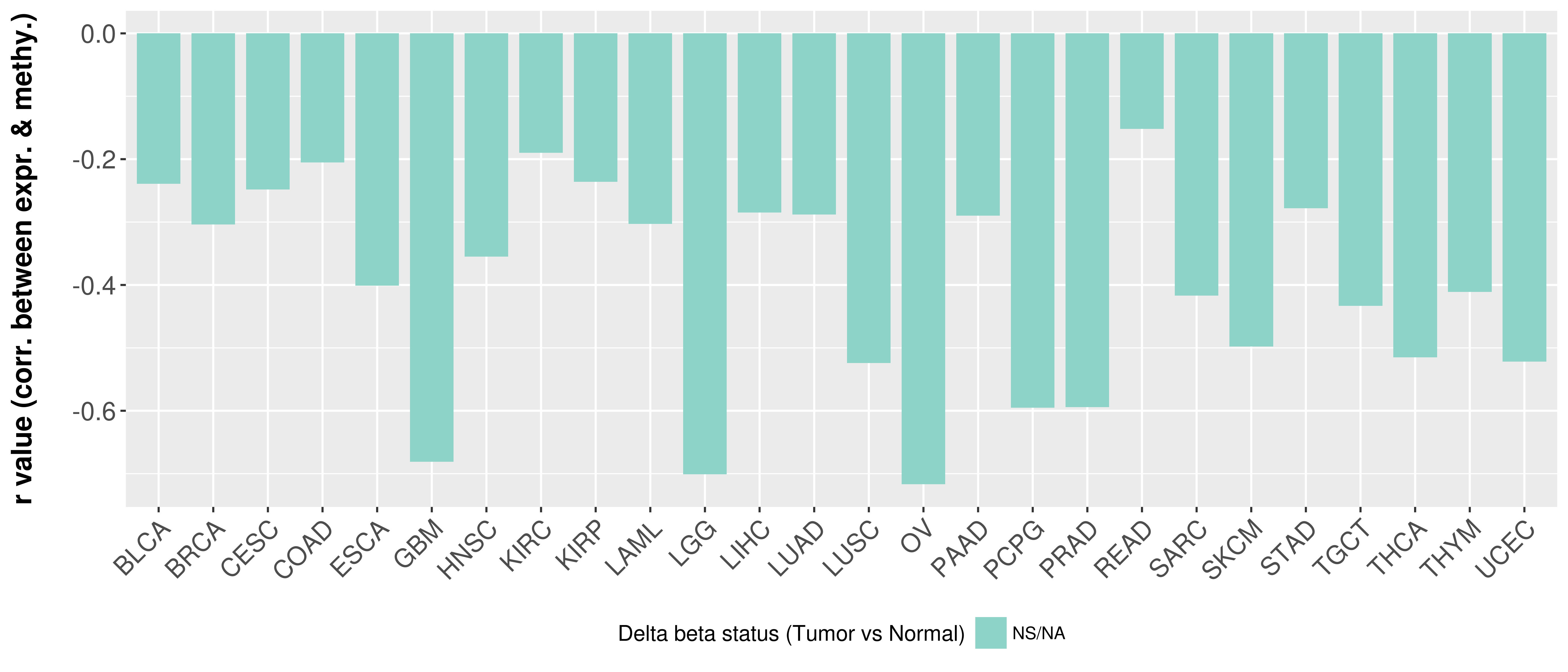
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
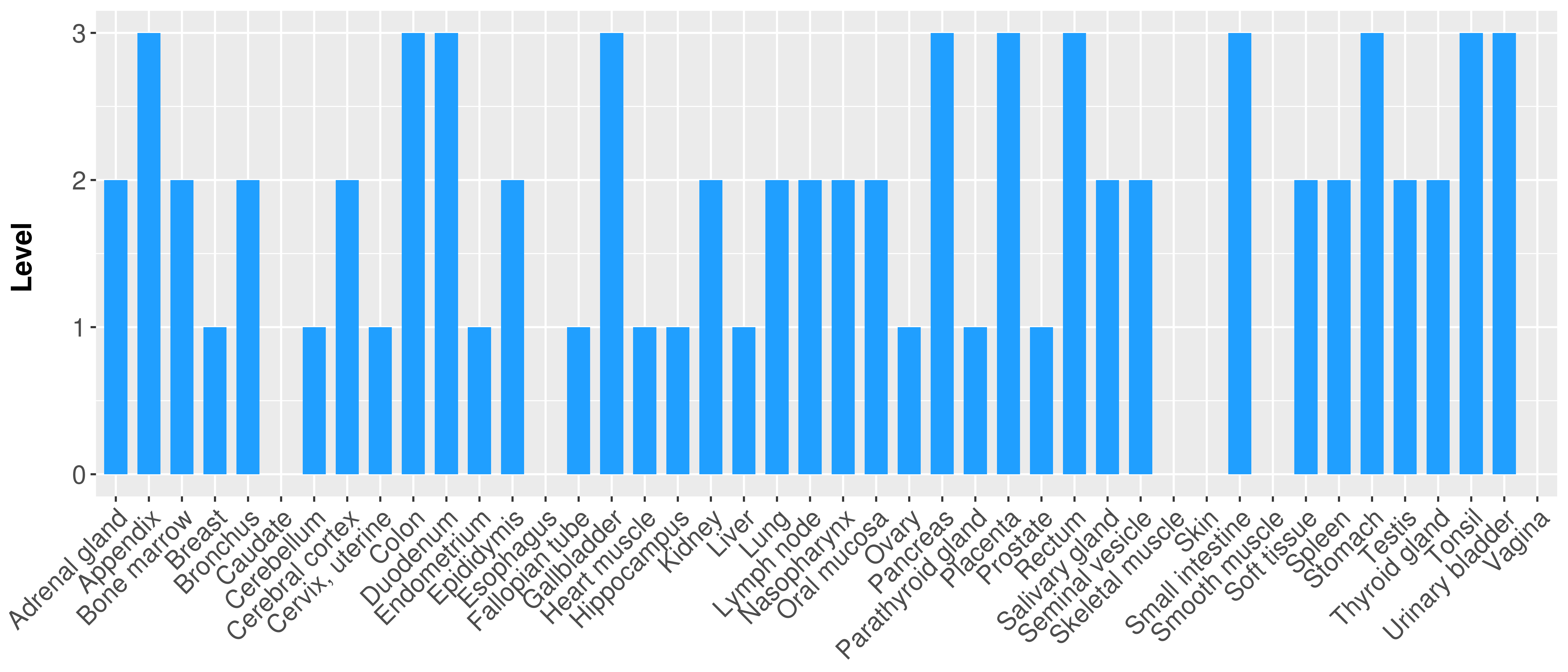
|
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
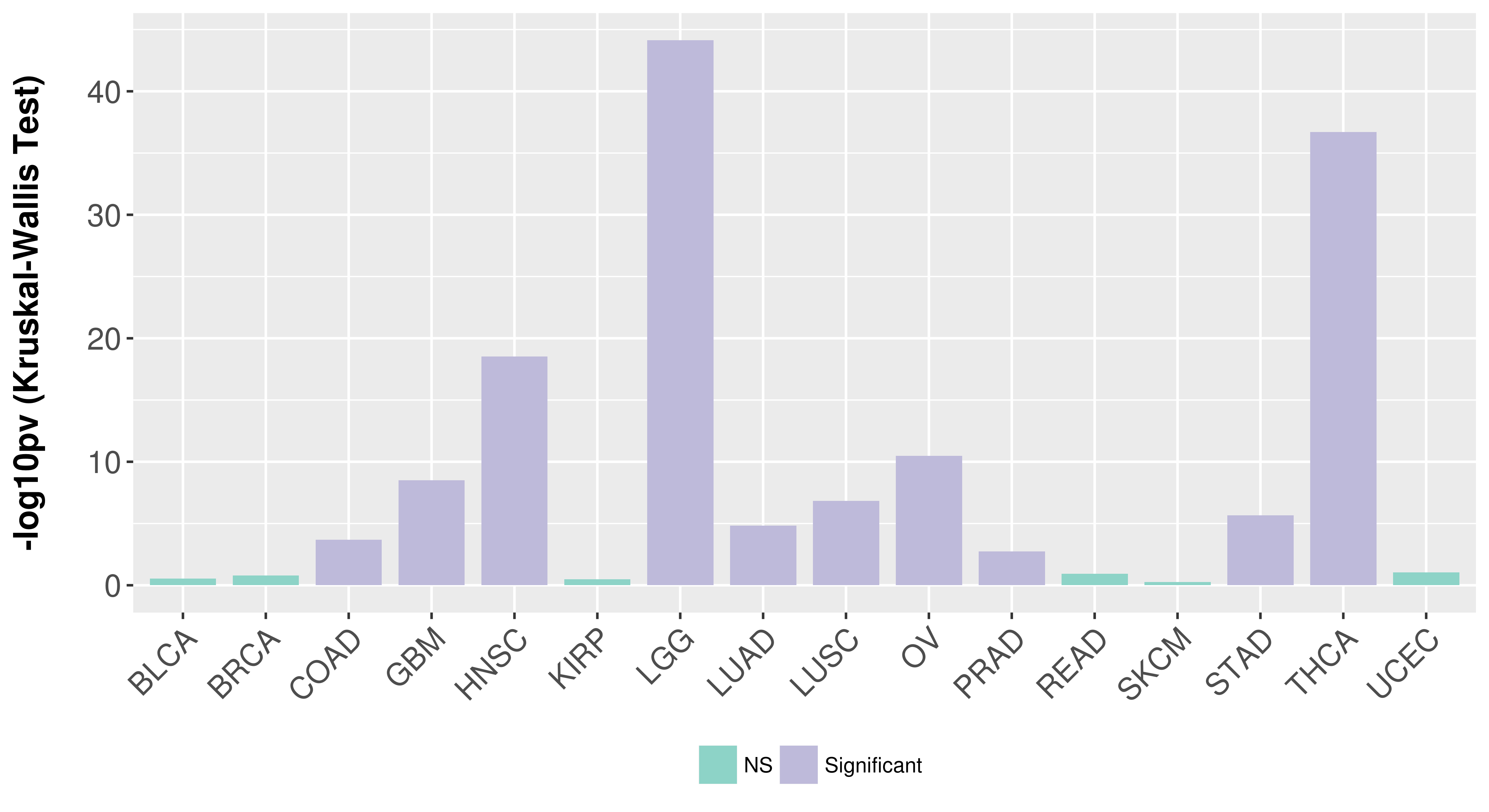
Association between expresson and subtype.
|
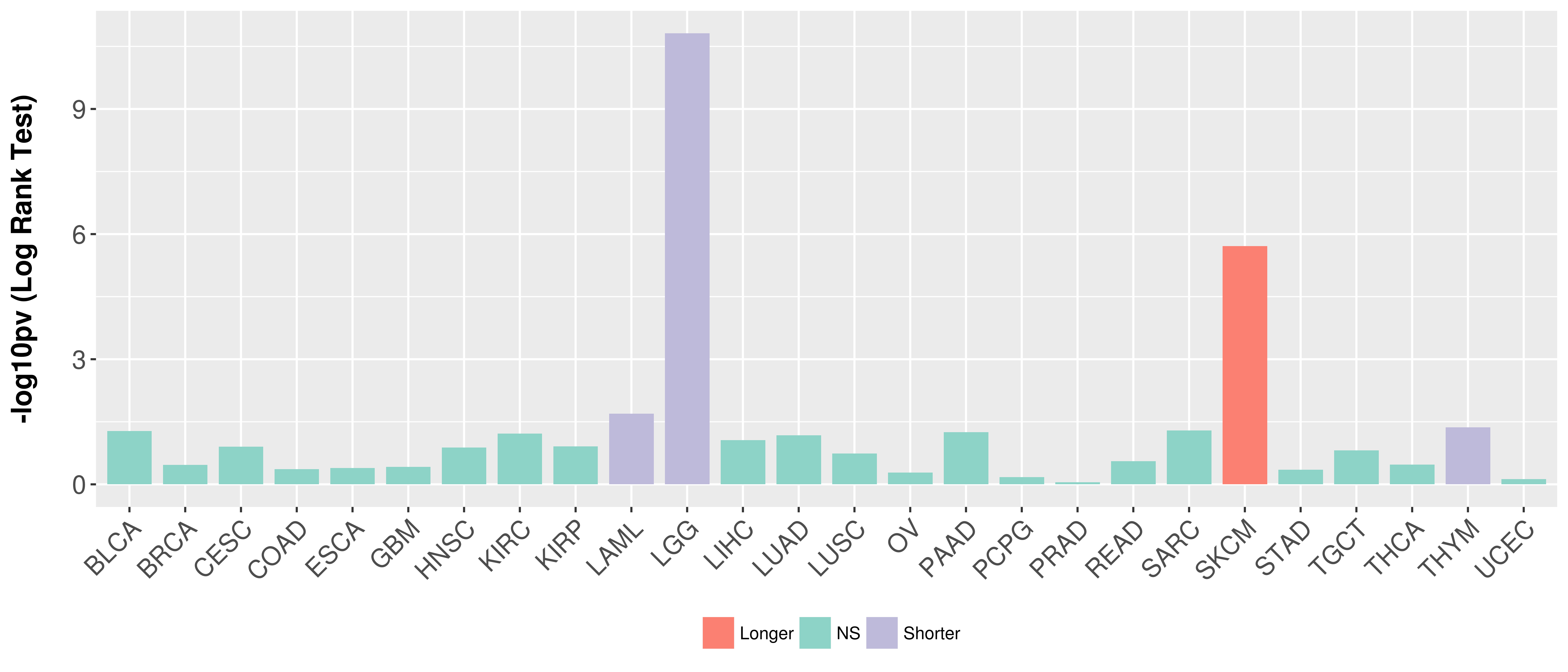
Overall survival analysis based on expression.
|

Association between expresson and stage.
|
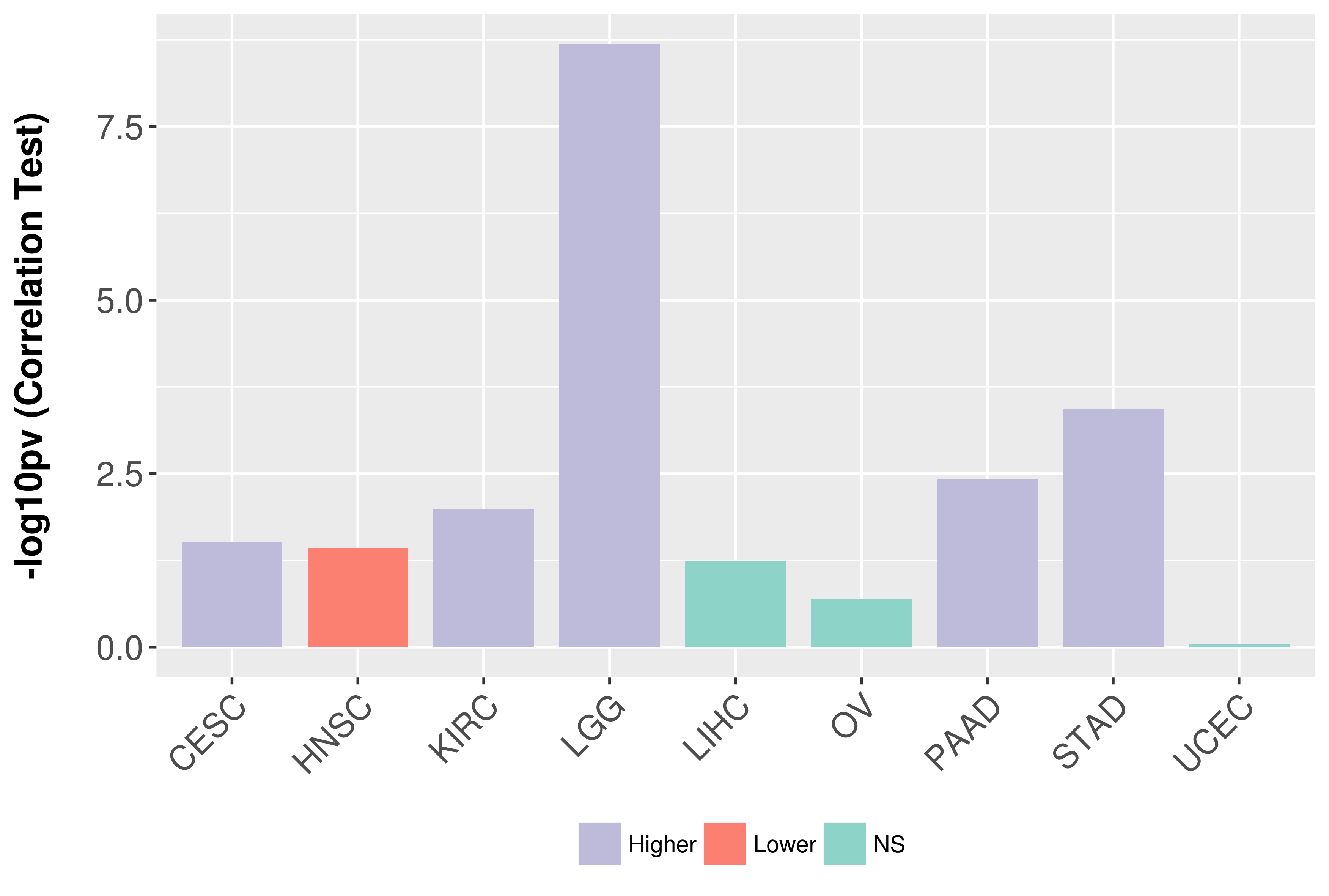
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for SP100. |
| Summary | |
|---|---|
| Symbol | SP100 |
| Name | SP100 nuclear antigen |
| Aliases | nuclear antigen Sp100; lysp100b; SP100-HMG nuclear autoantigen; nuclear dot-associated Sp100 protein; speckl ...... |
| Location | 2q37.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for SP100. |