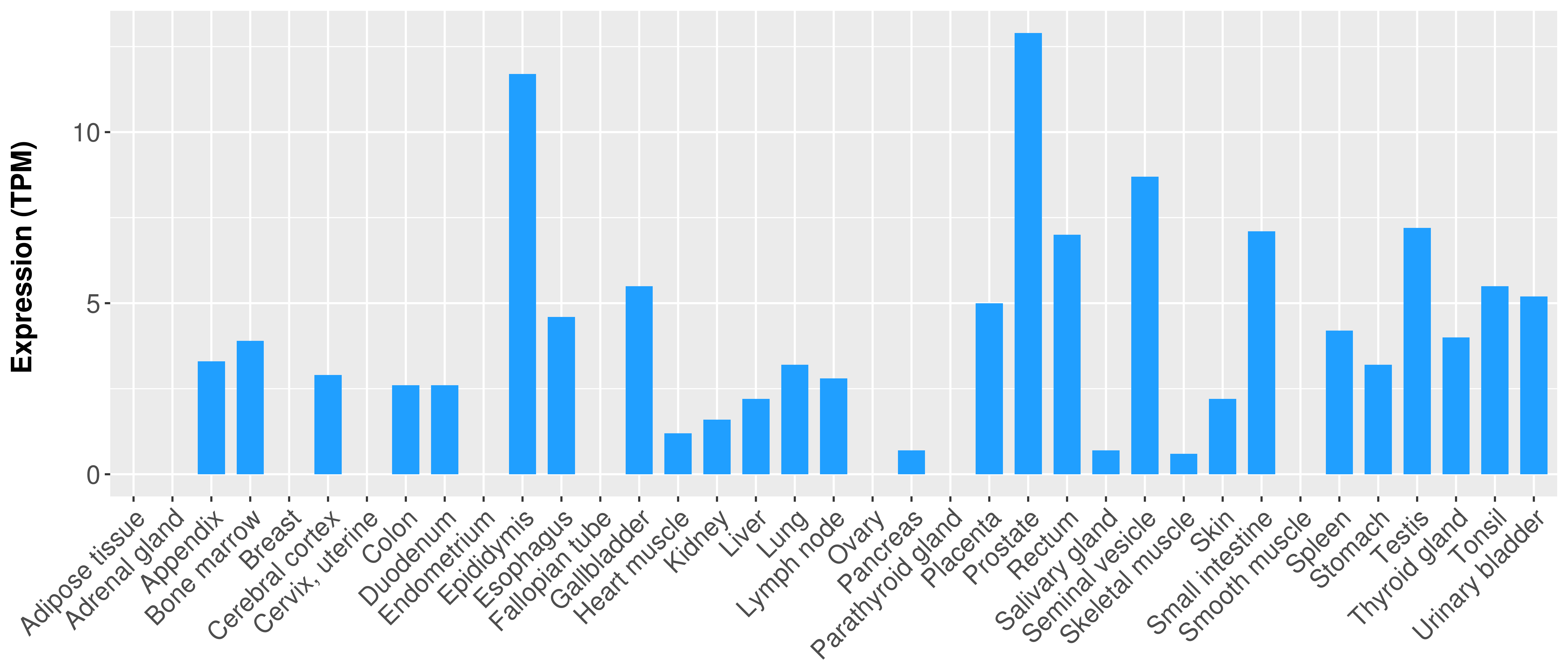
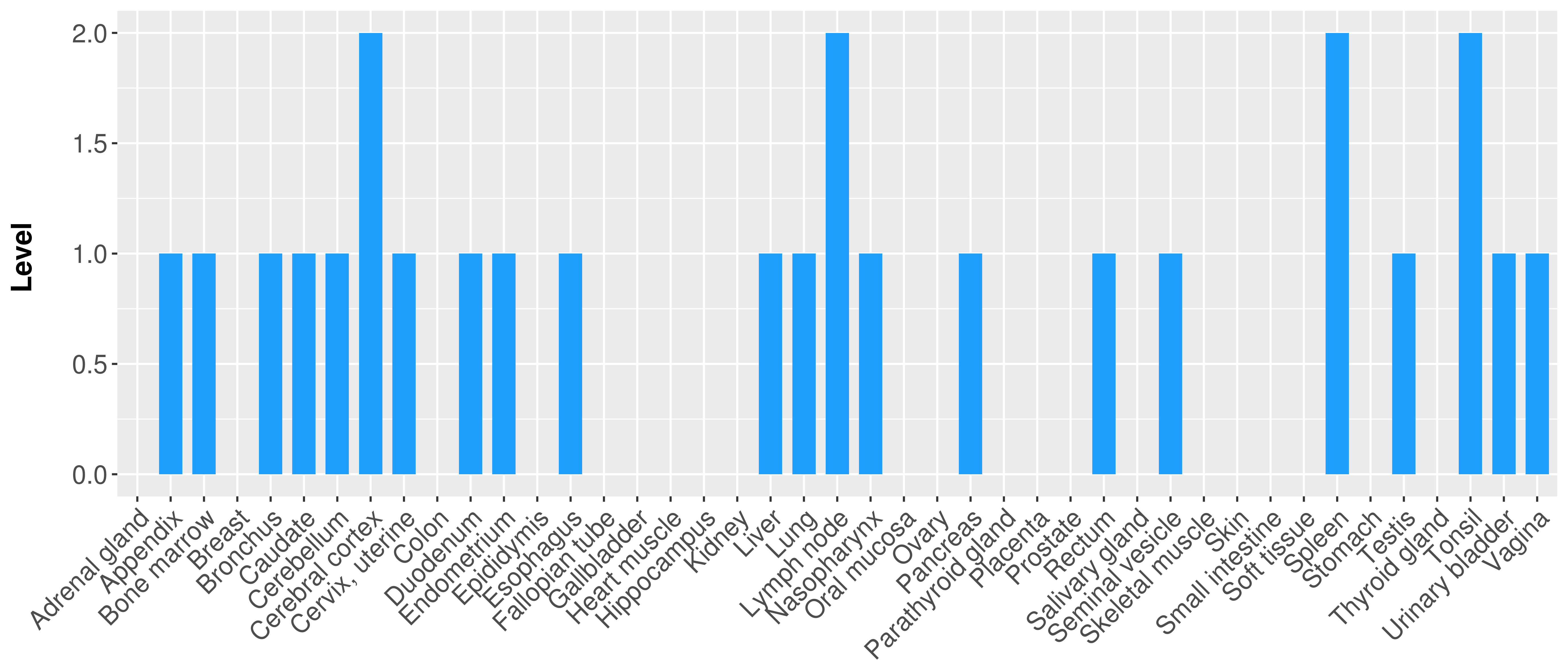
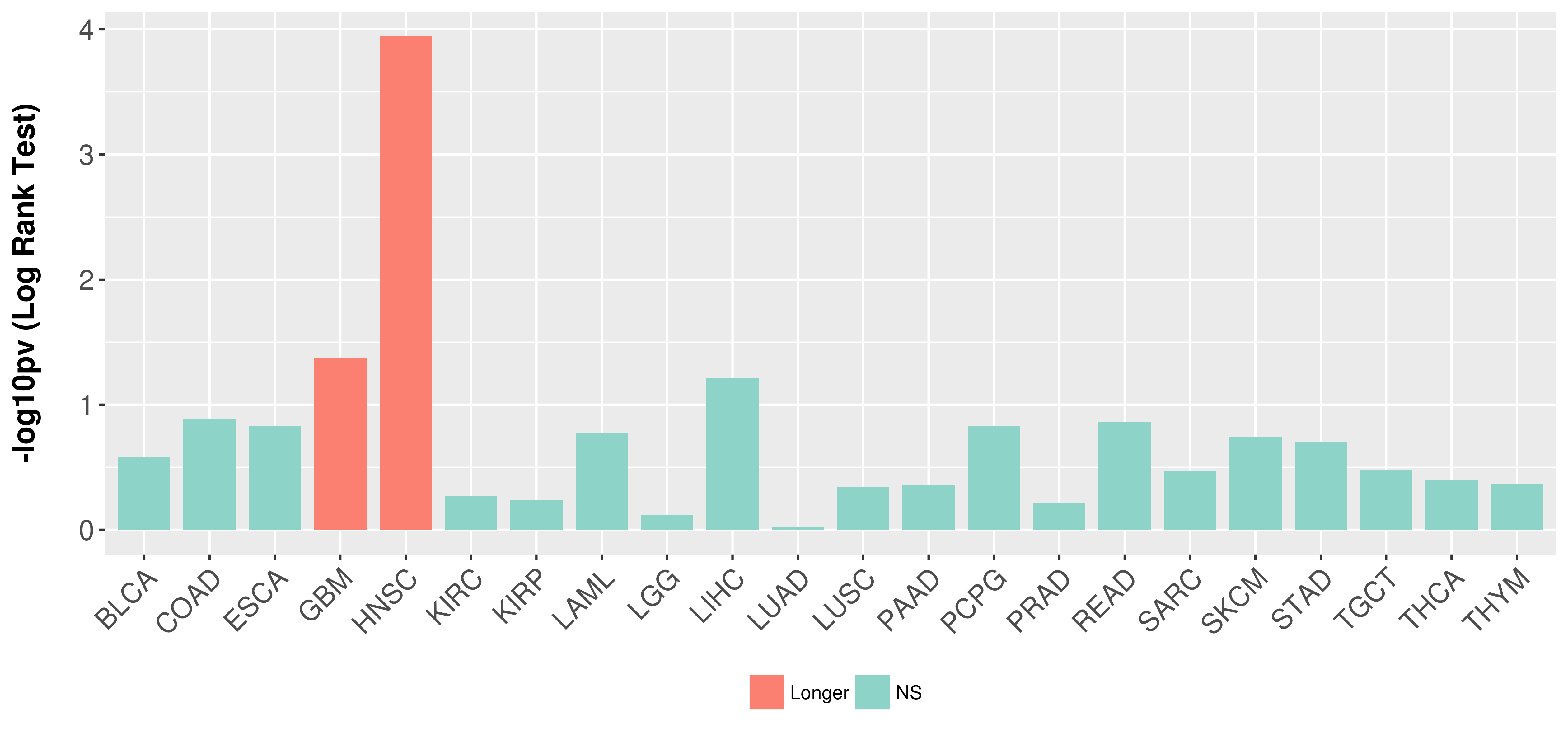
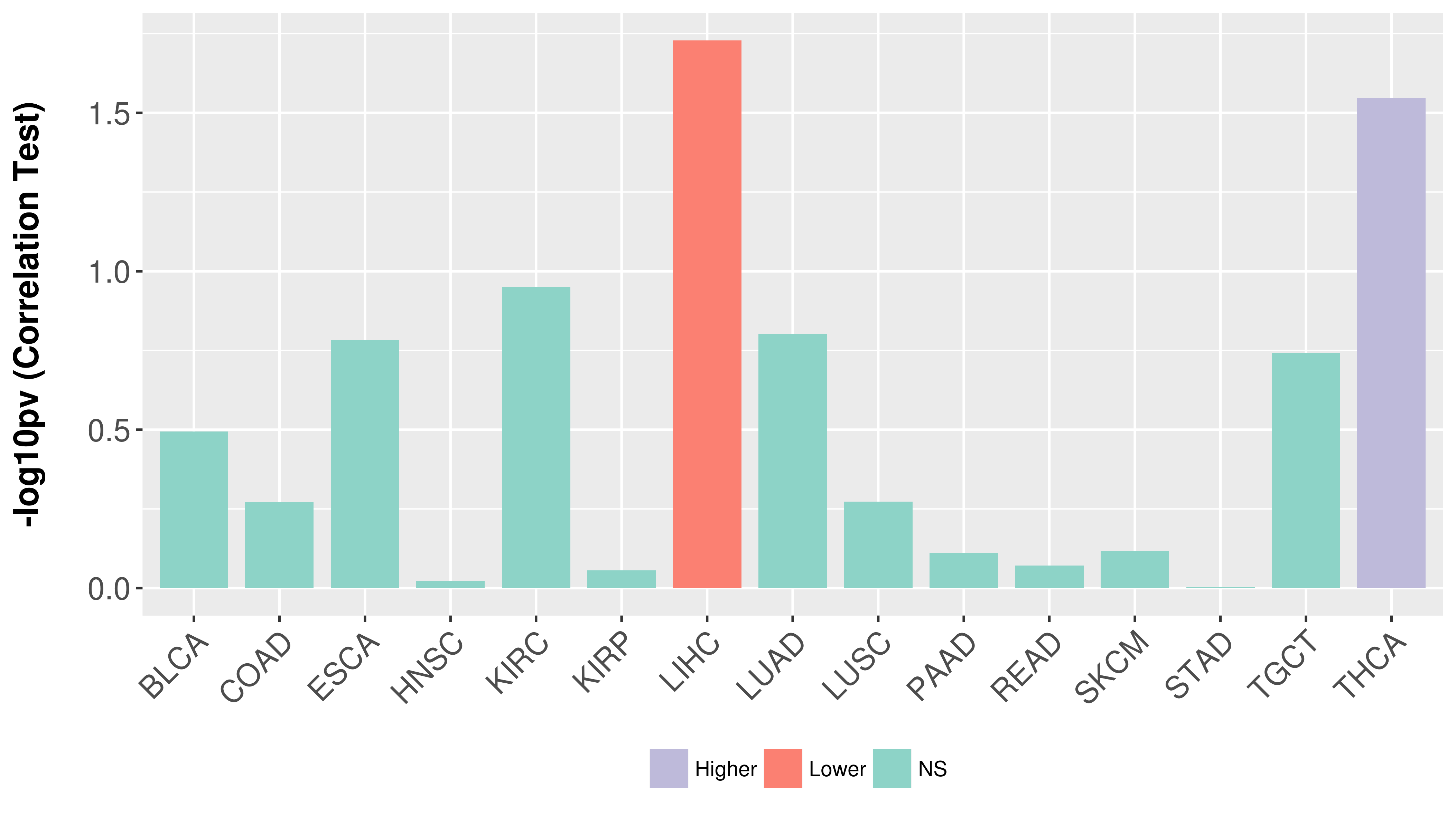
| COSM5418763 | c.112delC | p.L38fs*1 | Deletion - Frameshift | Haematopoietic_and_lymphoid_tissue
|
| COSM4902349 | c.3059G>T | p.R1020M | Substitution - Missense | Skin
|
| COSM4111710 | c.536A>G | p.D179G | Substitution - Missense | Stomach
|
| COSM1756705 | c.2029C>T | p.P677S | Substitution - Missense | Urinary_tract
|
| COSM5431620 | c.4003G>T | p.D1335Y | Substitution - Missense | Oesophagus
|
| COSM4645721 | c.3300C>T | p.C1100C | Substitution - coding silent | Large_intestine
|
| COSM3974026 | c.527T>C | p.V176A | Substitution - Missense | Central_nervous_system
|
| COSM3391911 | c.2822A>G | p.N941S | Substitution - Missense | Pancreas
|
| COSM6005447 | c.3924T>C | p.C1308C | Substitution - coding silent | Prostate
|
| COSM5757583 | c.1504G>A | p.V502I | Substitution - Missense | Large_intestine
|
| COSM3660039 | c.3659T>G | p.L1220R | Substitution - Missense | Liver
|
| COSM4740048 | c.3029A>G | p.N1010S | Substitution - Missense | Large_intestine
|
| COSM1756702 | c.2479C>T | p.Q827* | Substitution - Nonsense | Urinary_tract
|
| COSM3974023 | c.3455T>C | p.V1152A | Substitution - Missense | Central_nervous_system
|
| COSM488715 | c.3170C>A | p.A1057E | Substitution - Missense | Kidney
|
| COSM1469995 | c.3909A>T | p.V1303V | Substitution - coding silent | Large_intestine
|
| COSM3584396 | c.1274T>A | p.L425* | Substitution - Nonsense | Skin
|
| COSM4789876 | c.922A>G | p.I308V | Substitution - Missense | Liver
|
| COSM316443 | c.1979G>T | p.G660V | Substitution - Missense | Soft_tissue
|
| COSM1293085 | c.1264C>T | p.P422S | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM4610649 | c.1178T>G | p.F393C | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1159056 | c.1777delC | p.H593fs*111 | Deletion - Frameshift | Pancreas
|
| COSM1626211 | c.430C>A | p.H144N | Substitution - Missense | Liver
|
| COSM4449551 | c.2674-1G>A | p.? | Unknown | Skin
|
| COSM1644109 | c.3786A>G | p.K1262K | Substitution - coding silent | Stomach
|
| COSM1741515 | c.3238C>T | p.Q1080* | Substitution - Nonsense | Urinary_tract
|
| COSM316443 | c.1979G>T | p.G660V | Substitution - Missense | Lung
|
| COSM1159056 | c.1777delC | p.H593fs*111 | Deletion - Frameshift | Pancreas
|
| COSM1315739 | c.2793C>G | p.F931L | Substitution - Missense | Urinary_tract
|
| COSM3380265 | c.2422A>G | p.K808E | Substitution - Missense | Pancreas
|
| COSM5120308 | c.3053C>A | p.S1018Y | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM3660036 | c.3671T>A | p.M1224K | Substitution - Missense | Liver
|
| COSM3720483 | c.2888G>T | p.G963V | Substitution - Missense | Haematopoietic_and_lymphoid_tissue
|
| COSM1190663 | c.2252A>T | p.H751L | Substitution - Missense | Lung
|
| COSM1469997 | c.1169G>T | p.R390I | Substitution - Missense | Large_intestine
|
| COSM1715916 | c.3272C>T | p.P1091L | Substitution - Missense | Skin
|
| COSM1745546 | c.92_93insAG | p.S31fs*9 | Insertion - Frameshift | Urinary_tract
|
| COSM3660039 | c.3659T>G | p.L1220R | Substitution - Missense | Liver
|
| COSM3584393 | c.3508G>A | p.G1170S | Substitution - Missense | Skin
|
| COSM5643478 | c.3692C>G | p.S1231* | Substitution - Nonsense | Oesophagus
|
| COSM360674 | c.129G>A | p.R43R | Substitution - coding silent | Lung
|
| COSM3660042 | c.1525C>A | p.P509T | Substitution - Missense | Liver
|
| COSM1469993 | c.3955T>G | p.F1319V | Substitution - Missense | Large_intestine
|
| COSM316441 | c.3430C>G | p.L1144V | Substitution - Missense | Lung
|
| COSM5724547 | c.1503C>A | p.S501R | Substitution - Missense | Skin
|
| COSM3144212 | c.3922T>C | p.C1308R | Substitution - Missense | Large_intestine
|
| COSM1756702 | c.2479C>T | p.Q827* | Substitution - Nonsense | Urinary_tract
|
| COSM3800799 | c.808C>T | p.L270F | Substitution - Missense | Urinary_tract
|
| COSM316441 | c.3430C>G | p.L1144V | Substitution - Missense | Soft_tissue
|
| COSM1315743 | c.124G>T | p.E42* | Substitution - Nonsense | Urinary_tract
|
| COSM758045 | c.1911T>C | p.N637N | Substitution - coding silent | Lung
|
| COSM4740045 | c.3655C>T | p.H1219Y | Substitution - Missense | Large_intestine
|
| COSM3426800 | c.915G>A | p.R305R | Substitution - coding silent | Large_intestine
|
| COSM4448963 | c.152+2T>C | p.? | Unknown | Skin
|
| COSM1756702 | c.2479C>T | p.Q827* | Substitution - Nonsense | Urinary_tract
|
| COSM3660036 | c.3671T>A | p.M1224K | Substitution - Missense | Liver
|
| COSM5476925 | c.1113C>T | p.C371C | Substitution - coding silent | Large_intestine
|
| COSM1315741 | c.1719G>A | p.L573L | Substitution - coding silent | Urinary_tract
|
| COSM3660042 | c.1525C>A | p.P509T | Substitution - Missense | Liver
|
| COSM758047 | c.3846G>T | p.E1282D | Substitution - Missense | Lung
|
| COSM5809717 | c.588A>T | p.P196P | Substitution - coding silent | Liver
|
| COSM1756702 | c.2479C>T | p.Q827* | Substitution - Nonsense | Urinary_tract
|
| COSM4429344 | c.2689G>A | p.D897N | Substitution - Missense | Oesophagus
|
| COSM4789876 | c.922A>G | p.I308V | Substitution - Missense | Liver
|
| COSM4654578 | c.2848C>T | p.Q950* | Substitution - Nonsense | Large_intestine
|
| COSM5047409 | c.906_907insT | p.S303fs*7 | Insertion - Frameshift | Oesophagus
|
| COSM1626213 | c.395G>T | p.G132V | Substitution - Missense | Liver
|