Browse TRIM28 in pancancer
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00628 PHD-finger PF00643 B-box zinc finger PF14634 zinc-RING finger domain |
||||||||||
| Function |
Nuclear corepressor for KRAB domain-containing zinc finger proteins (KRAB-ZFPs). Mediates gene silencing by recruiting CHD3, a subunit of the nucleosome remodeling and deacetylation (NuRD) complex, and SETDB1 (which specifically methylates histone H3 at 'Lys-9' (H3K9me)) to the promoter regions of KRAB target genes. Enhances transcriptional repression by coordinating the increase in H3K9me, the decrease in histone H3 'Lys-9 and 'Lys-14' acetylation (H3K9ac and H3K14ac, respectively) and the disposition of HP1 proteins to silence gene expression. Recruitment of SETDB1 induces heterochromatinization. May play a role as a coactivator for CEBPB and NR3C1 in the transcriptional activation of ORM1. Also corepressor for ERBB4. Inhibits E2F1 activity by stimulating E2F1-HDAC1 complex formation and inhibiting E2F1 acetylation. May serve as a partial backup to prevent E2F1-mediated apoptosis in the absence of RB1. Important regulator of CDKN1A/p21(CIP1). Has E3 SUMO-protein ligase activity toward itself via its PHD-type zinc finger. Also specifically sumoylates IRF7, thereby inhibiting its transactivation activity. Ubiquitinates p53/TP53 leading to its proteosomal degradation; the function is enhanced by MAGEC2 and MAGEA2, and possibly MAGEA3 and MAGEA6. Mediates the nuclear localization of KOX1, ZNF268 and ZNF300 transcription factors. In association with isoform 2 of ZFP90, is required for the transcriptional repressor activity of FOXP3 and the suppressive function of regulatory T-cells (Treg) (PubMed:23543754). Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells (PubMed:24623306). Also required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs) (PubMed:24623306). Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing (PubMed:24623306). The SETDB1-TRIM28-ZNF274 complex may play a role in recruiting ATRX to the 3'-exons of zinc-finger coding genes with atypical chromatin signatures to establish or maintain/protect H3K9me3 at these transcriptionally active regions (PubMed:27029610). Acts as a corepressor for ZFP568 (By similarity). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001701 in utero embryonic development GO:0001837 epithelial to mesenchymal transition GO:0001890 placenta development GO:0001892 embryonic placenta development GO:0003401 axis elongation GO:0006282 regulation of DNA repair GO:0006304 DNA modification GO:0006305 DNA alkylation GO:0006306 DNA methylation GO:0006342 chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006352 DNA-templated transcription, initiation GO:0006367 transcription initiation from RNA polymerase II promoter GO:0006606 protein import into nucleus GO:0006913 nucleocytoplasmic transport GO:0007265 Ras protein signal transduction GO:0007565 female pregnancy GO:0007566 embryo implantation GO:0016458 gene silencing GO:0016925 protein sumoylation GO:0017038 protein import GO:0018205 peptidyl-lysine modification GO:0019058 viral life cycle GO:0019076 viral release from host cell GO:0019079 viral genome replication GO:0031935 regulation of chromatin silencing GO:0031937 positive regulation of chromatin silencing GO:0032259 methylation GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0034504 protein localization to nucleus GO:0035510 DNA dealkylation GO:0039692 single stranded viral RNA replication via double stranded DNA intermediate GO:0039694 viral RNA genome replication GO:0039703 RNA replication GO:0040029 regulation of gene expression, epigenetic GO:0042306 regulation of protein import into nucleus GO:0042307 positive regulation of protein import into nucleus GO:0042990 regulation of transcription factor import into nucleus GO:0042991 transcription factor import into nucleus GO:0042993 positive regulation of transcription factor import into nucleus GO:0043045 DNA methylation involved in embryo development GO:0043388 positive regulation of DNA binding GO:0043414 macromolecule methylation GO:0043900 regulation of multi-organism process GO:0043901 negative regulation of multi-organism process GO:0043903 regulation of symbiosis, encompassing mutualism through parasitism GO:0044033 multi-organism metabolic process GO:0044706 multi-multicellular organism process GO:0044728 DNA methylation or demethylation GO:0044744 protein targeting to nucleus GO:0045069 regulation of viral genome replication GO:0045071 negative regulation of viral genome replication GO:0045091 regulation of single stranded viral RNA replication via double stranded DNA intermediate GO:0045739 positive regulation of DNA repair GO:0045814 negative regulation of gene expression, epigenetic GO:0045869 negative regulation of single stranded viral RNA replication via double stranded DNA intermediate GO:0046777 protein autophosphorylation GO:0046822 regulation of nucleocytoplasmic transport GO:0046824 positive regulation of nucleocytoplasmic transport GO:0048525 negative regulation of viral process GO:0048568 embryonic organ development GO:0048608 reproductive structure development GO:0048762 mesenchymal cell differentiation GO:0050792 regulation of viral process GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051054 positive regulation of DNA metabolic process GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051101 regulation of DNA binding GO:0051169 nuclear transport GO:0051170 nuclear import GO:0051222 positive regulation of protein transport GO:0051259 protein oligomerization GO:0060026 convergent extension GO:0060028 convergent extension involved in axis elongation GO:0060485 mesenchyme development GO:0060560 developmental growth involved in morphogenesis GO:0060669 embryonic placenta morphogenesis GO:0060968 regulation of gene silencing GO:0061458 reproductive system development GO:0070988 demethylation GO:0071514 genetic imprinting GO:0080111 DNA demethylation GO:0090308 regulation of methylation-dependent chromatin silencing GO:0090309 positive regulation of methylation-dependent chromatin silencing GO:0090316 positive regulation of intracellular protein transport GO:1900180 regulation of protein localization to nucleus GO:1900182 positive regulation of protein localization to nucleus GO:1901535 regulation of DNA demethylation GO:1901536 negative regulation of DNA demethylation GO:1901538 changes to DNA methylation involved in embryo development GO:1902186 regulation of viral release from host cell GO:1902187 negative regulation of viral release from host cell GO:1902275 regulation of chromatin organization GO:1902593 single-organism nuclear import GO:1903533 regulation of protein targeting GO:1903829 positive regulation of cellular protein localization GO:1903900 regulation of viral life cycle GO:1903901 negative regulation of viral life cycle GO:1904589 regulation of protein import GO:1904591 positive regulation of protein import GO:1904951 positive regulation of establishment of protein localization GO:1905269 positive regulation of chromatin organization GO:2000653 regulation of genetic imprinting GO:2001020 regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus |
| Molecular Function |
GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001104 RNA polymerase II transcription cofactor activity GO:0001105 RNA polymerase II transcription coactivator activity GO:0001190 transcriptional activator activity, RNA polymerase II transcription factor binding GO:0001191 transcriptional repressor activity, RNA polymerase II transcription factor binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0003714 transcription corepressor activity GO:0004842 ubiquitin-protein transferase activity GO:0016874 ligase activity GO:0019787 ubiquitin-like protein transferase activity GO:0031625 ubiquitin protein ligase binding GO:0035851 Krueppel-associated box domain binding GO:0044389 ubiquitin-like protein ligase binding GO:0070087 chromo shadow domain binding GO:0098811 transcriptional repressor activity, RNA polymerase II activating transcription factor binding GO:1990841 promoter-specific chromatin binding |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000791 euchromatin GO:0000792 heterochromatin GO:0005667 transcription factor complex GO:0005719 nuclear euchromatin GO:0005720 nuclear heterochromatin GO:0044454 nuclear chromosome part GO:0044798 nuclear transcription factor complex GO:0090575 RNA polymerase II transcription factor complex |
| KEGG | - |
| Reactome |
R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway |
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for TRIM28. |
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
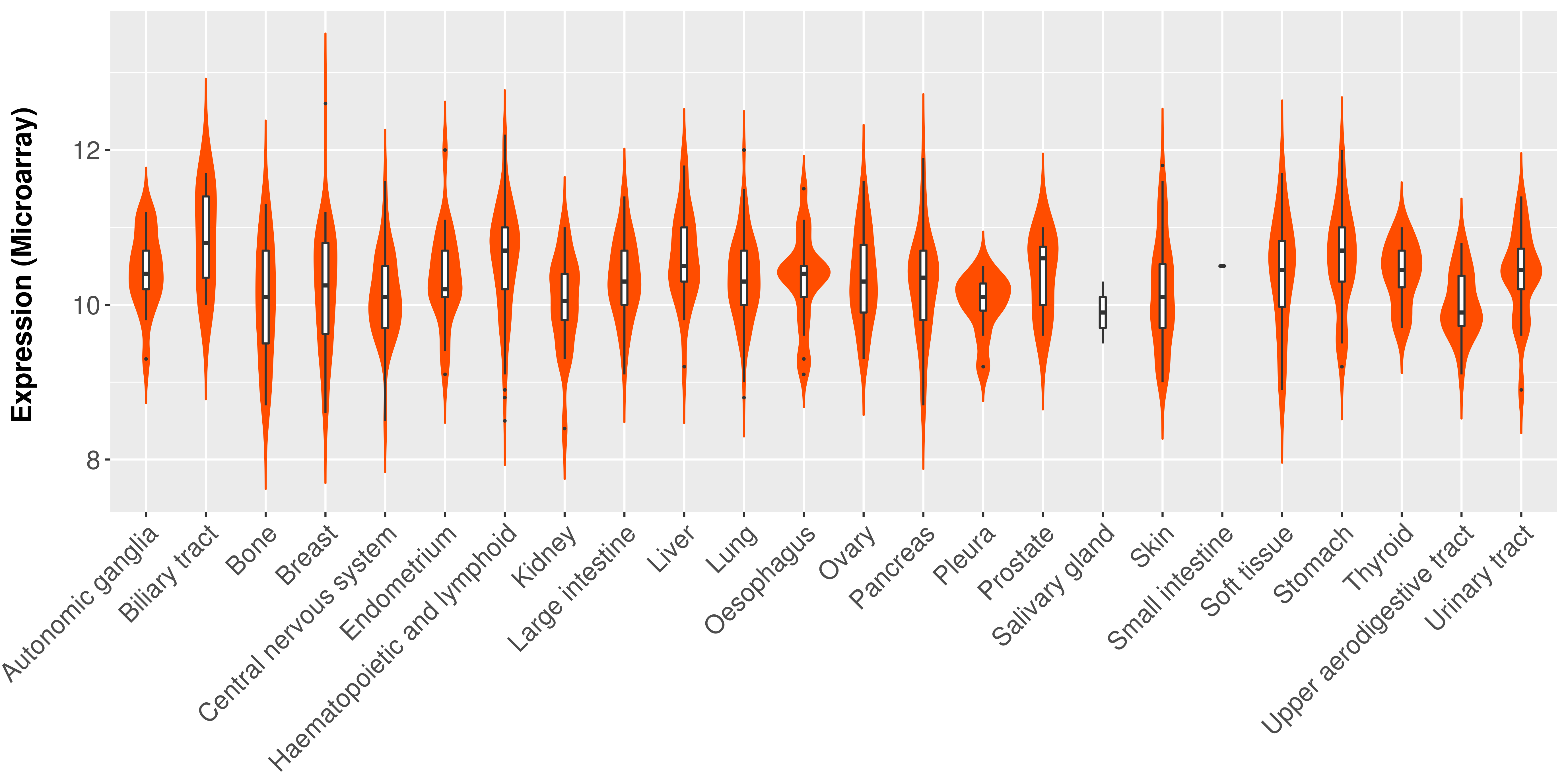
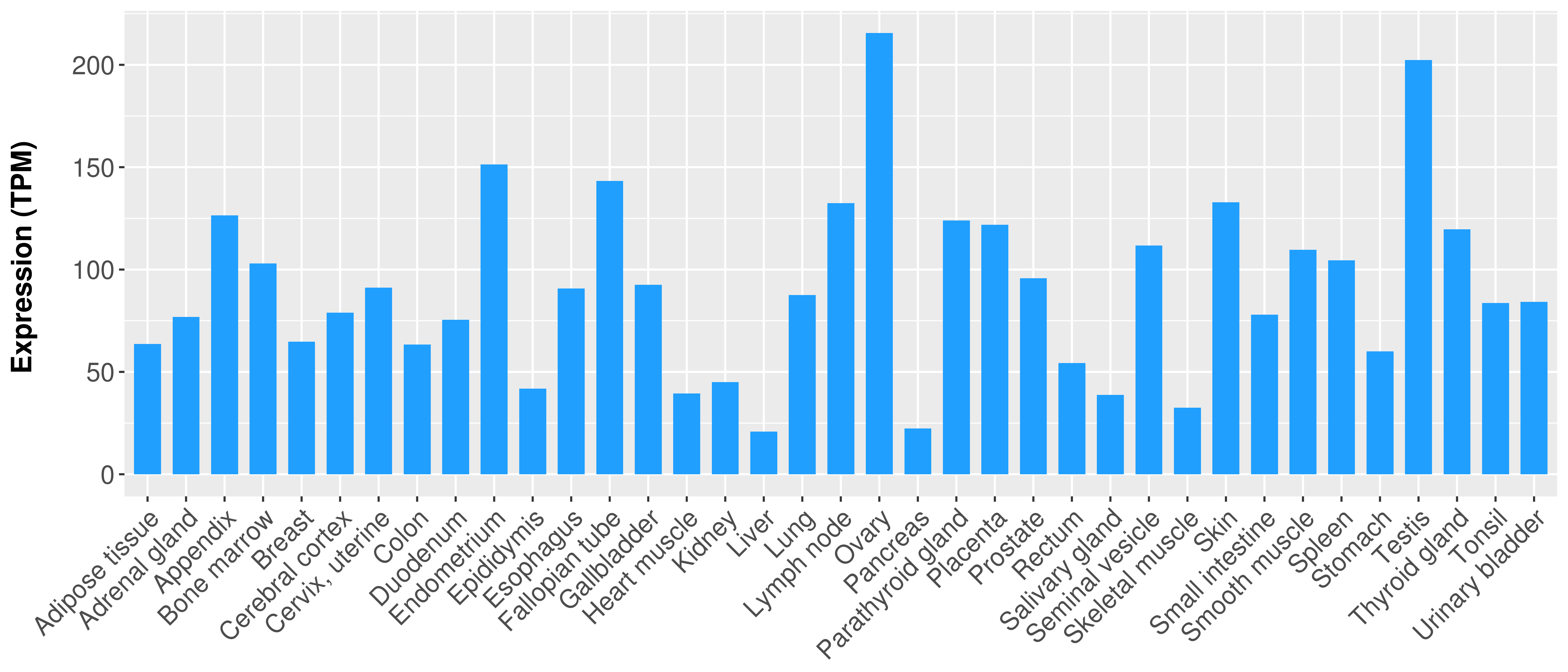
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
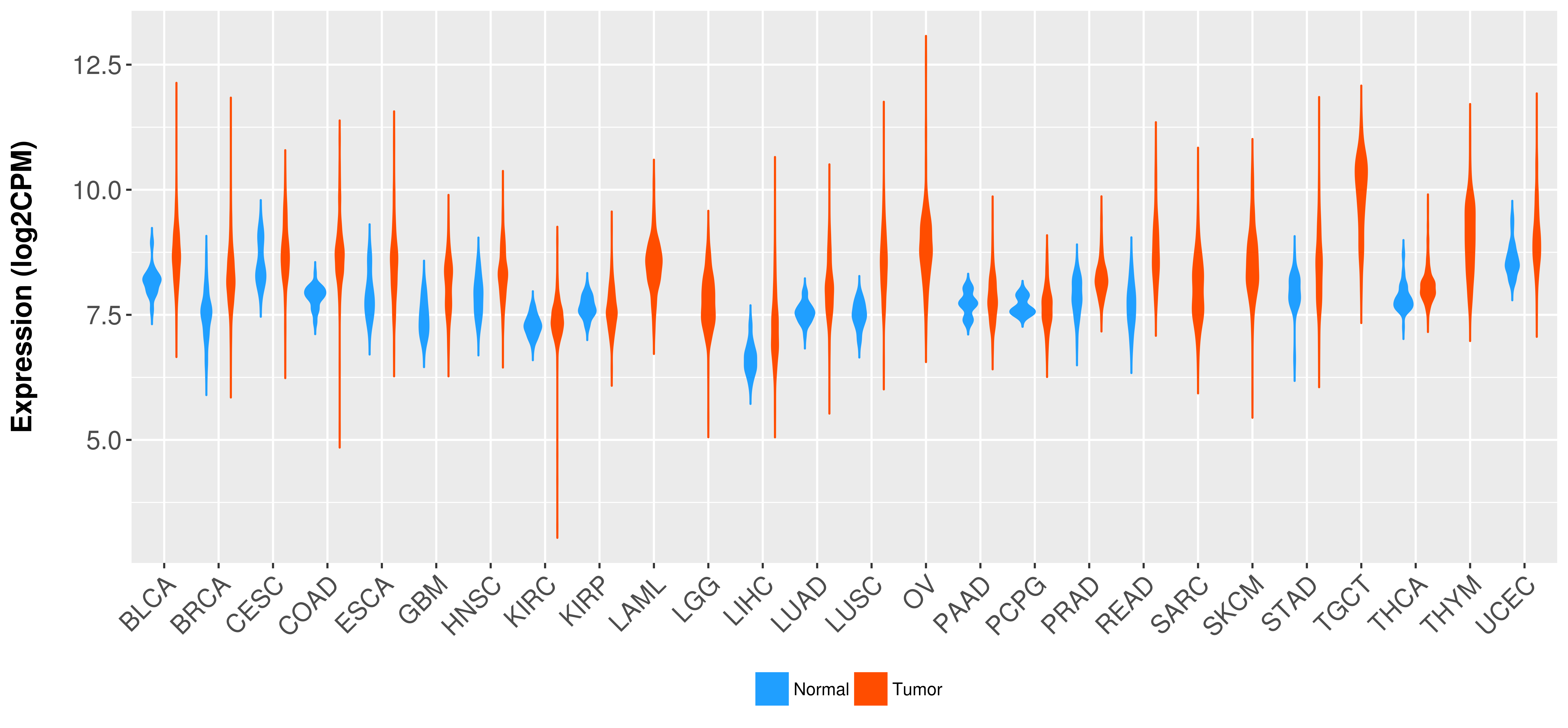
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
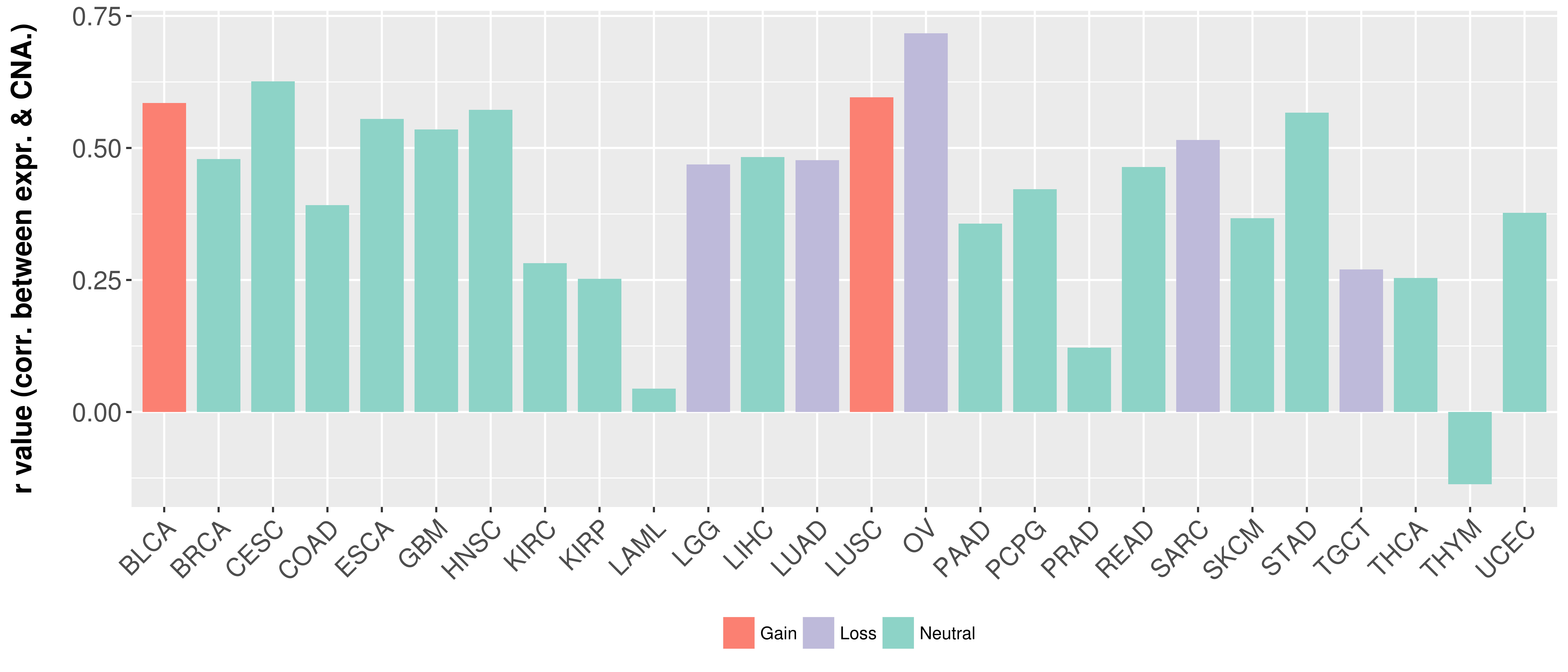
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |

Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
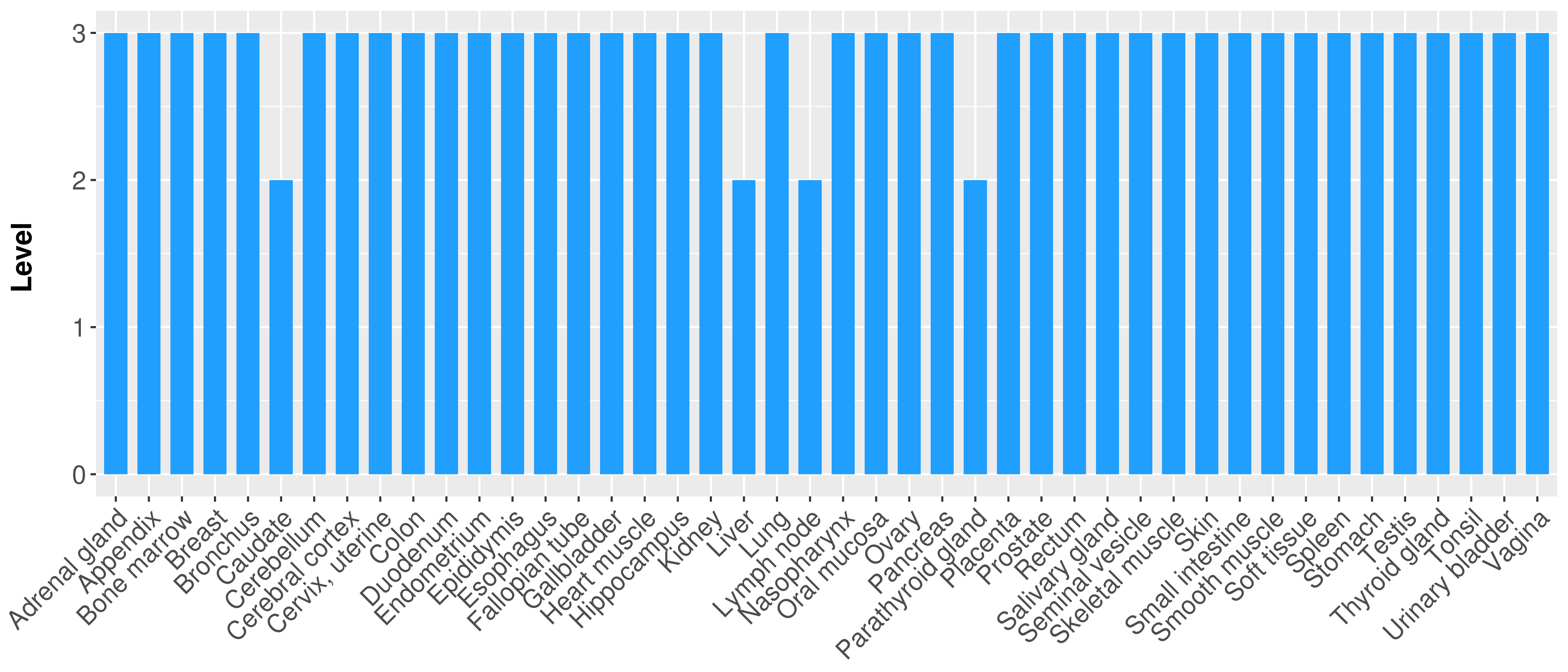
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
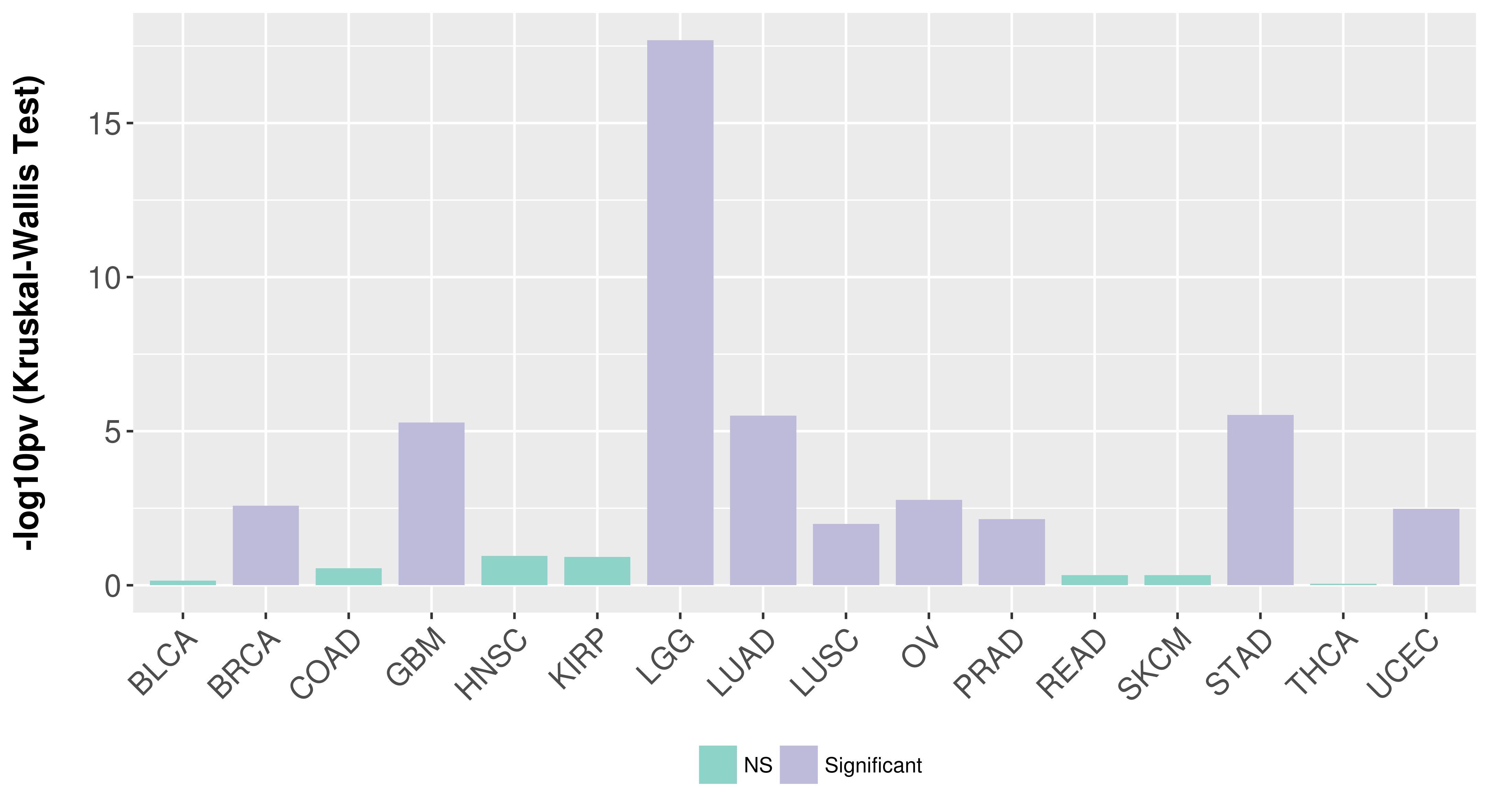
Association between expresson and subtype.
|
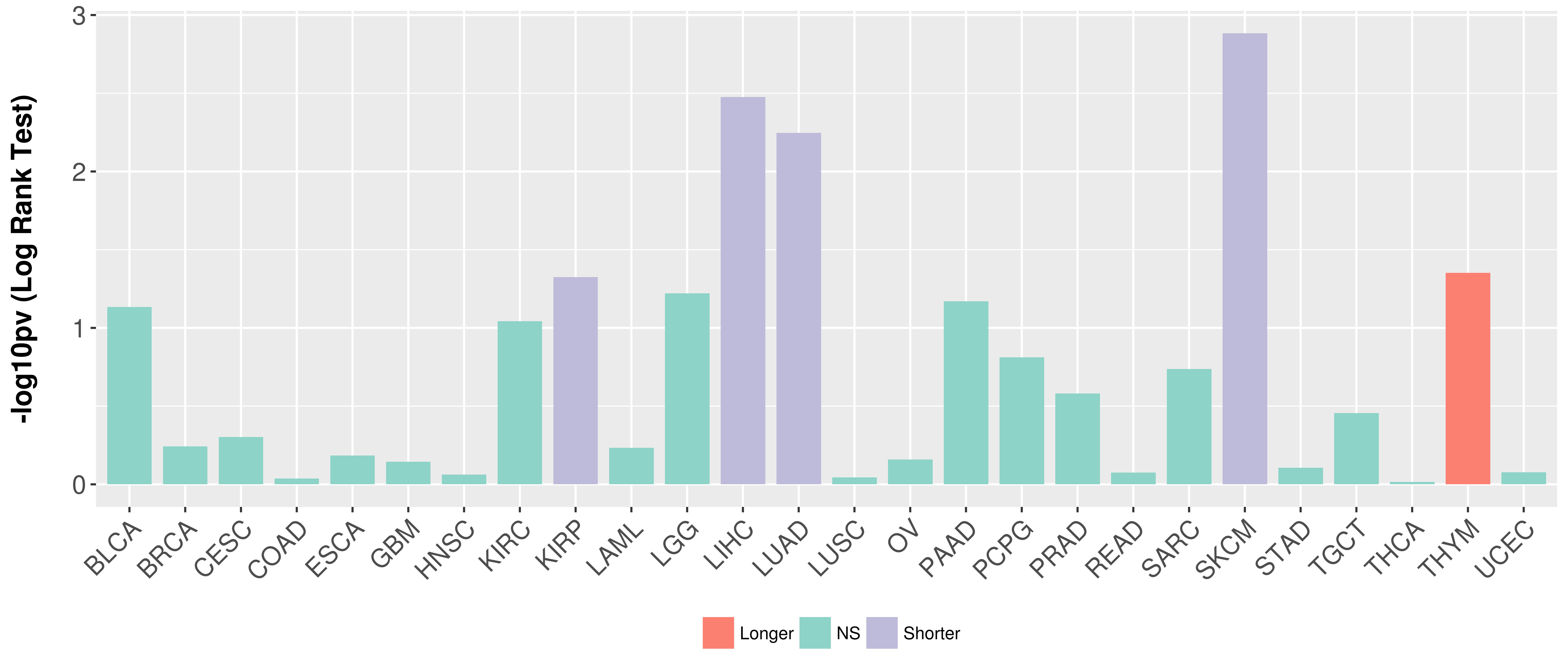
Overall survival analysis based on expression.
|
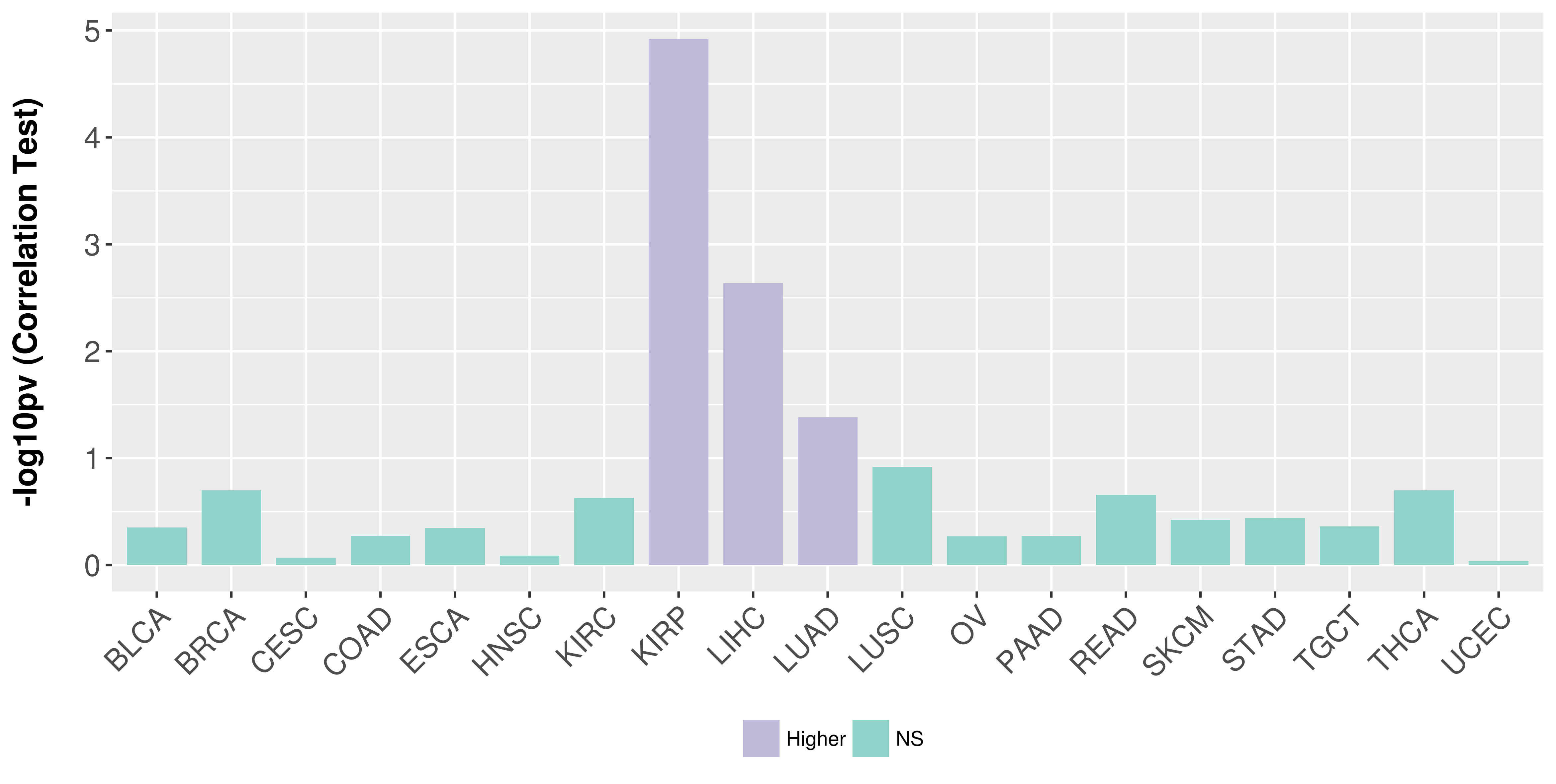
Association between expresson and stage.
|
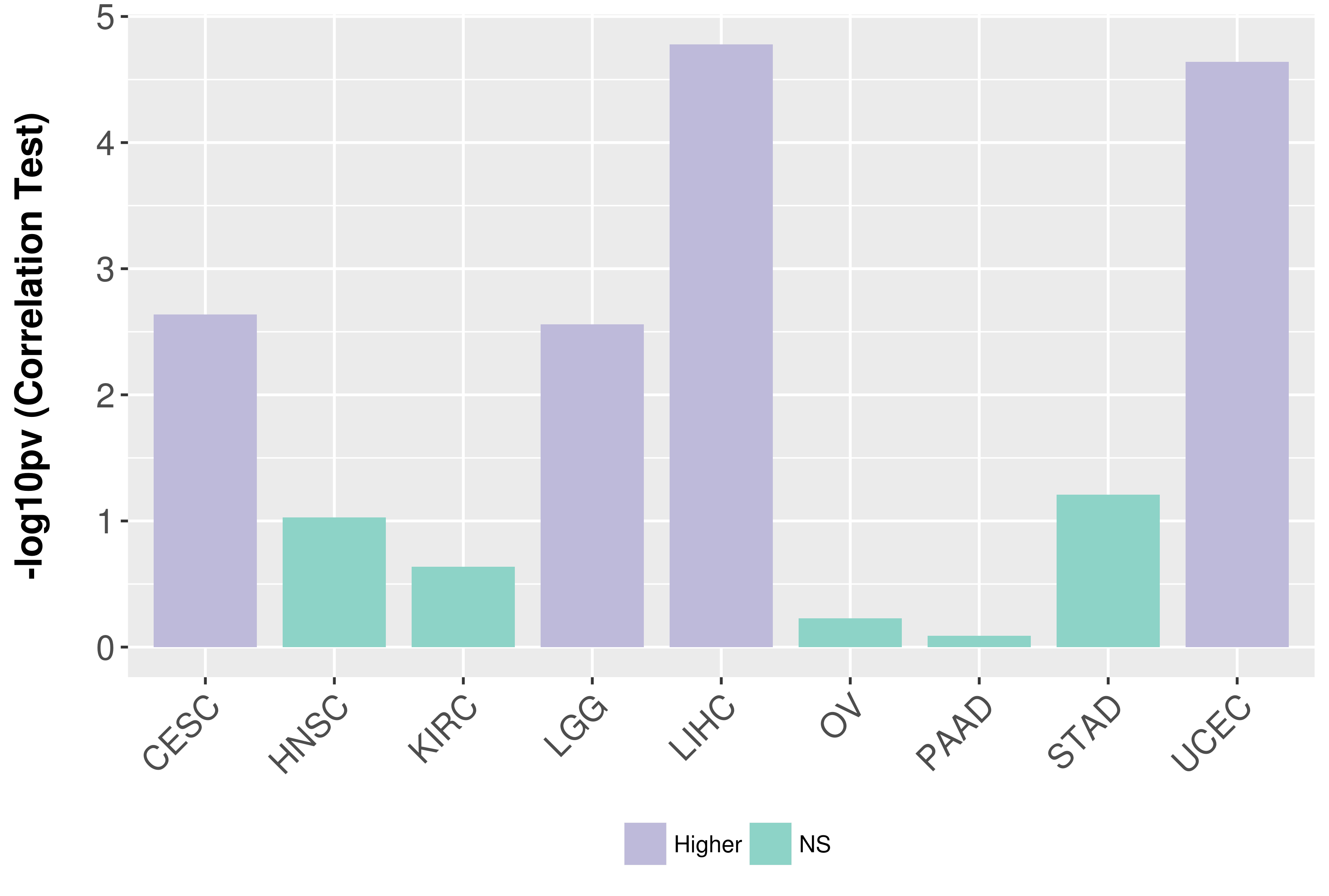
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for TRIM28. |
| Summary | |
|---|---|
| Symbol | TRIM28 |
| Name | tripartite motif containing 28 |
| Aliases | TIF1B; TF1B; RNF96; PPP1R157; protein phosphatase 1, regulatory subunit 157; tripartite motif-containing 28; ...... |
| Location | 19q13.4 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|