Browse TDRD12 in pancancer
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00270 DEAD/DEAH box helicase PF00567 Tudor domain |
||||||||||
| Function |
Probable ATP-binding RNA helicase required during spermatogenesis to repress transposable elements and preventing their mobilization, which is essential for the germline integrity. Acts via the piRNA metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and governs the methylation and subsequent repression of transposons. Involved in the secondary piRNAs metabolic process. Acts via the PET complex, a multiprotein complex required during the secondary piRNAs metabolic process for the PIWIL2 slicing-triggered loading of PIWIL4 piRNAs. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006304 DNA modification GO:0006305 DNA alkylation GO:0006306 DNA methylation GO:0006626 protein targeting to mitochondrion GO:0006839 mitochondrial transport GO:0007126 meiotic nuclear division GO:0007140 male meiosis GO:0007283 spermatogenesis GO:0009566 fertilization GO:0010501 RNA secondary structure unwinding GO:0010821 regulation of mitochondrion organization GO:0010822 positive regulation of mitochondrion organization GO:0016458 gene silencing GO:0031047 gene silencing by RNA GO:0032259 methylation GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0034587 piRNA metabolic process GO:0043046 DNA methylation involved in gamete generation GO:0043414 macromolecule methylation GO:0044728 DNA methylation or demethylation GO:0048232 male gamete generation GO:0051222 positive regulation of protein transport GO:0051321 meiotic cell cycle GO:0070585 protein localization to mitochondrion GO:0072655 establishment of protein localization to mitochondrion GO:0090316 positive regulation of intracellular protein transport GO:1903046 meiotic cell cycle process GO:1903214 regulation of protein targeting to mitochondrion GO:1903533 regulation of protein targeting GO:1903747 regulation of establishment of protein localization to mitochondrion GO:1903749 positive regulation of establishment of protein localization to mitochondrion GO:1903829 positive regulation of cellular protein localization GO:1903955 positive regulation of protein targeting to mitochondrion GO:1904951 positive regulation of establishment of protein localization |
| Molecular Function |
GO:0003724 RNA helicase activity GO:0004004 ATP-dependent RNA helicase activity GO:0004386 helicase activity GO:0008026 ATP-dependent helicase activity GO:0008186 RNA-dependent ATPase activity GO:0016887 ATPase activity GO:0042623 ATPase activity, coupled GO:0070035 purine NTP-dependent helicase activity |
| Cellular Component |
GO:1990923 PET complex |
| KEGG | - |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-211000: Gene Silencing by RNA R-HSA-5601884: PIWI-interacting RNA (piRNA) biogenesis |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record for TDRD12. |
| There is no record. |
| There is no record for TDRD12. |
| There is no record for TDRD12. |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
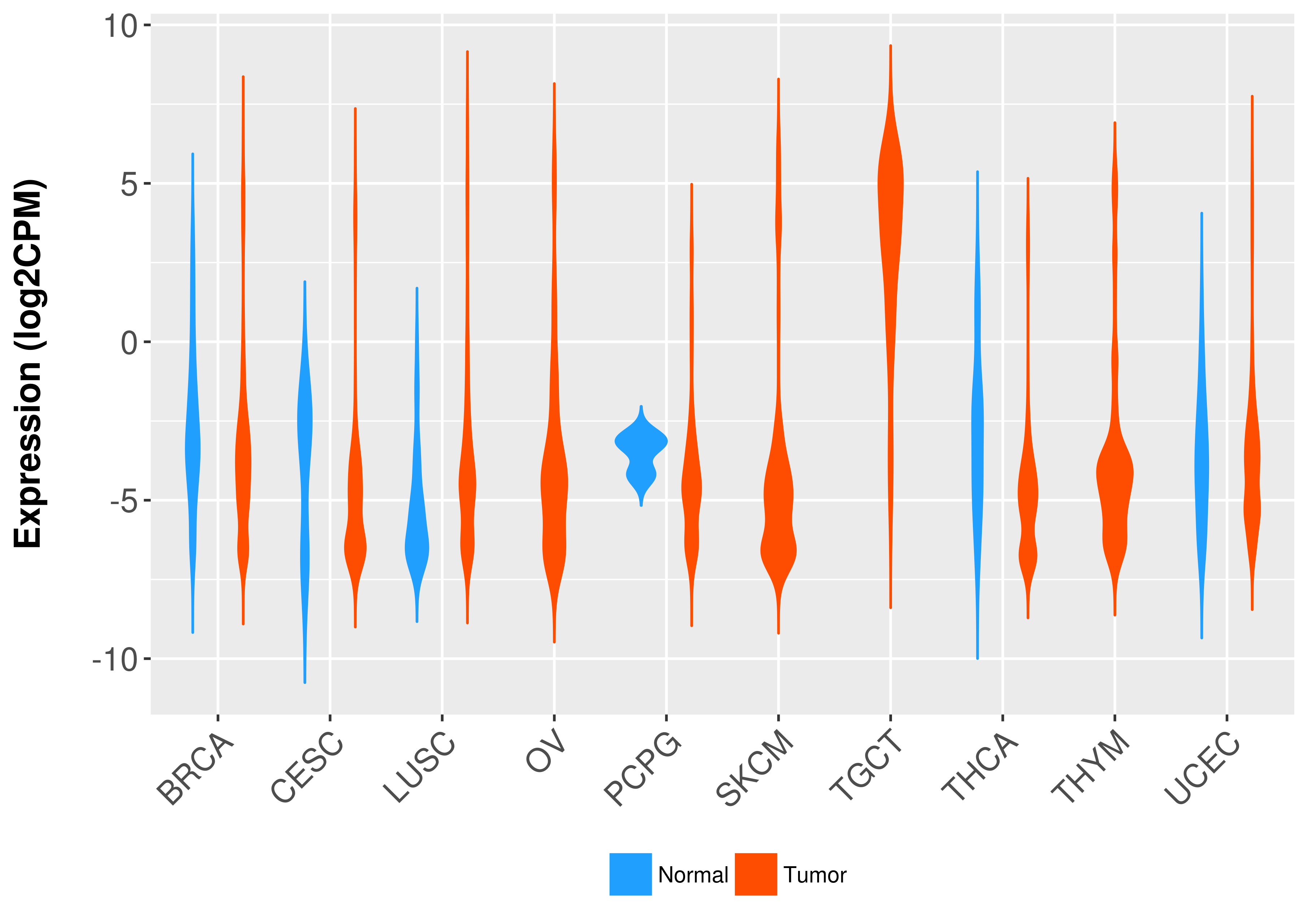
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
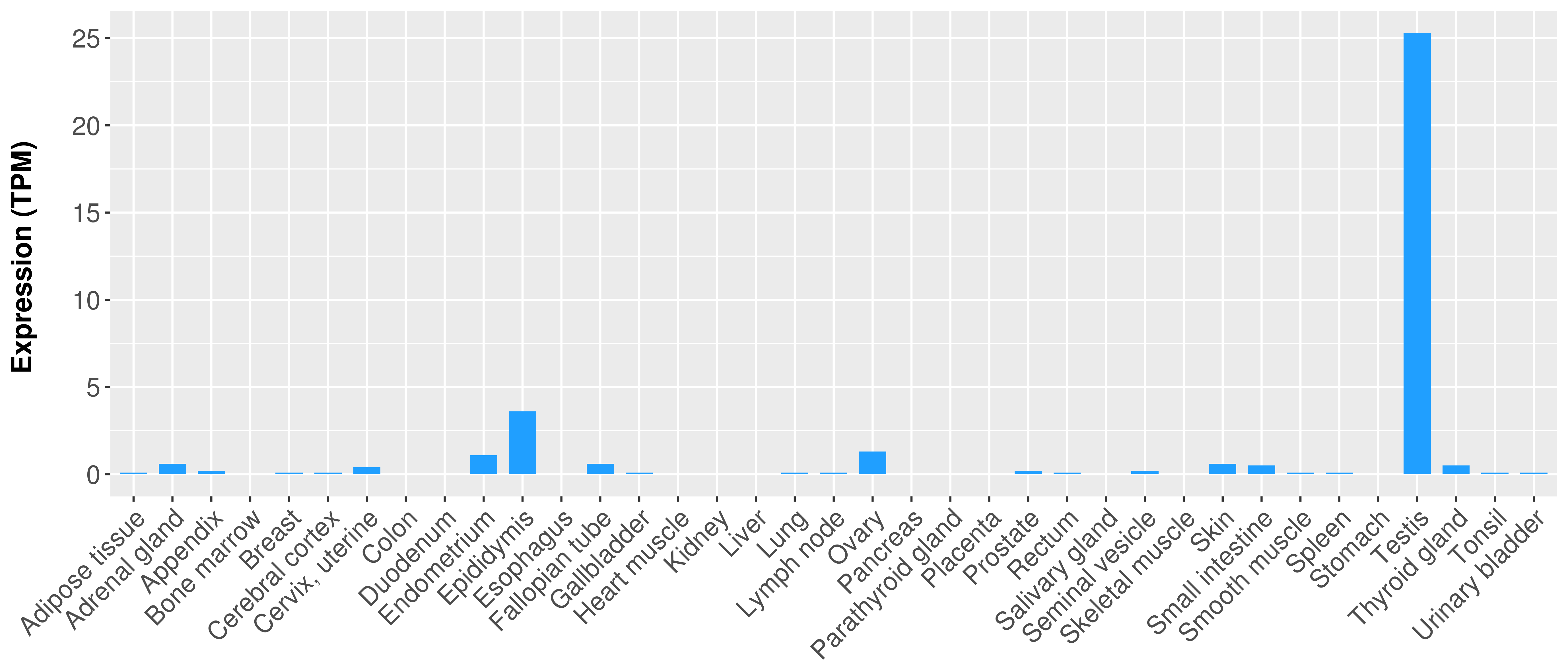
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
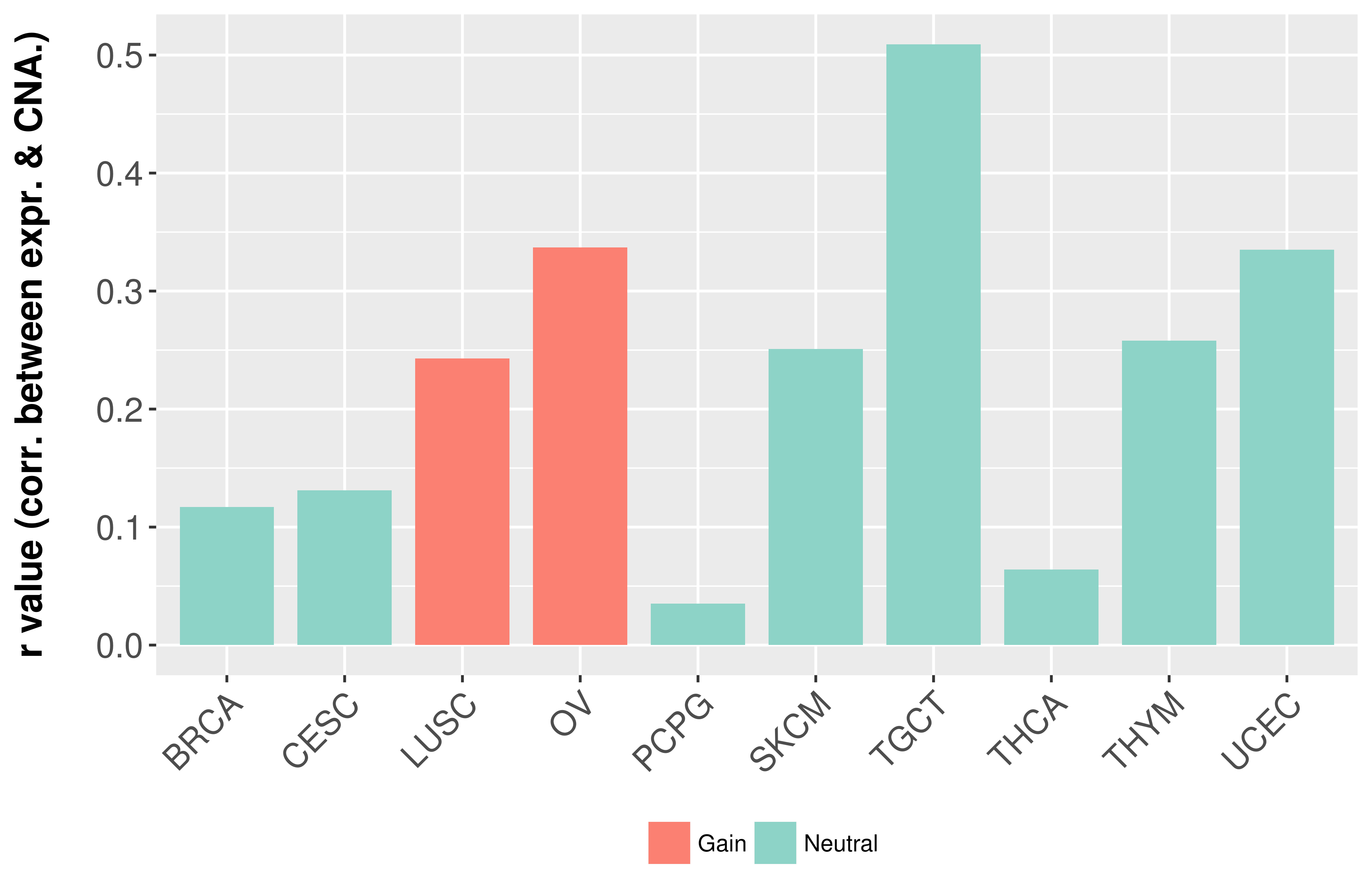
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
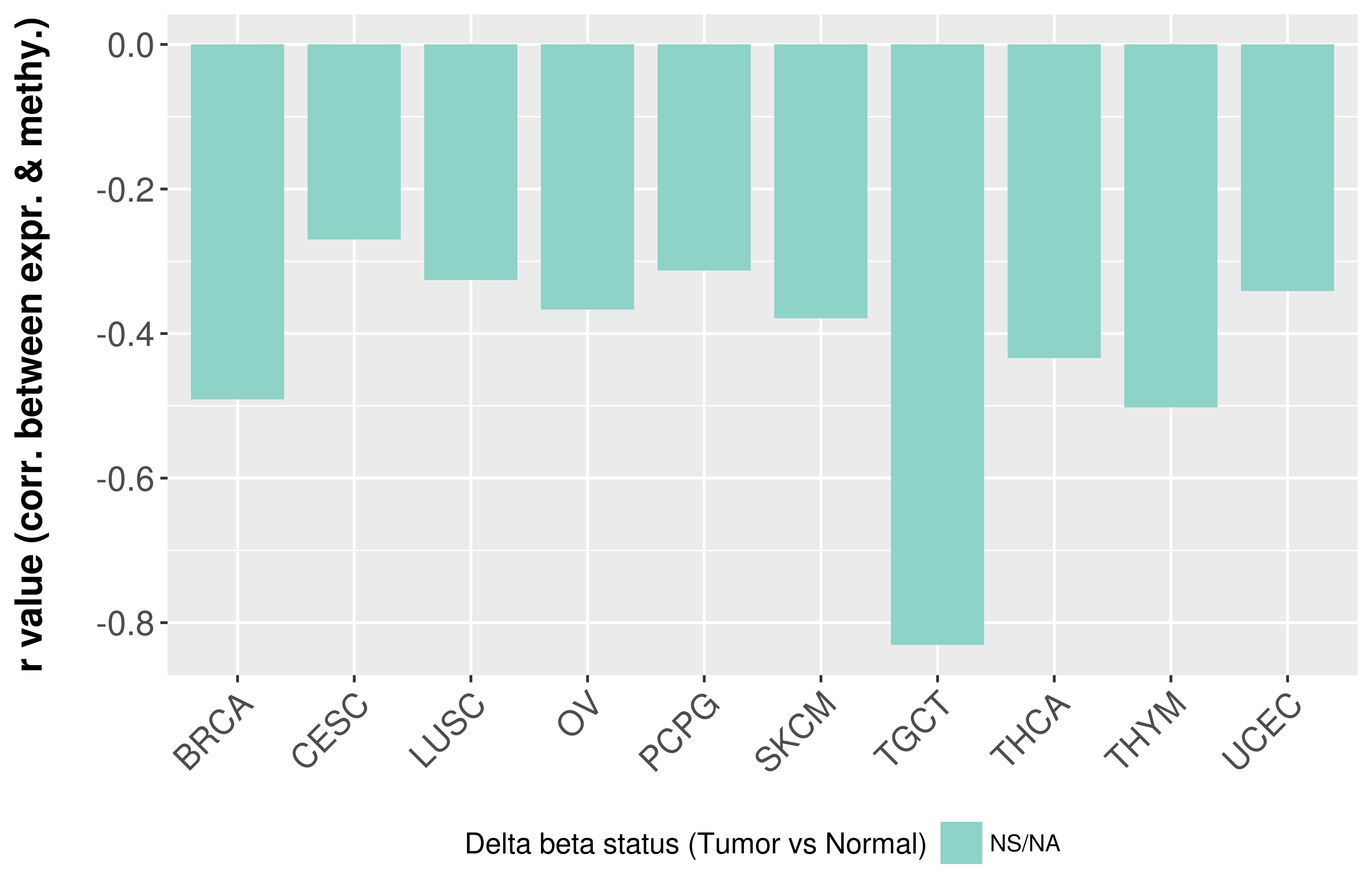
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
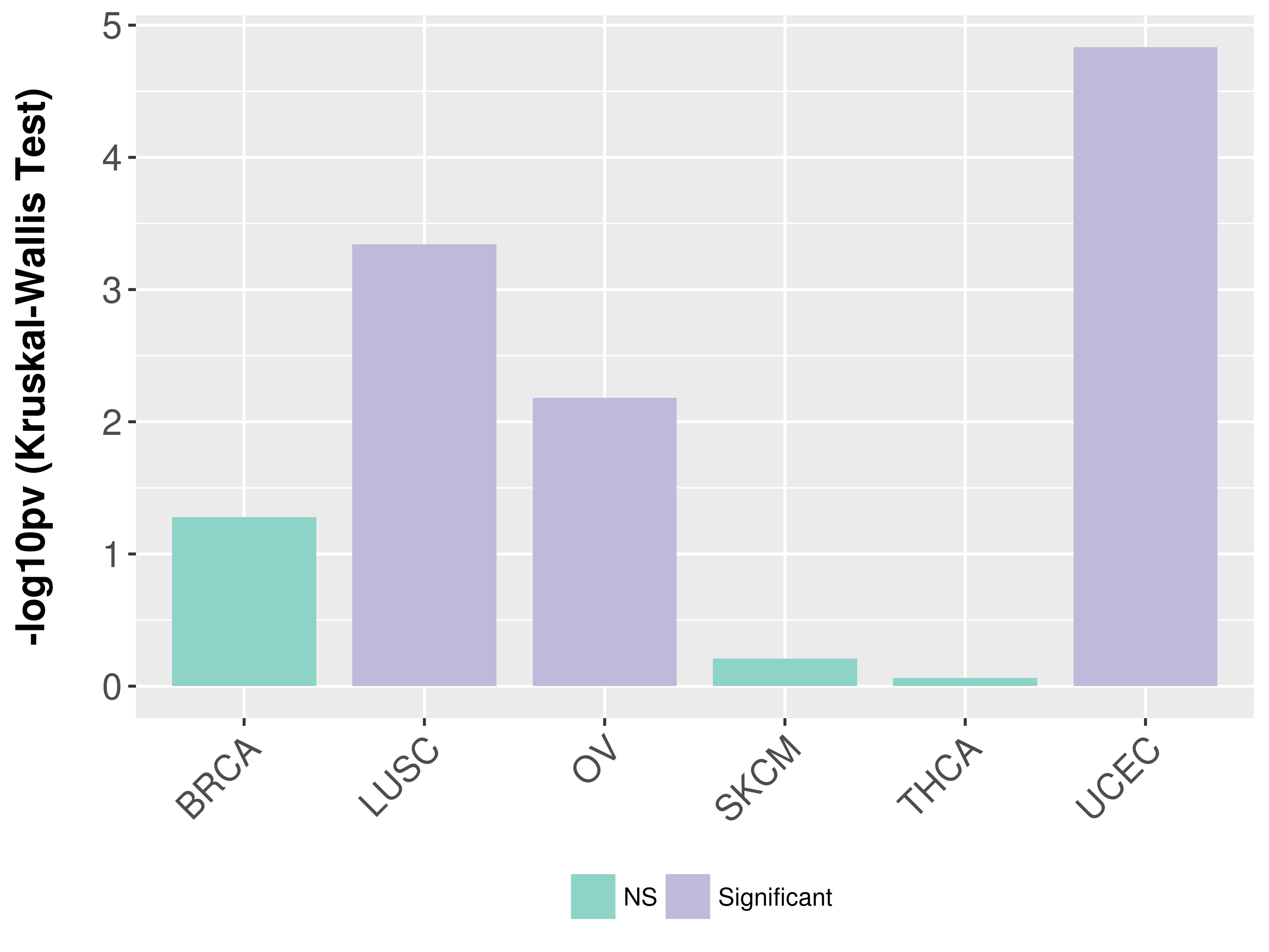
Association between expresson and subtype.
|
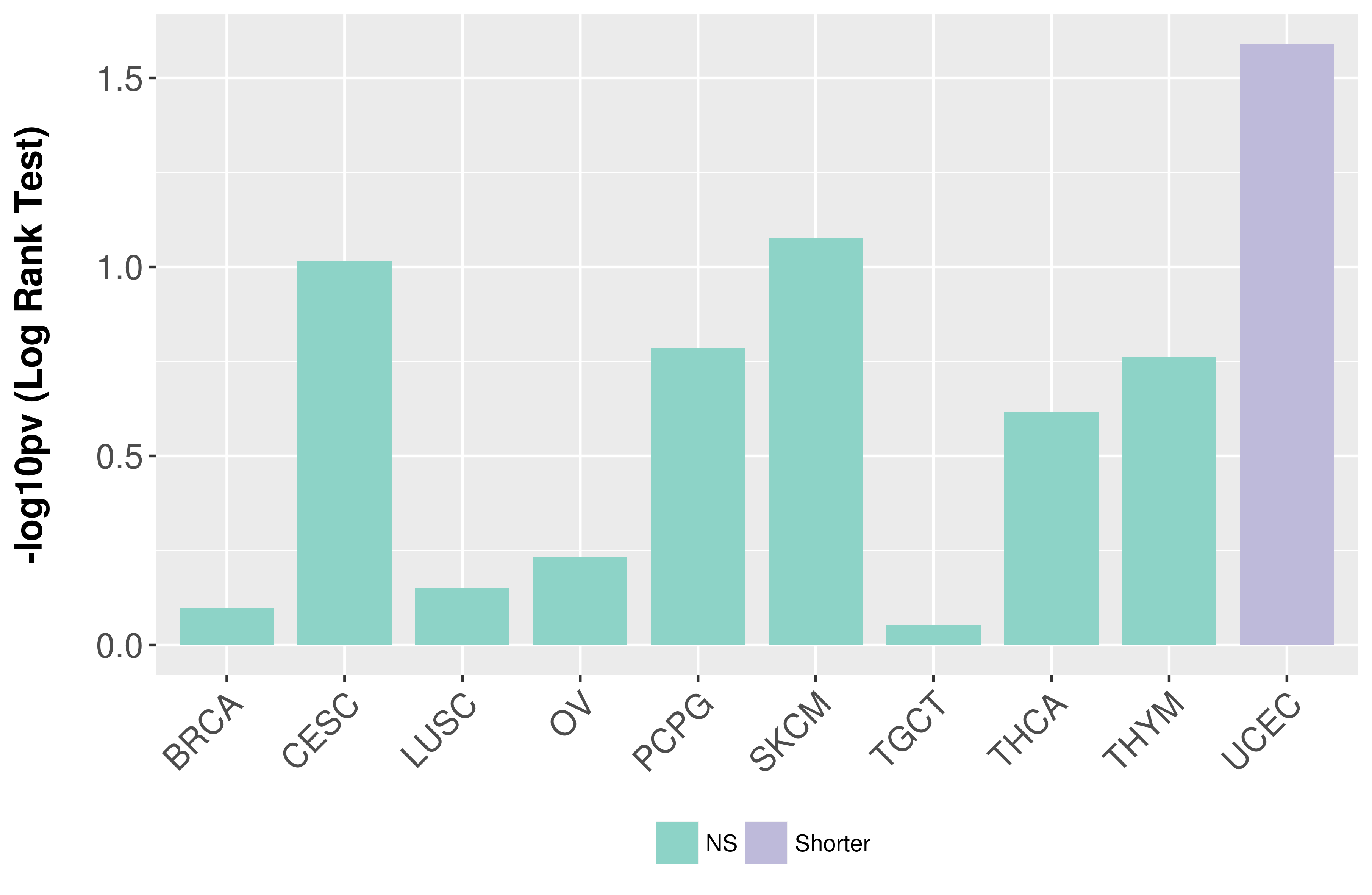
Overall survival analysis based on expression.
|
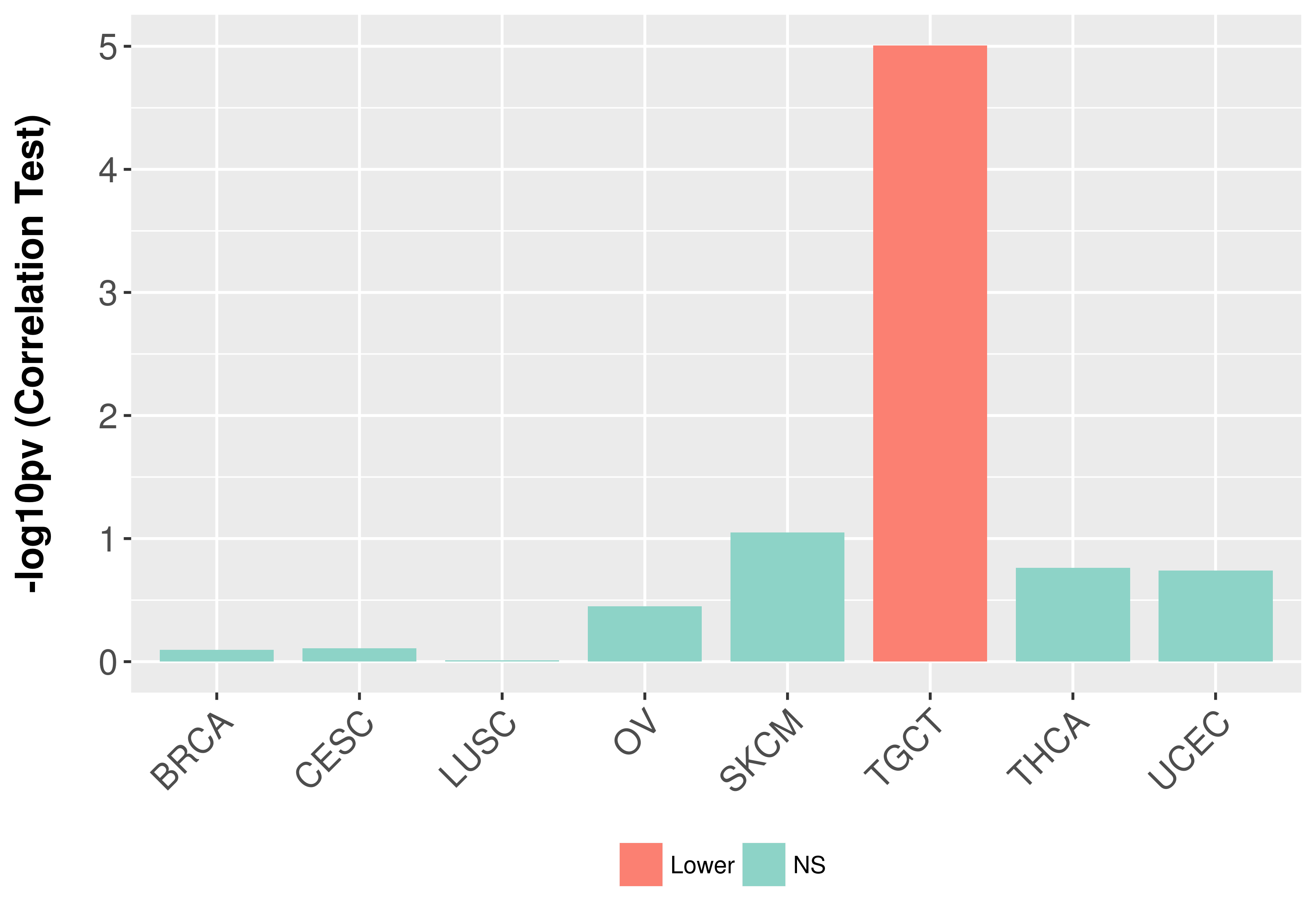
Association between expresson and stage.
|
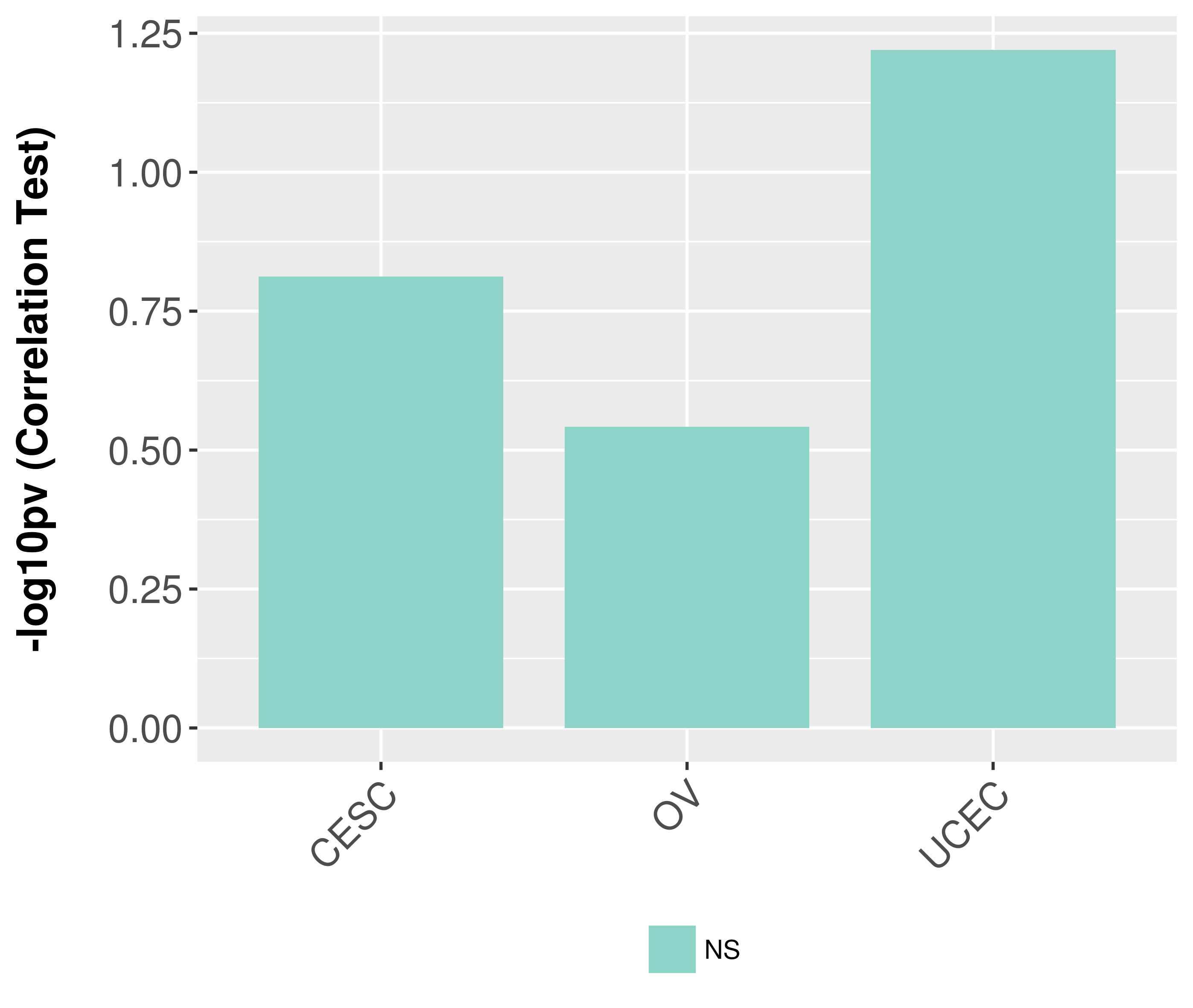
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for TDRD12. |
| Summary | |
|---|---|
| Symbol | TDRD12 |
| Name | tudor domain containing 12 |
| Aliases | ECAT8; FLJ13072; ES cell associated transcript 8; ES cell-associated transcript 8 protein; tudor domain-cont ...... |
| Location | 19q13.11 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for TDRD12. |
| There is no record for TDRD12. |