Browse TDG in pancancer
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF03167 Uracil DNA glycosylase superfamily |
||||||||||
| Function |
DNA glycosylase that plays a key role in active DNA demethylation: specifically recognizes and binds 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in the context of CpG sites and mediates their excision through base-excision repair (BER) to install an unmethylated cytosine. Cannot remove 5-hydroxymethylcytosine (5hmC). According to an alternative model, involved in DNA demethylation by mediating DNA glycolase activity toward 5-hydroxymethyluracil (5hmU) produced by deamination of 5hmC. Also involved in DNA repair by acting as a thymine-DNA glycosylase that mediates correction of G/T mispairs to G/C pairs: in the DNA of higher eukaryotes, hydrolytic deamination of 5-methylcytosine to thymine leads to the formation of G/T mismatches. Its role in the repair of canonical base damage is however minor compared to its role in DNA demethylation. It is capable of hydrolyzing the carbon-nitrogen bond between the sugar-phosphate backbone of the DNA and a mispaired thymine. In addition to the G/T, it can remove thymine also from C/T and T/T mispairs in the order G/T >> C/T > T/T. It has no detectable activity on apyrimidinic sites and does not catalyze the removal of thymine from A/T pairs or from single-stranded DNA. It can also remove uracil and 5-bromouracil from mispairs with guanine. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006220 pyrimidine nucleotide metabolic process GO:0006244 pyrimidine nucleotide catabolic process GO:0006284 base-excision repair GO:0006285 base-excision repair, AP site formation GO:0006298 mismatch repair GO:0006304 DNA modification GO:0009166 nucleotide catabolic process GO:0009219 pyrimidine deoxyribonucleotide metabolic process GO:0009223 pyrimidine deoxyribonucleotide catabolic process GO:0009262 deoxyribonucleotide metabolic process GO:0009264 deoxyribonucleotide catabolic process GO:0009394 2'-deoxyribonucleotide metabolic process GO:0016925 protein sumoylation GO:0018205 peptidyl-lysine modification GO:0019439 aromatic compound catabolic process GO:0019692 deoxyribose phosphate metabolic process GO:0032091 negative regulation of protein binding GO:0034655 nucleobase-containing compound catabolic process GO:0035510 DNA dealkylation GO:0035511 oxidative DNA demethylation GO:0035561 regulation of chromatin binding GO:0035562 negative regulation of chromatin binding GO:0040029 regulation of gene expression, epigenetic GO:0043393 regulation of protein binding GO:0044270 cellular nitrogen compound catabolic process GO:0044728 DNA methylation or demethylation GO:0045008 depyrimidination GO:0046386 deoxyribose phosphate catabolic process GO:0046434 organophosphate catabolic process GO:0046700 heterocycle catabolic process GO:0051098 regulation of binding GO:0051100 negative regulation of binding GO:0070988 demethylation GO:0070989 oxidative demethylation GO:0072527 pyrimidine-containing compound metabolic process GO:0072529 pyrimidine-containing compound catabolic process GO:0080111 DNA demethylation GO:1901136 carbohydrate derivative catabolic process GO:1901292 nucleoside phosphate catabolic process GO:1901361 organic cyclic compound catabolic process GO:1901565 organonitrogen compound catabolic process |
| Molecular Function |
GO:0000700 mismatch base pair DNA N-glycosylase activity GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001104 RNA polymerase II transcription cofactor activity GO:0003684 damaged DNA binding GO:0004844 uracil DNA N-glycosylase activity GO:0005080 protein kinase C binding GO:0008134 transcription factor binding GO:0008263 pyrimidine-specific mismatch base pair DNA N-glycosylase activity GO:0016798 hydrolase activity, acting on glycosyl bonds GO:0016799 hydrolase activity, hydrolyzing N-glycosyl compounds GO:0019104 DNA N-glycosylase activity GO:0030983 mismatched DNA binding GO:0032182 ubiquitin-like protein binding GO:0032183 SUMO binding GO:0043566 structure-specific DNA binding GO:0097506 deaminated base DNA N-glycosylase activity |
| Cellular Component |
GO:0016604 nuclear body GO:0016605 PML body |
| KEGG |
hsa03410 Base excision repair |
| Reactome |
R-HSA-73884: Base Excision Repair R-HSA-73929: Base-Excision Repair, AP Site Formation R-HSA-110329: Cleavage of the damaged pyrimidine R-HSA-73894: DNA Repair R-HSA-73928: Depyrimidination R-HSA-110357: Displacement of DNA glycosylase by APEX1 R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression R-HSA-392499: Metabolism of proteins R-HSA-597592: Post-translational protein modification R-HSA-110328: Recognition and association of DNA glycosylase with site containing an affected pyrimidine R-HSA-73933: Resolution of Abasic Sites (AP sites) R-HSA-3108232: SUMO E3 ligases SUMOylate target proteins R-HSA-2990846: SUMOylation R-HSA-3108214: SUMOylation of DNA damage response and repair proteins R-HSA-5221030: TET1,2,3 and TDG demethylate DNA |
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
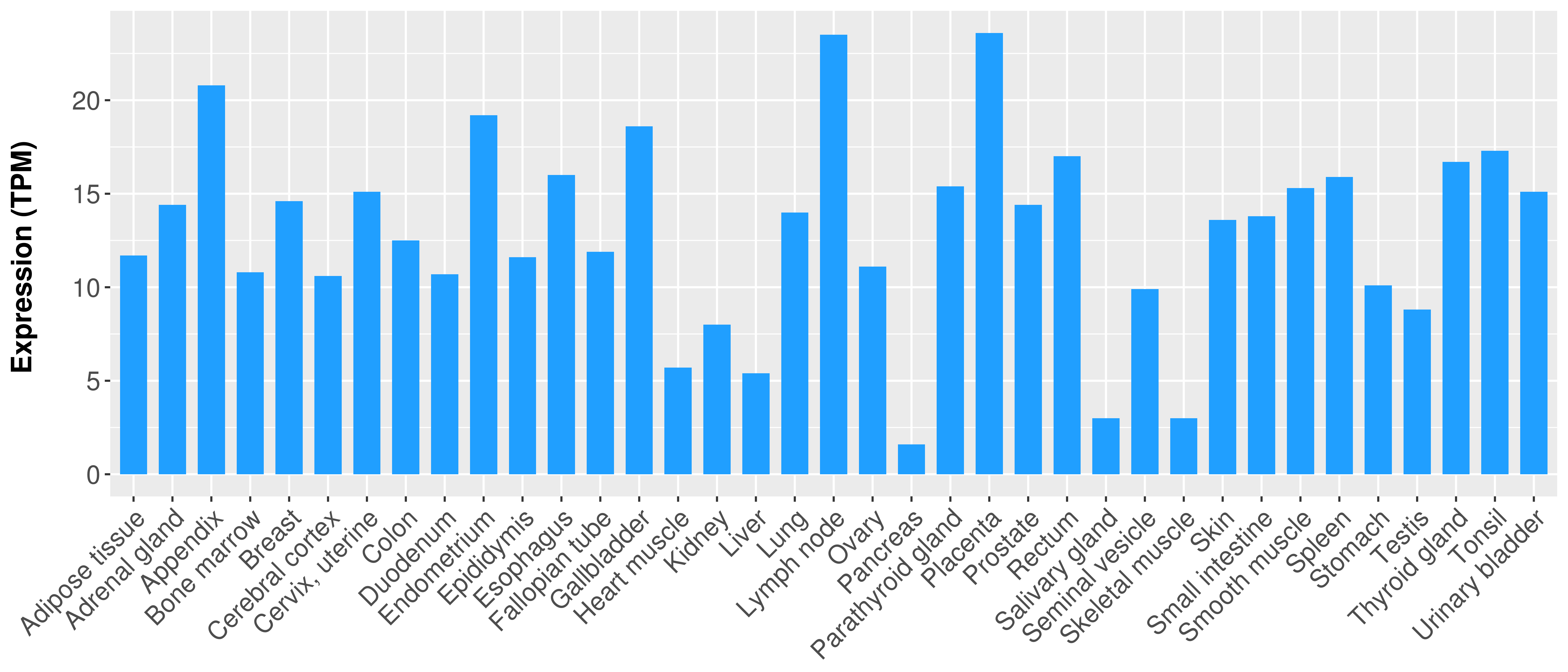
|
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
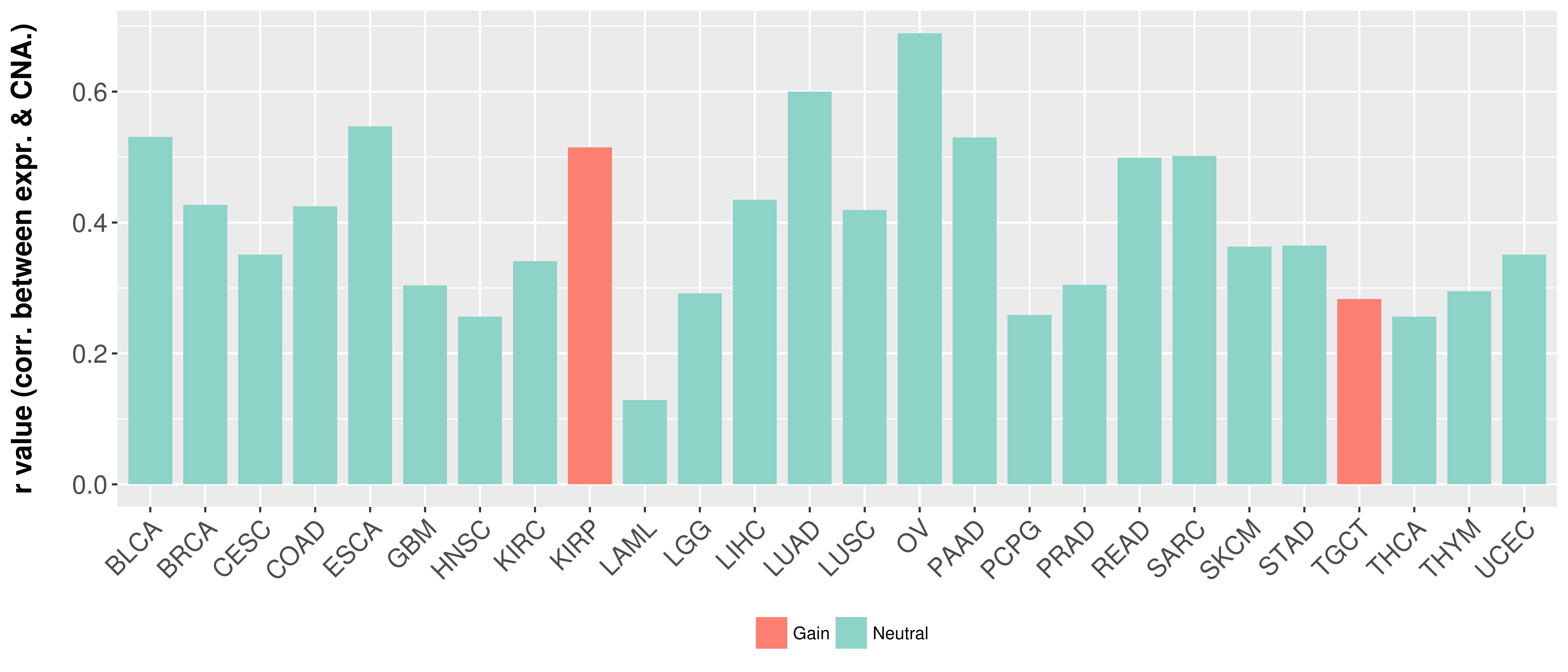
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |

Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
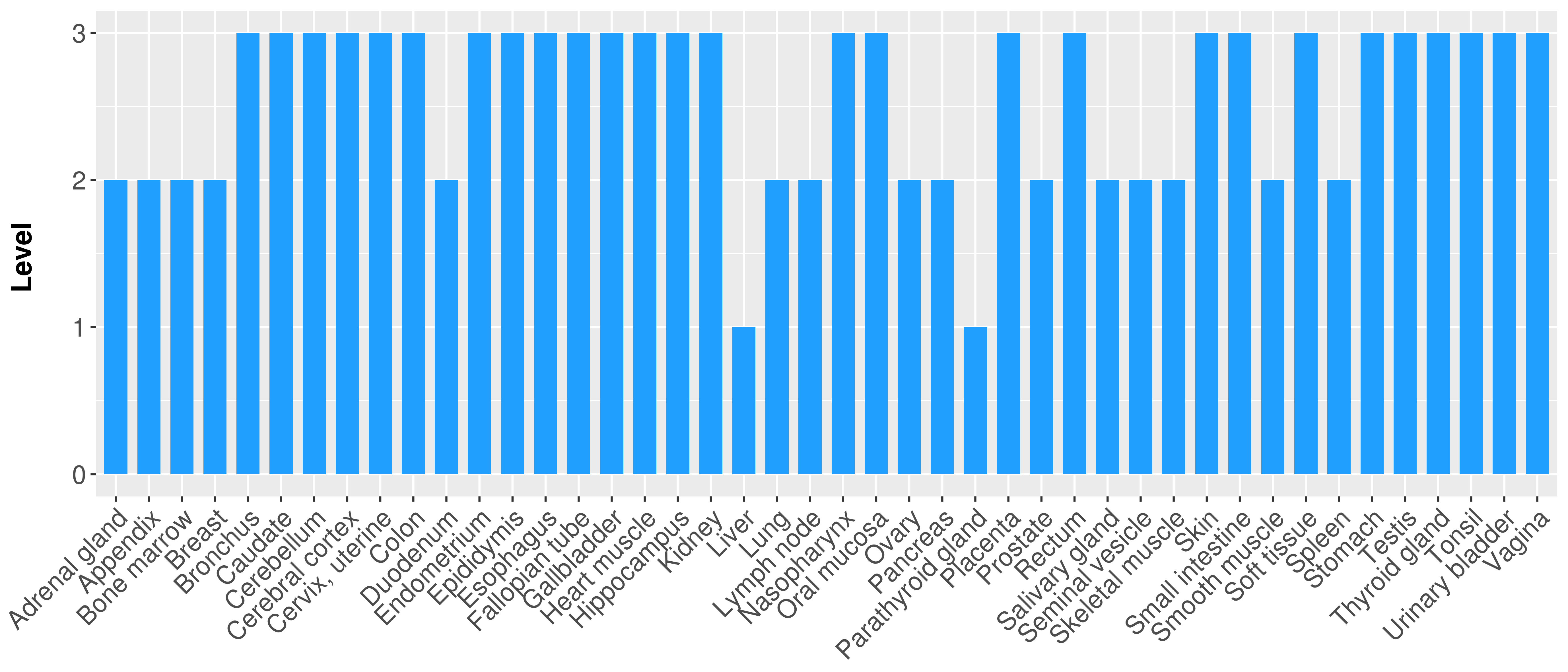
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
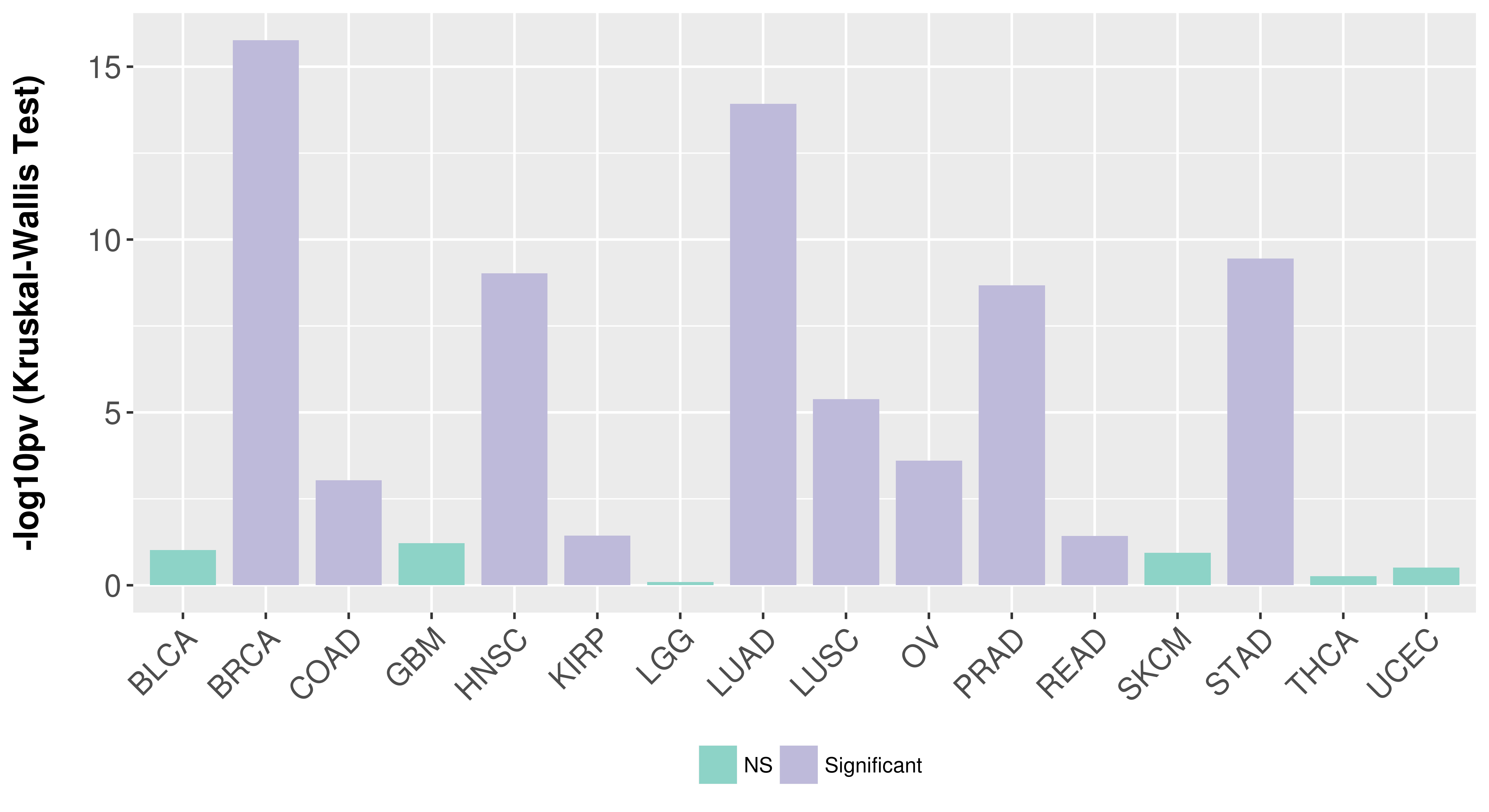
Association between expresson and subtype.
|
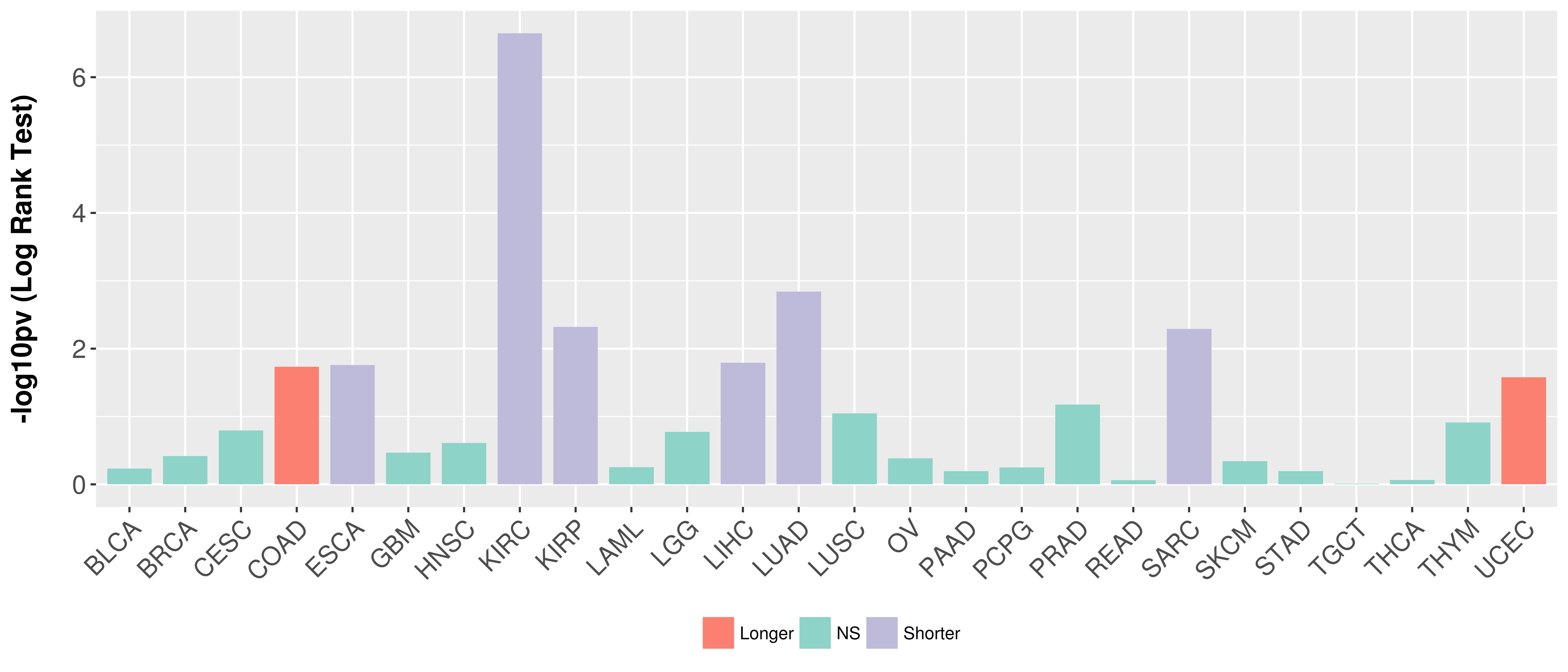
Overall survival analysis based on expression.
|
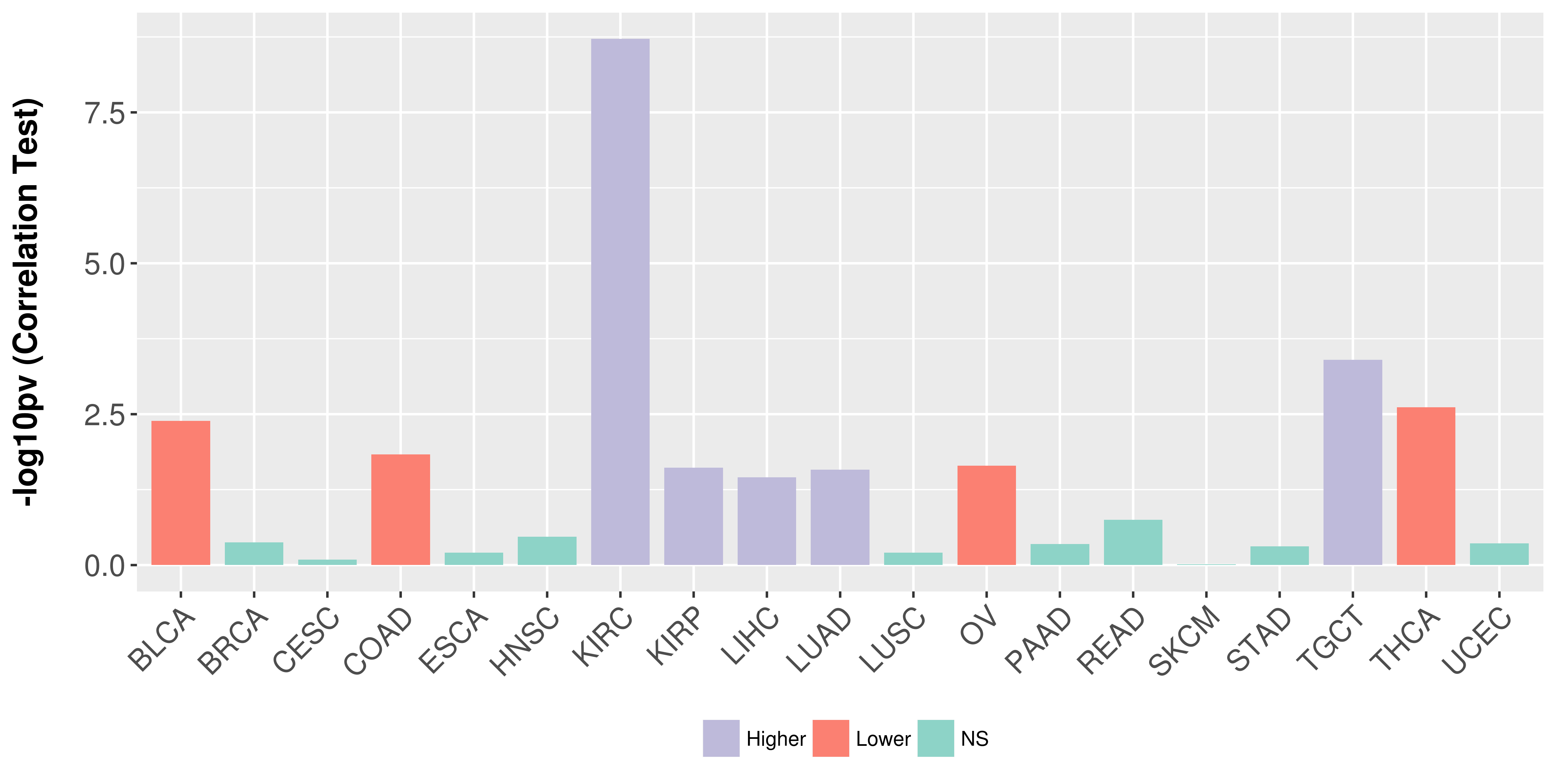
Association between expresson and stage.
|
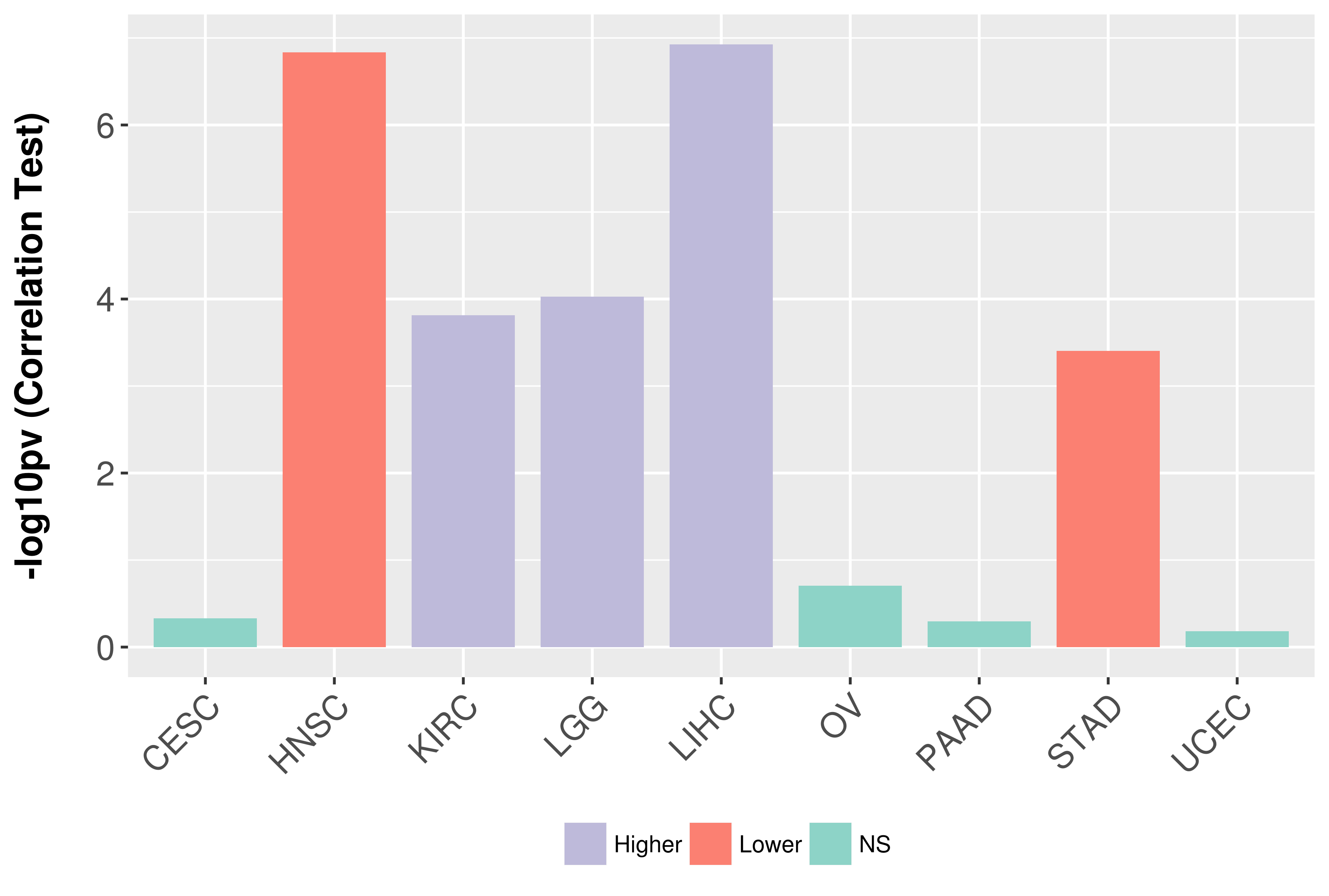
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for TDG. |
| Summary | |
|---|---|
| Symbol | TDG |
| Name | thymine DNA glycosylase |
| Aliases | G/T mismatch-specific thymine DNA glycosylase; hTDG; thymine-DNA glycosylase |
| Location | 12q23.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|