Browse UHRF1 in pancancer
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00628 PHD-finger PF02182 SAD/SRA domain PF12148 Tandem tudor domain within UHRF1 PF00240 Ubiquitin family |
||||||||||
| Function |
Multidomain protein that acts as a key epigenetic regulator by bridging DNA methylation and chromatin modification. Specifically recognizes and binds hemimethylated DNA at replication forks via its YDG domain and recruits DNMT1 methyltransferase to ensure faithful propagation of the DNA methylation patterns through DNA replication. In addition to its role in maintenance of DNA methylation, also plays a key role in chromatin modification: through its tudor-like regions and PHD-type zinc fingers, specifically recognizes and binds histone H3 trimethylated at 'Lys-9' (H3K9me3) and unmethylated at 'Arg-2' (H3R2me0), respectively, and recruits chromatin proteins. Enriched in pericentric heterochromatin where it recruits different chromatin modifiers required for this chromatin replication. Also localizes to euchromatic regions where it negatively regulates transcription possibly by impacting DNA methylation and histone modifications. Has E3 ubiquitin-protein ligase activity by mediating the ubiquitination of target proteins such as histone H3 and PML. It is still unclear how E3 ubiquitin-protein ligase activity is related to its role in chromatin in vivo. May be involved in DNA repair. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006342 chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006513 protein monoubiquitination GO:0010216 maintenance of DNA methylation GO:0010390 histone monoubiquitination GO:0010911 regulation of isomerase activity GO:0010912 positive regulation of isomerase activity GO:0016458 gene silencing GO:0016570 histone modification GO:0016574 histone ubiquitination GO:0031935 regulation of chromatin silencing GO:0032781 positive regulation of ATPase activity GO:0040029 regulation of gene expression, epigenetic GO:0042787 protein ubiquitination involved in ubiquitin-dependent protein catabolic process GO:0043462 regulation of ATPase activity GO:0045814 negative regulation of gene expression, epigenetic GO:0051052 regulation of DNA metabolic process GO:0051865 protein autoubiquitination GO:0060968 regulation of gene silencing GO:0090308 regulation of methylation-dependent chromatin silencing GO:1902275 regulation of chromatin organization GO:2000371 regulation of DNA topoisomerase (ATP-hydrolyzing) activity GO:2000373 positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity |
| Molecular Function |
GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0003682 chromatin binding GO:0004842 ubiquitin-protein transferase activity GO:0008327 methyl-CpG binding GO:0016874 ligase activity GO:0019787 ubiquitin-like protein transferase activity GO:0031491 nucleosome binding GO:0031493 nucleosomal histone binding GO:0035064 methylated histone binding GO:0042393 histone binding GO:0044729 hemi-methylated DNA-binding GO:0061630 ubiquitin protein ligase activity GO:0061659 ubiquitin-like protein ligase activity |
| Cellular Component |
GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0000791 euchromatin GO:0000792 heterochromatin GO:0005657 replication fork GO:0005720 nuclear heterochromatin GO:0016363 nuclear matrix GO:0034399 nuclear periphery GO:0044454 nuclear chromosome part |
| KEGG | - |
| Reactome |
R-HSA-5334118: DNA methylation R-HSA-212165: Epigenetic regulation of gene expression R-HSA-74160: Gene Expression |
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record for UHRF1. |
| There is no record. |
|
| There is no record for UHRF1. |
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
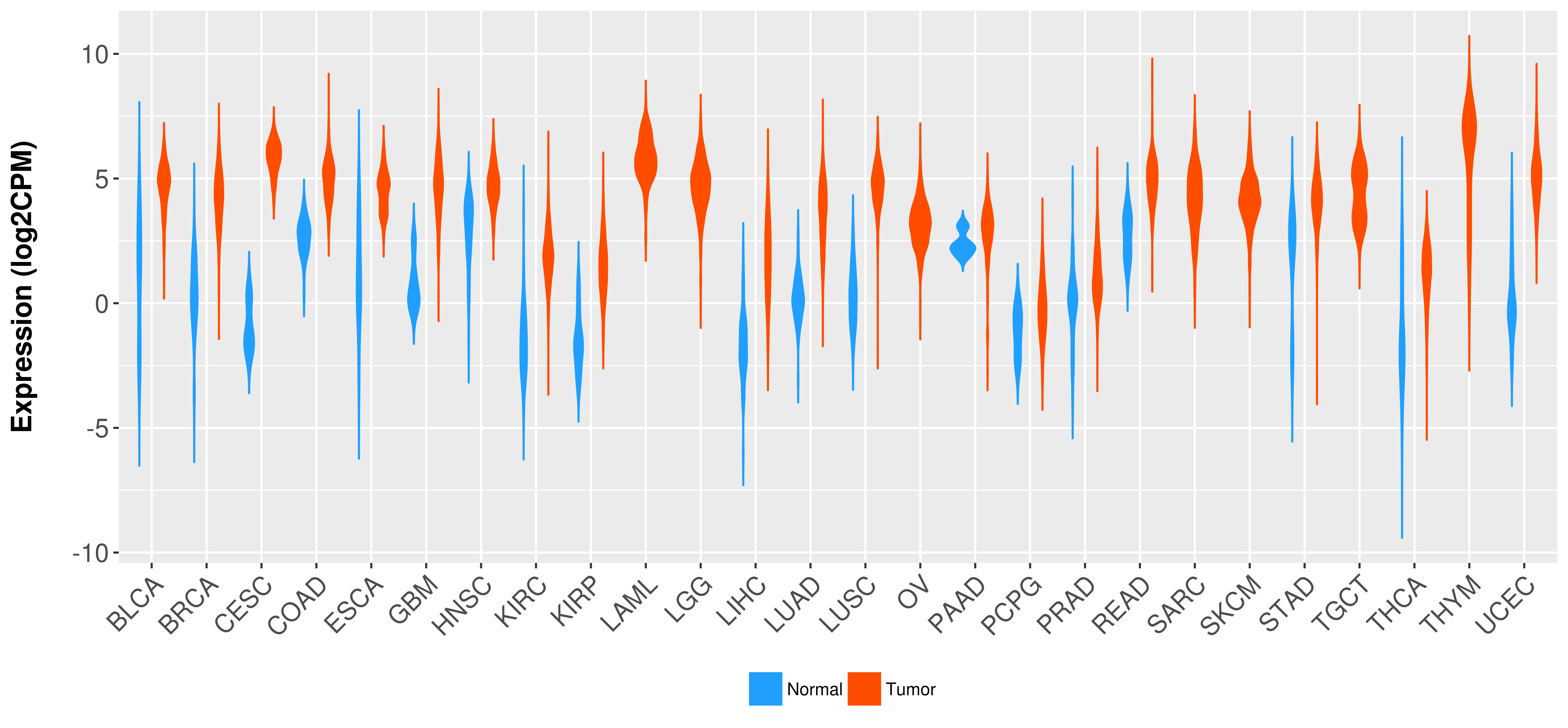
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
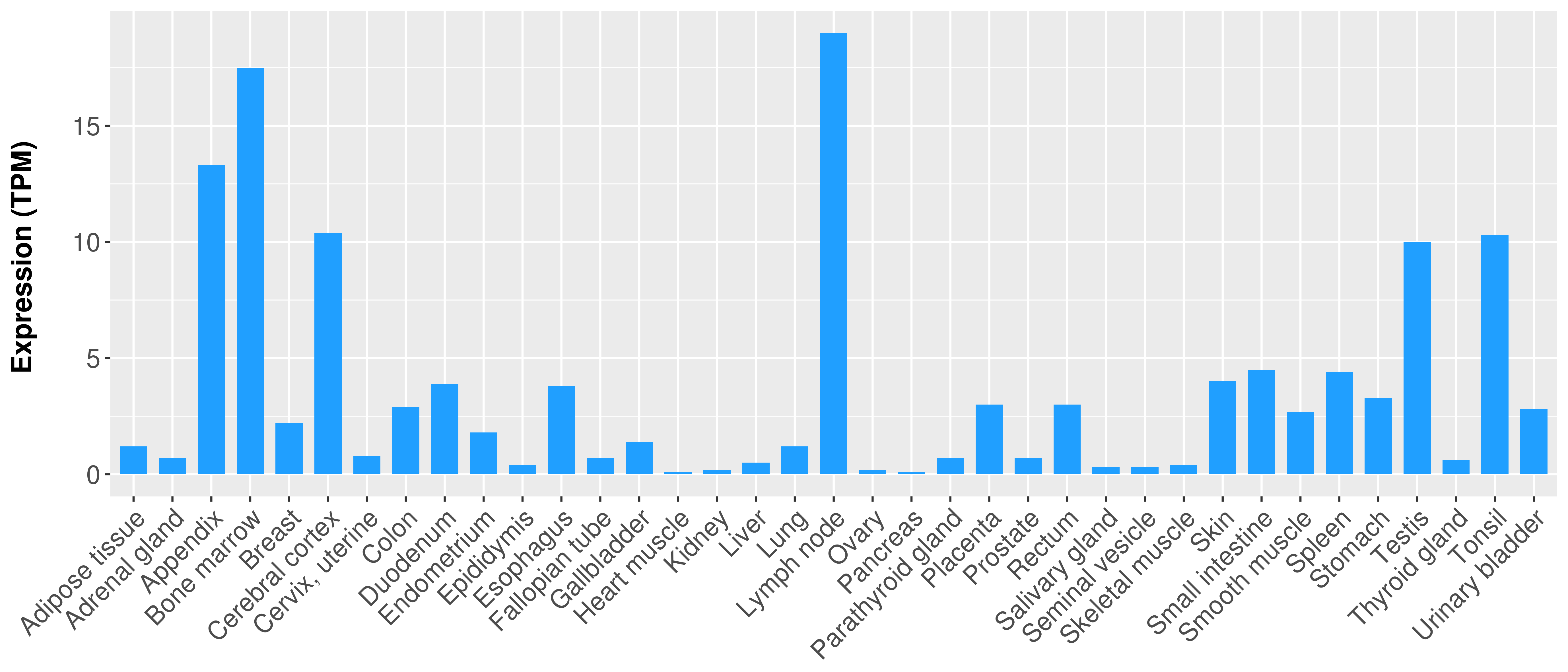
|
|
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
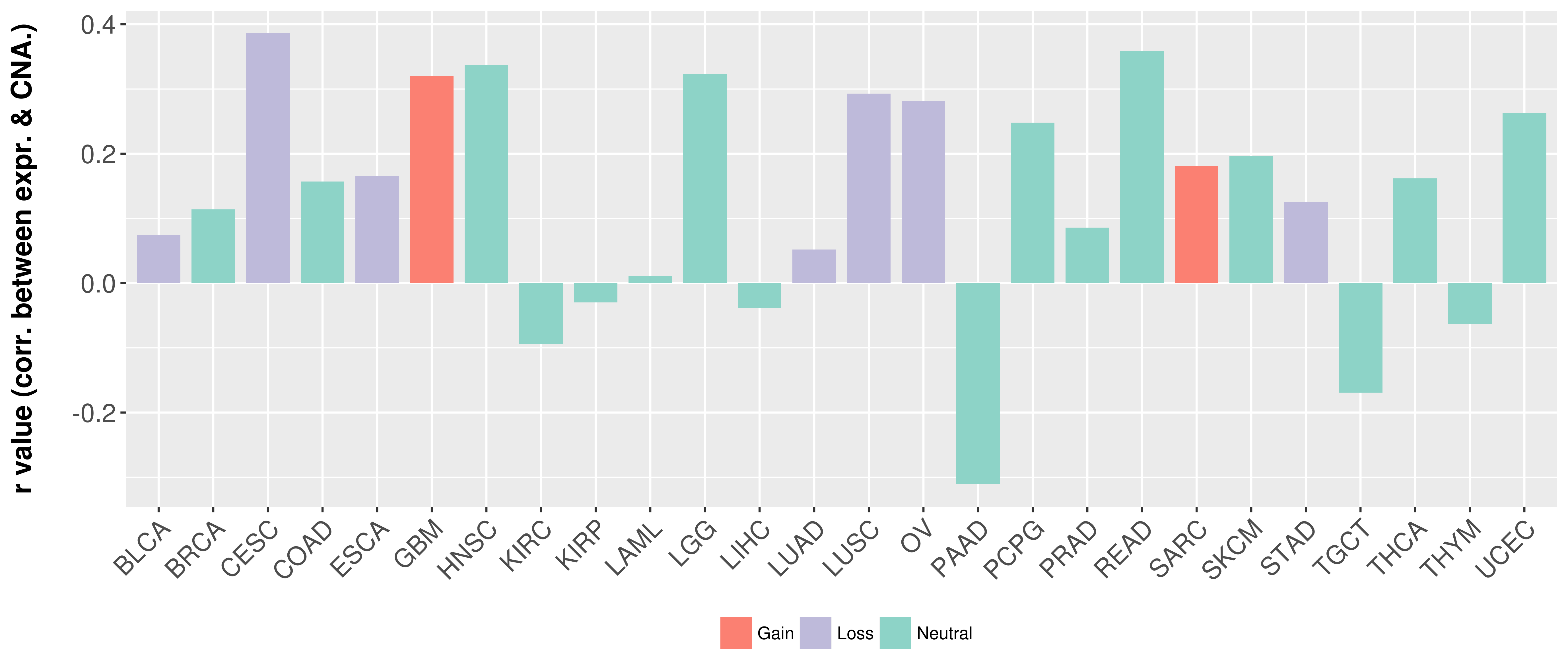
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
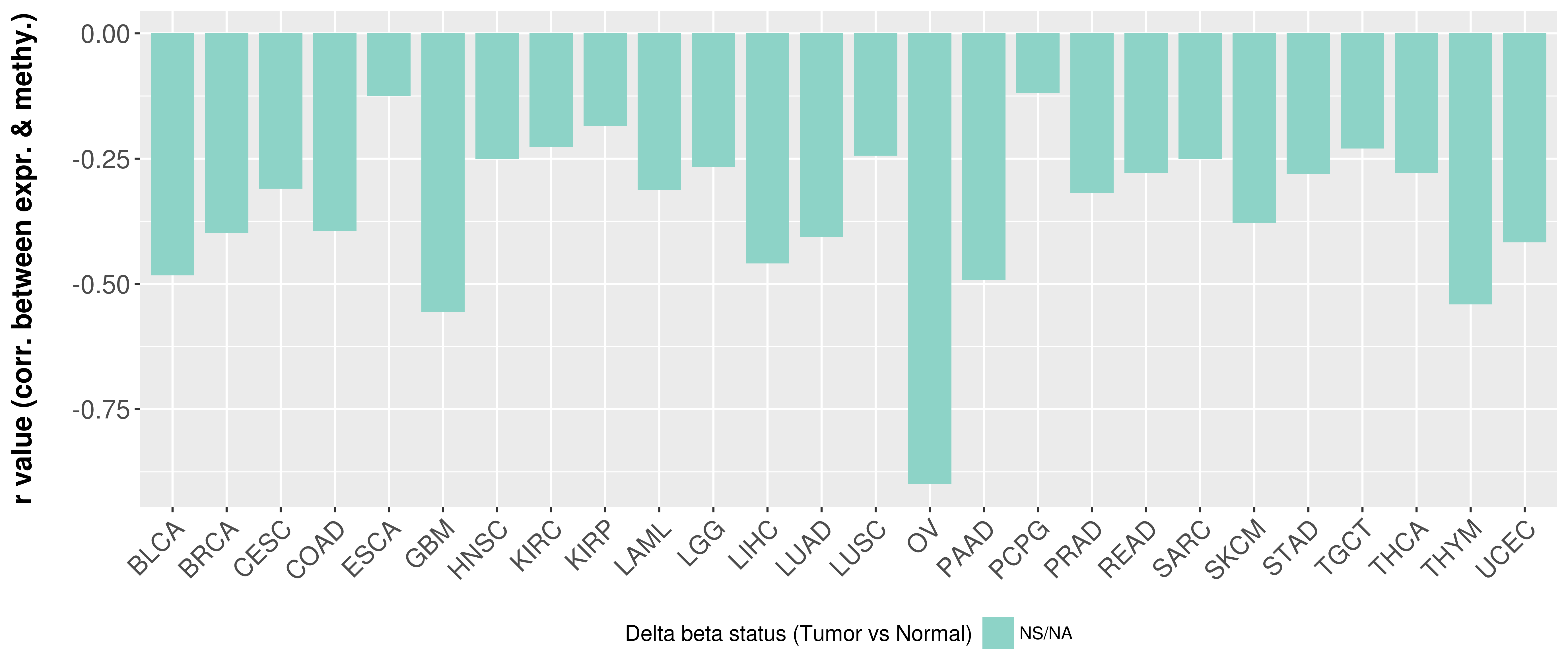
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
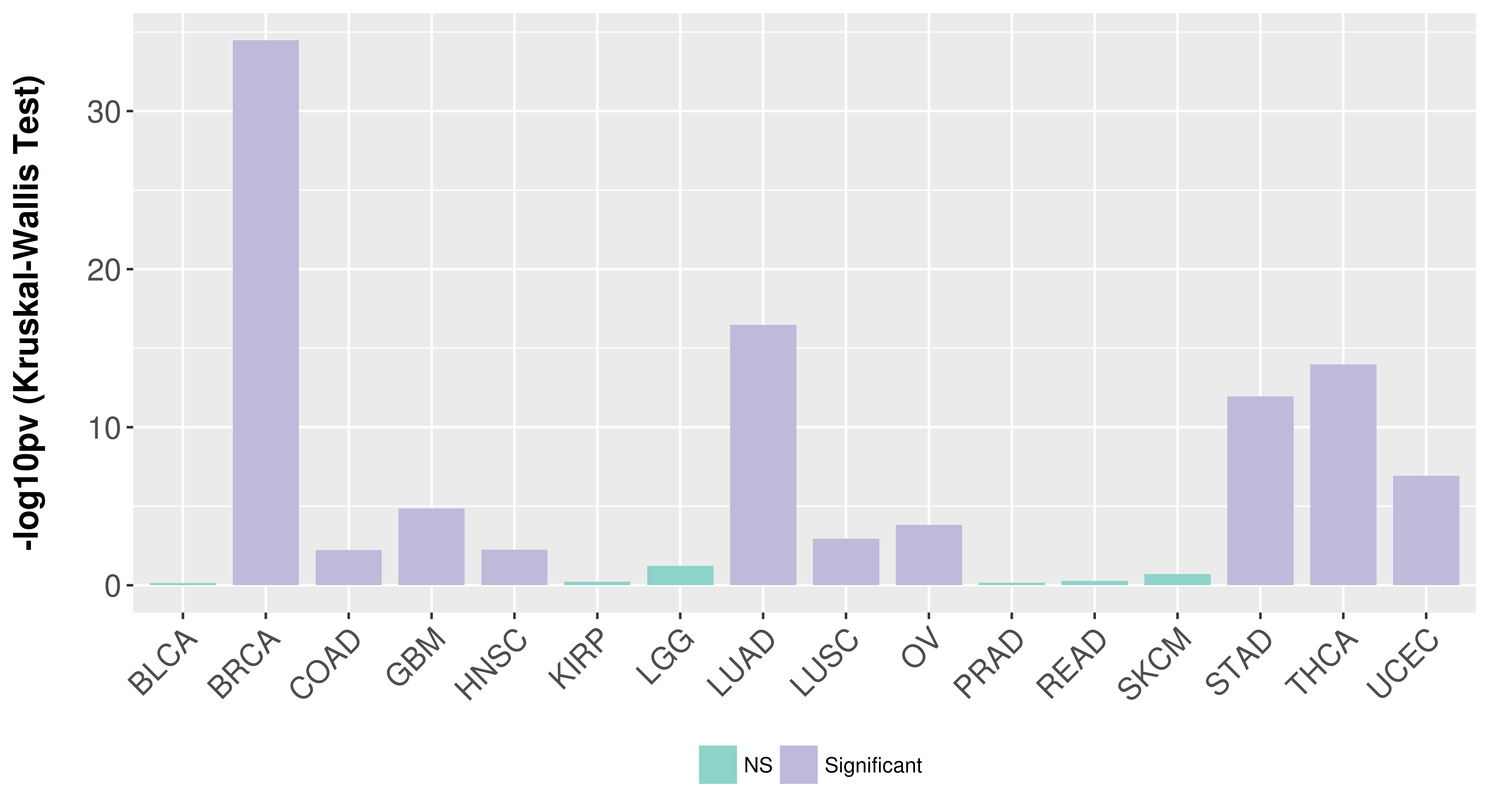
Association between expresson and subtype.
|
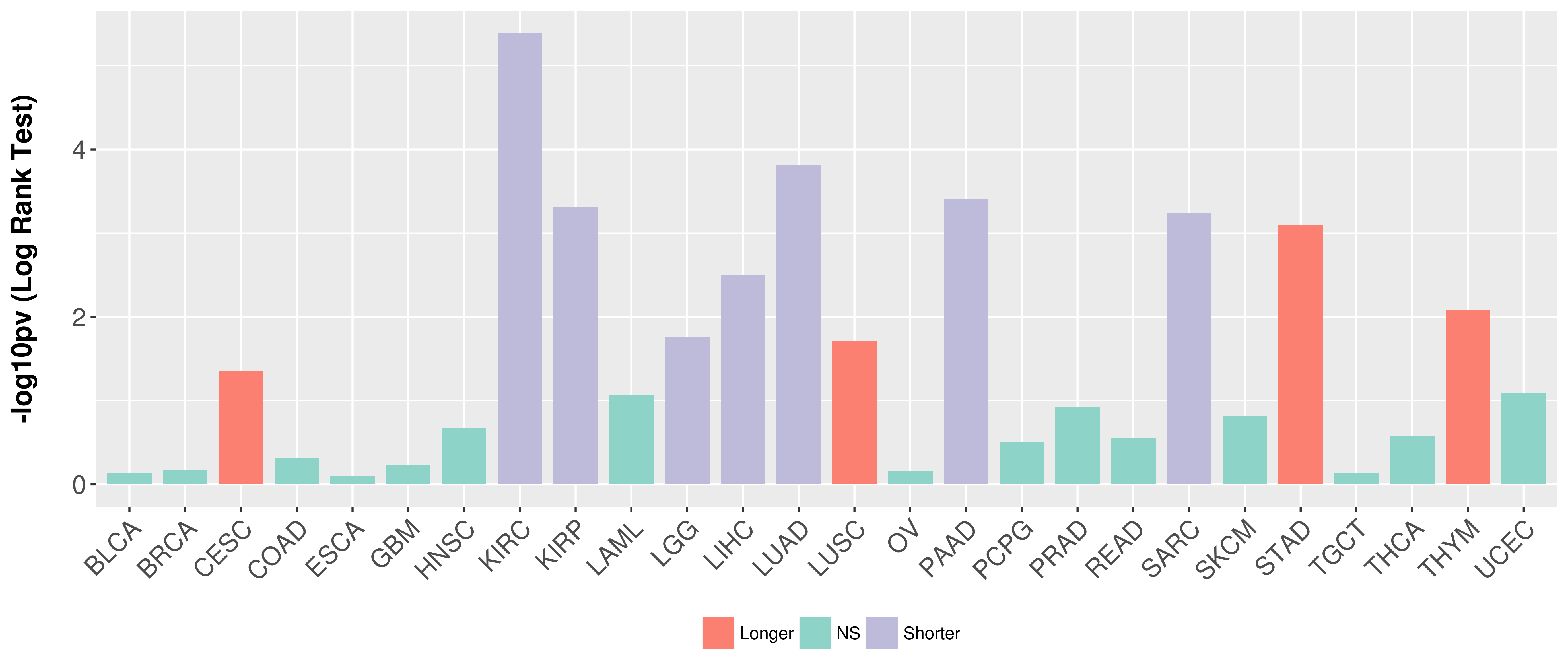
Overall survival analysis based on expression.
|
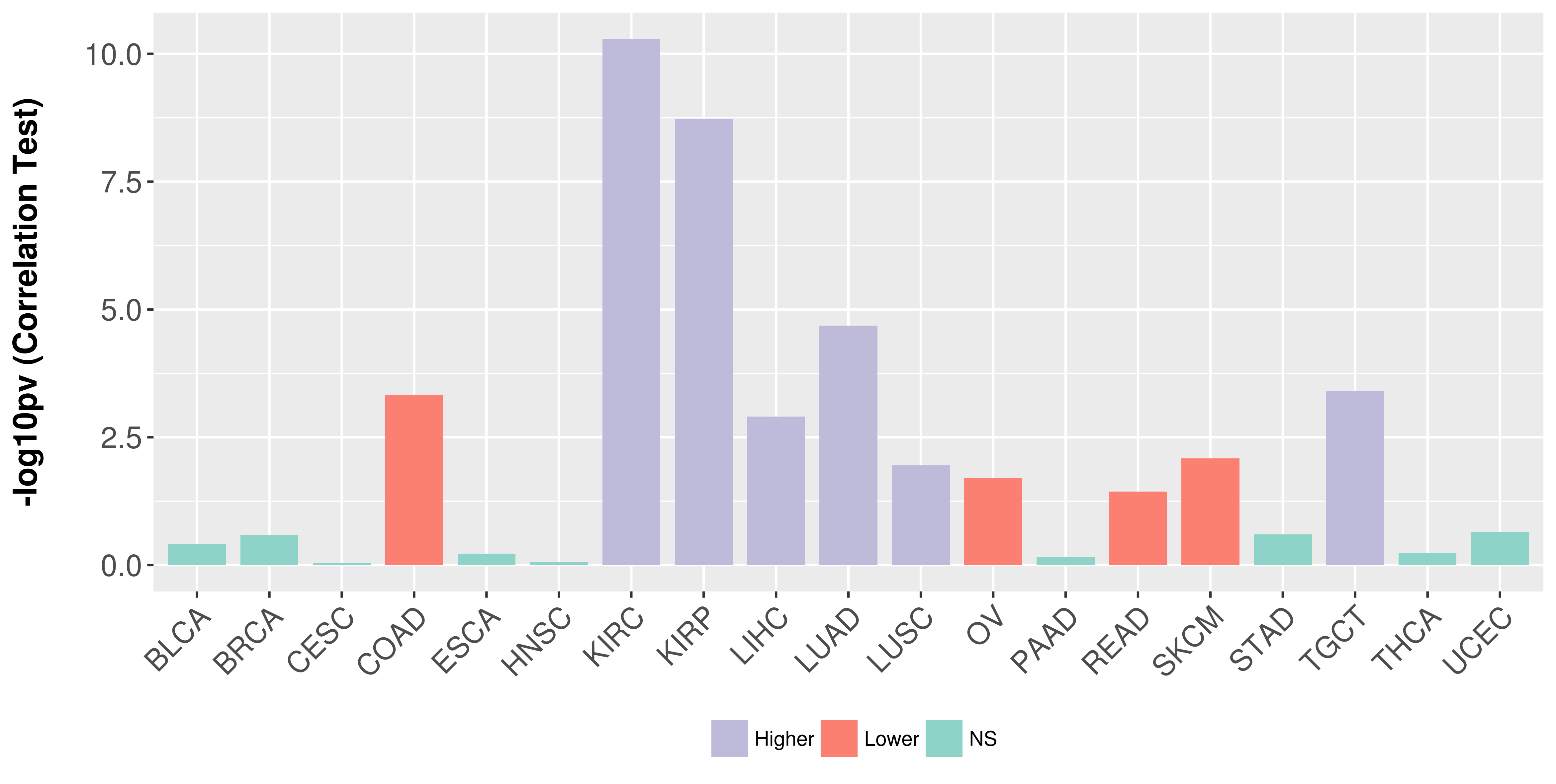
Association between expresson and stage.
|
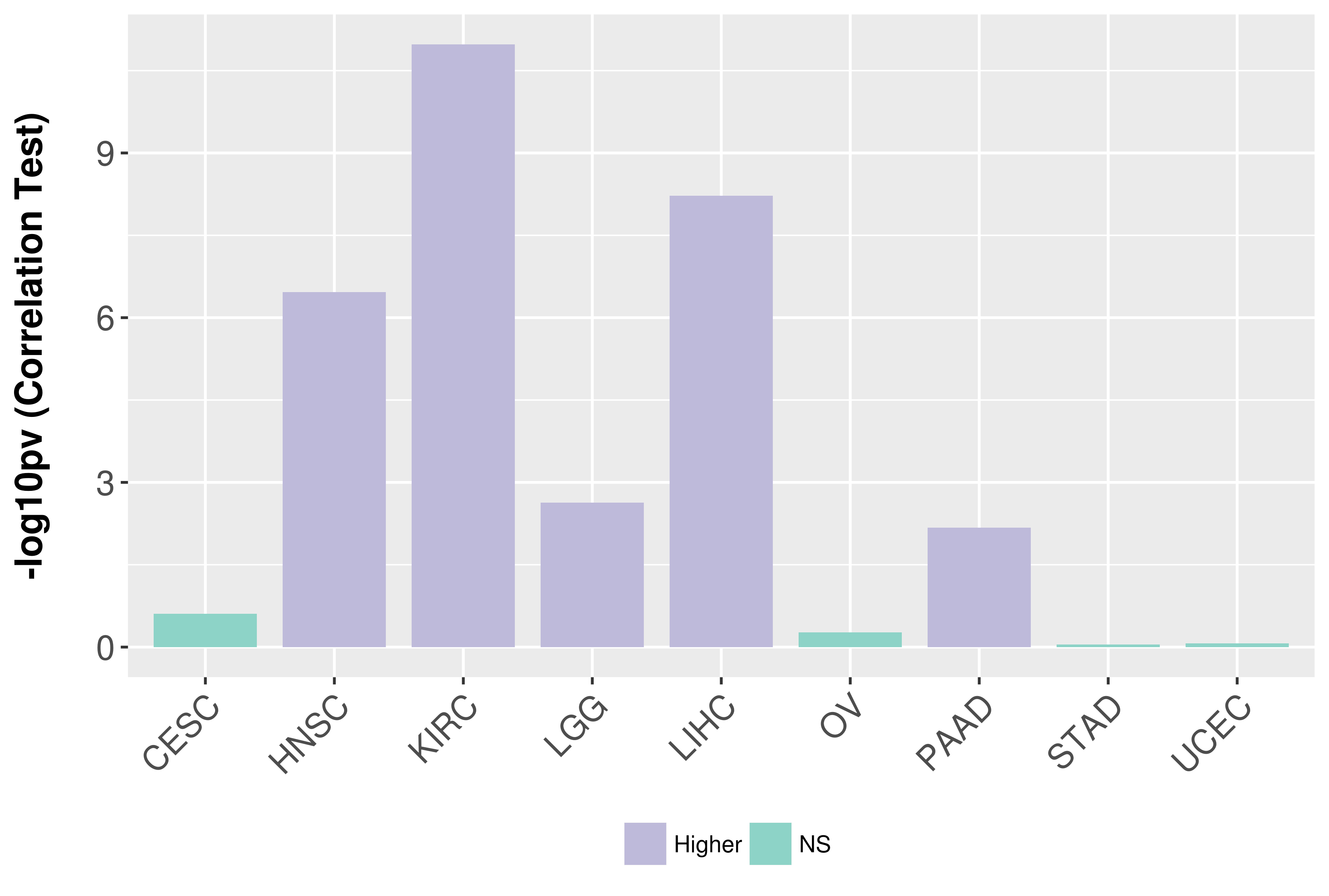
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for UHRF1. |
| Summary | |
|---|---|
| Symbol | UHRF1 |
| Name | ubiquitin like with PHD and ring finger domains 1 |
| Aliases | Np95; FLJ21925; RNF106; TDRD22; hNP95; hUHRF1; huNp95; RING finger protein 106; inverted CCAAT box-binding p ...... |
| Location | 19p13.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|