microRNA information: hsa-miR-126-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-126-5p | miRbase |
| Accession: | MIMAT0000444 | miRbase |
| Precursor name: | hsa-mir-126 | miRbase |
| Precursor accession: | MI0000471 | miRbase |
| Symbol: | MIR126 | HGNC |
| RefSeq ID: | NR_029695 | GenBank |
| Sequence: | CAUUAUUACUUUUGGUACGCG |
Reported expression in cancers: hsa-miR-126-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-126-5p | acute myeloid leukemia | upregulation | "In this manner we identified multiple differential ......" | 24477595 | |
| hsa-miR-126-5p | breast cancer | downregulation | "Expression of spliced exon7-8 and excised mature m ......" | 21249429 | Northern blot |
| hsa-miR-126-5p | breast cancer | downregulation | "MiR-126 expression was investigated in forty cases ......" | 26261534 | qPCR |
| hsa-miR-126-5p | cervical and endocervical cancer | downregulation | "miR-126 is an endothelial-specific microRNA essent ......" | 24037526 | |
| hsa-miR-126-5p | cervical and endocervical cancer | downregulation | "In cervical cancer one of the most common malignan ......" | 24377569 | qPCR |
| hsa-miR-126-5p | colon cancer | downregulation | "Additionally using reporter constructs we show tha ......" | 18663744 | |
| hsa-miR-126-5p | colon cancer | downregulation | "However the underlying mechanisms of miR-126 in co ......" | 27517626 | |
| hsa-miR-126-5p | colorectal cancer | downregulation | "Epigenetic silencing of miR 126 contributes to tum ......" | 23900443 | qPCR |
| hsa-miR-126-5p | colorectal cancer | downregulation | "MiR-126 has been shown to be down-regulated in CRC ......" | 24312276 | |
| hsa-miR-126-5p | esophageal cancer | downregulation | "Using next-generation sequencing NGS-based miRNA p ......" | 25512445 | RNA-Seq; in situ hybridization |
| hsa-miR-126-5p | esophageal cancer | downregulation | "MicroRNA 126 is down regulated in human esophageal ......" | 26191164 | |
| hsa-miR-126-5p | gastric cancer | downregulation | "miR 126 functions as a tumour suppressor in human ......" | 20619534 | |
| hsa-miR-126-5p | gastric cancer | downregulation | "MiR-126 expression was investigated in GC tissue s ......" | 25027343 | qPCR |
| hsa-miR-126-5p | kidney renal cell cancer | downregulation | "Low expression of miR 126 is a prognostic marker f ......" | 25572155 | |
| hsa-miR-126-5p | kidney renal cell cancer | downregulation | "Conversely decreased expression of miR-106b miR-99 ......" | 26416448 | |
| hsa-miR-126-5p | liver cancer | deregulation | "Array-based miRNA profiling was performed on HCC c ......" | 18555017 | Microarray |
| hsa-miR-126-5p | liver cancer | downregulation | "Decreased expression of miR 126 correlates with me ......" | 23378255 | Microarray; qPCR |
| hsa-miR-126-5p | liver cancer | upregulation | "Hepatic miR 126 is a potential plasma biomarker fo ......" | 26756996 | |
| hsa-miR-126-5p | lung cancer | downregulation | "In the consistently reported down-regulated microR ......" | 22672859 | |
| hsa-miR-126-5p | lung cancer | downregulation | "let 7b and miR 126 are down regulated in tumor tis ......" | 23029111 | qPCR |
| hsa-miR-126-5p | lung squamous cell cancer | downregulation | "Down regulation of microRNA 126 and microRNA 133b ......" | 26823832 | qPCR |
| hsa-miR-126-5p | lymphoma | downregulation | "Clinical and epidemiological data suggest that chr ......" | 22307176 | Reverse transcription PCR; in situ hybridization |
| hsa-miR-126-5p | pancreatic cancer | downregulation | "Loss of miR 126 is crucial to pancreatic cancer pr ......" | 22845403 | |
| hsa-miR-126-5p | prostate cancer | downregulation | "Association of microRNA 126 expression with clinic ......" | 24350576 | Reverse transcription PCR; qPCR |
| hsa-miR-126-5p | sarcoma | upregulation | "The discovery of microRNAs miRNAs provides a new a ......" | 24384842 | |
| hsa-miR-126-5p | sarcoma | downregulation | "miR-126 has been reported to be downregulated in O ......" | 25213697 | |
| hsa-miR-126-5p | sarcoma | deregulation | "The expression level of miR-126 was determined by ......" | 26194657 | qPCR |
| hsa-miR-126-5p | thyroid cancer | downregulation | "microRNA-126 miR-126 has been reported to play tum ......" | 26239517 | |
| hsa-miR-126-5p | thyroid cancer | downregulation | "The objectives of this study are to investigate th ......" | 26384552 | |
| hsa-miR-126-5p | thyroid cancer | downregulation | "MicroRNA-126 miR-126 has previously been reported ......" | 27175968 | qPCR |
Reported cancer pathway affected by hsa-miR-126-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-126-5p | acute myeloid leukemia | Apoptosis pathway | "Attenuation of microRNA 126 expression that drives ......" | 24477595 | |
| hsa-miR-126-5p | bladder cancer | PI3K/Akt signaling pathway; cell cycle pathway; Apoptosis pathway | "MiR 126 regulates proliferation and invasion in th ......" | 27578985 | Luciferase; Western blot; Flow cytometry; Colony formation; Transwell assay |
| hsa-miR-126-5p | breast cancer | PI3K/Akt signaling pathway | "Endothelial specific intron derived miR 126 is dow ......" | 21249429 | Western blot; Luciferase |
| hsa-miR-126-5p | colon cancer | PI3K/Akt signaling pathway | "The noncoding RNA miR 126 suppresses the growth of ......" | 18663744 | |
| hsa-miR-126-5p | colon cancer | cell cycle pathway | "MiR 126 suppresses colon cancer cell proliferation ......" | 23615712 | |
| hsa-miR-126-5p | colon cancer | Apoptosis pathway | "miR 126 inhibits colon cancer proliferation and in ......" | 23981989 | |
| hsa-miR-126-5p | colorectal cancer | cell cycle pathway; Apoptosis pathway | "MicroRNA 126 functions as a tumor suppressor in co ......" | 24189753 | Transwell assay; Western blot; Luciferase |
| hsa-miR-126-5p | colorectal cancer | cell cycle pathway; Apoptosis pathway | "Down regulation of miR 126 is associated with colo ......" | 24312276 | Luciferase |
| hsa-miR-126-5p | colorectal cancer | Apoptosis pathway | "Deregulation of miR 126 expression in colorectal c ......" | 26455548 | |
| hsa-miR-126-5p | esophageal cancer | PI3K/Akt signaling pathway | "Using next-generation sequencing NGS-based miRNA p ......" | 25512445 | RNAi |
| hsa-miR-126-5p | esophageal cancer | PI3K/Akt signaling pathway | "MicroRNA 126 is down regulated in human esophageal ......" | 26191164 | Luciferase |
| hsa-miR-126-5p | gastric cancer | cell cycle pathway | "miR 126 functions as a tumour suppressor in human ......" | 20619534 | |
| hsa-miR-126-5p | gastric cancer | cell cycle pathway | "MicroRNA 126 inhibits cell proliferation in gastri ......" | 26054677 | |
| hsa-miR-126-5p | kidney renal cell cancer | Apoptosis pathway | "Low expression of miR 126 is a prognostic marker f ......" | 25572155 | |
| hsa-miR-126-5p | liver cancer | Apoptosis pathway; cell cycle pathway | "miR 126 inhibits cell proliferation and induces ce ......" | 25585946 | Luciferase |
| hsa-miR-126-5p | lung cancer | cell cycle pathway | "MiR 126 restoration down regulate VEGF and inhibit ......" | 19223090 | Luciferase; Flow cytometry |
| hsa-miR-126-5p | lung squamous cell cancer | Apoptosis pathway | "Expression profile of miRNA in these two groups wa ......" | 20728239 | |
| hsa-miR-126-5p | lung squamous cell cancer | cell cycle pathway | "miR 126 inhibits proliferation of small cell lung ......" | 21439283 | |
| hsa-miR-126-5p | lung squamous cell cancer | PI3K/Akt signaling pathway | "miR 126 enhances the sensitivity of non small cell ......" | 22510476 | |
| hsa-miR-126-5p | lung squamous cell cancer | cell cycle pathway; Apoptosis pathway | "Matrine induces cell cycle arrest and apoptosis wi ......" | 27665734 | Western blot |
| hsa-miR-126-5p | prostate cancer | Apoptosis pathway | "The effects of AX2 on mTOR PTEN PI3K and Akt at th ......" | 26645890 | Western blot; Luciferase; Flow cytometry |
| hsa-miR-126-5p | sarcoma | Apoptosis pathway | "miR 126 functions as a tumor suppressor in osteosa ......" | 24384842 | Luciferase |
| hsa-miR-126-5p | sarcoma | Apoptosis pathway; cell cycle pathway | "Overexpression of miR 126 sensitizes osteosarcoma ......" | 25510179 | Flow cytometry; MTT assay |
| hsa-miR-126-5p | sarcoma | cell cycle pathway | "MicroRNA 126 Overexpression Inhibits Proliferation ......" | 26319109 | Transwell assay; Wound Healing Assay |
| hsa-miR-126-5p | thyroid cancer | cell cycle pathway; Apoptosis pathway | "miR 126 inhibits papillary thyroid carcinoma growt ......" | 26239517 | Colony formation |
| hsa-miR-126-5p | thyroid cancer | cell cycle pathway; Apoptosis pathway | "Interactive role of miR 126 on VEGF A and progress ......" | 27067785 | Western blot |
Reported cancer prognosis affected by hsa-miR-126-5p
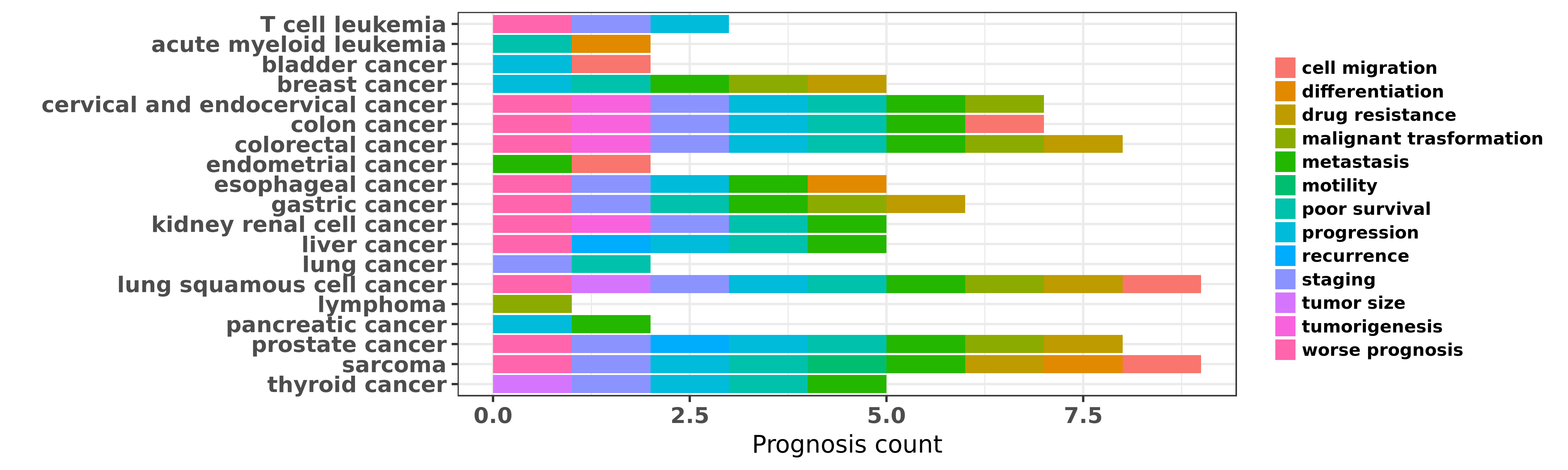
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-126-5p | T cell leukemia | progression; worse prognosis; staging | "Impact of miR 155 and miR 126 as novel biomarkers ......" | 22884882 | |
| hsa-miR-126-5p | acute myeloid leukemia | differentiation | "We studied miRNA expression of leukemic blasts of ......" | 20425795 | |
| hsa-miR-126-5p | acute myeloid leukemia | poor survival | "Attenuation of microRNA 126 expression that drives ......" | 24477595 | |
| hsa-miR-126-5p | bladder cancer | progression | "MicroRNA 126 inhibits invasion in bladder cancer v ......" | 24823697 | |
| hsa-miR-126-5p | bladder cancer | cell migration | "MiR 126 regulates proliferation and invasion in th ......" | 27578985 | Luciferase; Western blot; Flow cytometry; Colony formation; Transwell assay |
| hsa-miR-126-5p | breast cancer | metastasis; poor survival | "Of these microRNAs miR-126 restoration reduces ove ......" | 18185580 | |
| hsa-miR-126-5p | breast cancer | metastasis | "The third identifies miR-335 miR-206 and miR-126 a ......" | 18373886 | |
| hsa-miR-126-5p | breast cancer | metastasis | "Genetic variants within miR 126 and miR 335 are no ......" | 21046227 | |
| hsa-miR-126-5p | breast cancer | drug resistance | "MiRNA microarray analysis identified 299 and 226 m ......" | 21399894 | |
| hsa-miR-126-5p | breast cancer | drug resistance | "The inhibition of cell proliferation was measured ......" | 21563499 | MTT assay |
| hsa-miR-126-5p | breast cancer | drug resistance | "Expression levels of miR-210 miR-21 miR-29a and mi ......" | 22370716 | |
| hsa-miR-126-5p | breast cancer | metastasis | "Real Time RT-PCR was performed to identify the miR ......" | 22524830 | |
| hsa-miR-126-5p | breast cancer | metastasis | "miR 126 and miR 126* repress recruitment of mesenc ......" | 23396050 | |
| hsa-miR-126-5p | breast cancer | poor survival | "Increased expression of miR 126 and miR 10a predic ......" | 23968733 | |
| hsa-miR-126-5p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-126-5p | breast cancer | progression | "We identified eight microRNAs miR-10a miR-10b miR- ......" | 27433802 | |
| hsa-miR-126-5p | cervical and endocervical cancer | tumorigenesis; progression; staging | "Repression of miR 126 and upregulation of adrenome ......" | 24037526 | |
| hsa-miR-126-5p | cervical and endocervical cancer | malignant trasformation | "miR 126 Suppresses the proliferation of cervical c ......" | 24377569 | MTT assay |
| hsa-miR-126-5p | cervical and endocervical cancer | worse prognosis; poor survival; staging; metastasis | "Decreased expression of microRNA 126 is associated ......" | 25551621 | |
| hsa-miR-126-5p | colon cancer | poor survival; staging | "We studied the impact of the expression of miR-17- ......" | 18521848 | |
| hsa-miR-126-5p | colon cancer | tumorigenesis | "The noncoding RNA miR 126 suppresses the growth of ......" | 18663744 | |
| hsa-miR-126-5p | colon cancer | metastasis; progression | "MiR 126 suppresses colon cancer cell proliferation ......" | 23615712 | |
| hsa-miR-126-5p | colon cancer | cell migration | "Expression of miR 126 suppresses migration and inv ......" | 23744532 | MTT assay; Western blot; Luciferase |
| hsa-miR-126-5p | colon cancer | metastasis; worse prognosis | "miR 126 inhibits colon cancer proliferation and in ......" | 23981989 | |
| hsa-miR-126-5p | colon cancer | staging | "The prognostic value of microRNA 126 and microvess ......" | 25199818 | |
| hsa-miR-126-5p | colon cancer | metastasis; poor survival | "Recently increasing studies have demonstrated that ......" | 26893826 | |
| hsa-miR-126-5p | colon cancer | metastasis; staging; worse prognosis | "MicroRNA 126 inhibits colon cancer cell proliferat ......" | 27517626 | |
| hsa-miR-126-5p | colorectal cancer | malignant trasformation; tumorigenesis | "Down regulation of miR 126 expression in colorecta ......" | 20680522 | |
| hsa-miR-126-5p | colorectal cancer | progression | "Epigenetic silencing of miR 126 contributes to tum ......" | 23900443 | Luciferase |
| hsa-miR-126-5p | colorectal cancer | progression | "MicroRNA 126 functions as a tumor suppressor in co ......" | 24189753 | Transwell assay; Western blot; Luciferase |
| hsa-miR-126-5p | colorectal cancer | worse prognosis; poor survival; staging; metastasis | "Low expression of microRNA 126 is associated with ......" | 24532280 | Western blot |
| hsa-miR-126-5p | colorectal cancer | metastasis; staging | "Differential expression of serum miR 126 miR 141 a ......" | 24653631 | |
| hsa-miR-126-5p | colorectal cancer | tumorigenesis | "Underexpression of miR 126 and miR 20b in heredita ......" | 24994098 | |
| hsa-miR-126-5p | colorectal cancer | drug resistance | "Changes in circulating microRNA 126 during treatme ......" | 25584492 | |
| hsa-miR-126-5p | colorectal cancer | metastasis; staging | "Intra tumoural vessel area estimated by expression ......" | 25592646 | |
| hsa-miR-126-5p | colorectal cancer | staging; poor survival; metastasis | "Deregulation of miR 126 expression in colorectal c ......" | 26455548 | |
| hsa-miR-126-5p | colorectal cancer | staging; worse prognosis | "Serological under expression of microRNA 21 microR ......" | 27142899 | |
| hsa-miR-126-5p | colorectal cancer | progression | "Of these 3 upregulated let-7b miR-1290 and miR-126 ......" | 27698796 | |
| hsa-miR-126-5p | endometrial cancer | cell migration; metastasis | "MicroRNA 126 inhibits the migration and invasion o ......" | 26893720 | Transwell assay; Western blot; Luciferase |
| hsa-miR-126-5p | esophageal cancer | staging; metastasis; differentiation | "In this study we selected 10 miRNAs and analyzed t ......" | 20309880 | |
| hsa-miR-126-5p | esophageal cancer | worse prognosis; progression | "Using next-generation sequencing NGS-based miRNA p ......" | 25512445 | RNAi |
| hsa-miR-126-5p | esophageal cancer | progression | "MicroRNA 126 is down regulated in human esophageal ......" | 26191164 | Luciferase |
| hsa-miR-126-5p | gastric cancer | poor survival | "Several microarray studies have reported microRNA ......" | 19951901 | |
| hsa-miR-126-5p | gastric cancer | staging; metastasis | "miR 126 functions as a tumour suppressor in human ......" | 20619534 | |
| hsa-miR-126-5p | gastric cancer | malignant trasformation | "MiR 126 inhibits the invasion of gastric cancer ce ......" | 25027343 | Transwell assay; Western blot |
| hsa-miR-126-5p | gastric cancer | staging | "MicroRNA 126 regulates migration and invasion of g ......" | 26464628 | Luciferase |
| hsa-miR-126-5p | gastric cancer | drug resistance | "MicroRNA 126 increases chemosensitivity in drug re ......" | 27622325 | |
| hsa-miR-126-5p | gastric cancer | worse prognosis | "Regulator of G protein signaling 3 targeted by miR ......" | 27754994 | Western blot; Luciferase |
| hsa-miR-126-5p | kidney renal cell cancer | poor survival | "Global miRNA expression profiles were obtained by ......" | 22492545 | |
| hsa-miR-126-5p | kidney renal cell cancer | poor survival; worse prognosis | "Combination of expression levels of miR 21 and miR ......" | 24428907 | |
| hsa-miR-126-5p | kidney renal cell cancer | poor survival | "Impact of miR 21 miR 126 and miR 221 as prognostic ......" | 25279769 | |
| hsa-miR-126-5p | kidney renal cell cancer | staging; poor survival; tumorigenesis | "Low expression of miR 126 is a prognostic marker f ......" | 25572155 | |
| hsa-miR-126-5p | kidney renal cell cancer | worse prognosis | "Conversely decreased expression of miR-106b miR-99 ......" | 26416448 | |
| hsa-miR-126-5p | kidney renal cell cancer | metastasis | "MicroRNA 126 inhibits tumor cell invasion and meta ......" | 27108693 | Luciferase; Western blot |
| hsa-miR-126-5p | liver cancer | recurrence | "18 miRNAs including 6 up-regulated and 12 down-reg ......" | 22552153 | |
| hsa-miR-126-5p | liver cancer | recurrence; worse prognosis; metastasis; poor survival | "Decreased expression of miR 126 correlates with me ......" | 23378255 | Colony formation |
| hsa-miR-126-5p | liver cancer | metastasis | "MiR 126 3p suppresses tumor metastasis and angioge ......" | 25240815 | Transwell assay; Western blot |
| hsa-miR-126-5p | liver cancer | progression | "miR 126 inhibits cell proliferation and induces ce ......" | 25585946 | Luciferase |
| hsa-miR-126-5p | lung cancer | poor survival | "let 7b and miR 126 are down regulated in tumor tis ......" | 23029111 | |
| hsa-miR-126-5p | lung cancer | staging | "The study contained 2 phases: first preliminary ma ......" | 25639977 | |
| hsa-miR-126-5p | lung squamous cell cancer | cell migration; drug resistance | "miR 126 inhibits non small cell lung cancer cells ......" | 20034472 | Western blot; Flow cytometry |
| hsa-miR-126-5p | lung squamous cell cancer | poor survival; staging | "Independent and tissue specific prognostic impact ......" | 21264844 | |
| hsa-miR-126-5p | lung squamous cell cancer | staging | "There was statistical difference in the serum leve ......" | 22009180 | |
| hsa-miR-126-5p | lung squamous cell cancer | metastasis | "The miRs were quantified by microarray hybridizati ......" | 22295063 | |
| hsa-miR-126-5p | lung squamous cell cancer | poor survival; drug resistance | "miR 126 enhances the sensitivity of non small cell ......" | 22510476 | |
| hsa-miR-126-5p | lung squamous cell cancer | poor survival; worse prognosis; staging; malignant trasformation | "MicroRNA 126 inhibits tumor cell growth and its ex ......" | 22900072 | Luciferase |
| hsa-miR-126-5p | lung squamous cell cancer | worse prognosis; poor survival; tumor size | "Expression of microRNA miR 126 and miR 200c is ass ......" | 25124149 | |
| hsa-miR-126-5p | lung squamous cell cancer | tumor size; staging | "Here we analyzed expression of miR-15a/16 miR-21 m ......" | 25384507 | |
| hsa-miR-126-5p | lung squamous cell cancer | poor survival; staging | "Prognostic value of the MicroRNA regulators Dicer ......" | 25525410 | |
| hsa-miR-126-5p | lung squamous cell cancer | metastasis; progression; worse prognosis; staging; poor survival | "Down regulation of microRNA 126 and microRNA 133b ......" | 26823832 | |
| hsa-miR-126-5p | lung squamous cell cancer | staging | "Diagnostic Value of Serum miR 182 miR 183 miR 210 ......" | 27093275 | |
| hsa-miR-126-5p | lymphoma | malignant trasformation | "Expression of miR 155 and miR 126 in situ in cutan ......" | 24033365 | |
| hsa-miR-126-5p | pancreatic cancer | progression; metastasis | "Loss of miR 126 is crucial to pancreatic cancer pr ......" | 22845403 | |
| hsa-miR-126-5p | prostate cancer | recurrence; malignant trasformation; worse prognosis; staging; metastasis; poor survival; progression | "Association of microRNA 126 expression with clinic ......" | 24350576 | |
| hsa-miR-126-5p | prostate cancer | metastasis | "Osteoblast derived WNT induced secreted protein 1 ......" | 25277191 | |
| hsa-miR-126-5p | prostate cancer | drug resistance | "Syndecan 1 responsive microRNA 126 and 149 regulat ......" | 25462564 | Cell Proliferation Assay |
| hsa-miR-126-5p | sarcoma | differentiation | "In order to investigate the involvement of miRNAs ......" | 23133552 | |
| hsa-miR-126-5p | sarcoma | progression | "miR 126 functions as a tumor suppressor in osteosa ......" | 24384842 | Luciferase |
| hsa-miR-126-5p | sarcoma | motility | "Naringin suppress chondrosarcoma migration through ......" | 24975661 | |
| hsa-miR-126-5p | sarcoma | staging; metastasis | "miR 126 inhibits cell growth invasion and migratio ......" | 25213697 | Luciferase |
| hsa-miR-126-5p | sarcoma | drug resistance | "Overexpression of miR 126 sensitizes osteosarcoma ......" | 25510179 | Flow cytometry; MTT assay |
| hsa-miR-126-5p | sarcoma | staging; metastasis; poor survival; worse prognosis | "Tissue microRNA 126 expression level predicts outc ......" | 26194657 | |
| hsa-miR-126-5p | sarcoma | cell migration | "MicroRNA 126 Overexpression Inhibits Proliferation ......" | 26319109 | Transwell assay; Wound Healing Assay |
| hsa-miR-126-5p | thyroid cancer | staging; metastasis; tumor size | "miR 126 inhibits papillary thyroid carcinoma growt ......" | 26239517 | Colony formation |
| hsa-miR-126-5p | thyroid cancer | metastasis | "miR 126 3p Inhibits Thyroid Cancer Cell Growth and ......" | 26244545 | |
| hsa-miR-126-5p | thyroid cancer | poor survival | "MicroRNA 126 suppresses proliferation of undiffere ......" | 26384552 | Colony formation; Western blot |
| hsa-miR-126-5p | thyroid cancer | progression | "Interactive role of miR 126 on VEGF A and progress ......" | 27067785 | Western blot |
Reported gene related to hsa-miR-126-5p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-126-5p | breast cancer | VEGFA | "Endothelial specific intron derived miR 126 is dow ......" | 21249429 |
| hsa-miR-126-5p | colorectal cancer | VEGFA | "Using both in silico prediction and immunoblotting ......" | 23900443 |
| hsa-miR-126-5p | gastric cancer | VEGFA | "Reduced miR 126 expression facilitates angiogenesi ......" | 25428912 |
| hsa-miR-126-5p | kidney renal cell cancer | VEGFA | "In addition we found a negative correlation of exp ......" | 22086373 |
| hsa-miR-126-5p | lung cancer | VEGFA | "In A549 cell line overexpression of microRNA-126 i ......" | 25902169 |
| hsa-miR-126-5p | lung cancer | VEGFA | "MiR 126 restoration down regulate VEGF and inhibit ......" | 19223090 |
| hsa-miR-126-5p | lung squamous cell cancer | VEGFA | "miR-126 has been related to tumor angiogenesis and ......" | 21264844 |
| hsa-miR-126-5p | thyroid cancer | VEGFA | "Interactive role of miR 126 on VEGF A and progress ......" | 27067785 |
| hsa-miR-126-5p | bladder cancer | ADAM9 | "MicroRNA 126 inhibits invasion in bladder cancer v ......" | 24823697 |
| hsa-miR-126-5p | breast cancer | ADAM9 | "MiR 126 regulated breast cancer cell invasion by t ......" | 26261534 |
| hsa-miR-126-5p | esophageal cancer | ADAM9 | "ADAM9 was identified as a key target of miR-126; E ......" | 25512445 |
| hsa-miR-126-5p | pancreatic cancer | ADAM9 | "ADAM9 is overexpressed in PDAC and also a direct t ......" | 22845403 |
| hsa-miR-126-5p | pancreatic cancer | ADAM9 | "MiR 126 acts as a tumor suppressor in pancreatic c ......" | 22064652 |
| hsa-miR-126-5p | sarcoma | ADAM9 | "miR 126 inhibits cell growth invasion and migratio ......" | 25213697 |
| hsa-miR-126-5p | sarcoma | ADAM9 | "Cells overexpressing microRNA-126 exhibited reduce ......" | 26319109 |
| hsa-miR-126-5p | breast cancer | EGFL7 | "Using Northern blot and real-time PCR we confirmed ......" | 21249429 |
| hsa-miR-126-5p | colon cancer | EGFL7 | "No correlation was found between miR-126 expressio ......" | 18521848 |
| hsa-miR-126-5p | colorectal cancer | EGFL7 | "The aim of the present descriptive study was to an ......" | 25592646 |
| hsa-miR-126-5p | colorectal cancer | EGFL7 | "Mechanistically we found that the silencing of miR ......" | 23900443 |
| hsa-miR-126-5p | esophageal cancer | EGFL7 | "Downregulation of miR-126 was due to promoter hype ......" | 25512445 |
| hsa-miR-126-5p | lung squamous cell cancer | EGFL7 | "Mir-34b was silenced by the DNA methylation of its ......" | 21702040 |
| hsa-miR-126-5p | lung squamous cell cancer | EGFL7 | "miR 126 inhibits non small cell lung cancer cells ......" | 20034472 |
| hsa-miR-126-5p | gastric cancer | CRK | "MiR 126 inhibits the invasion of gastric cancer ce ......" | 25027343 |
| hsa-miR-126-5p | gastric cancer | CRK | "Mechanistically we identified the adaptor protein ......" | 20619534 |
| hsa-miR-126-5p | lung squamous cell cancer | CRK | "The overexpression of mir-34b and mir-126 decrease ......" | 21702040 |
| hsa-miR-126-5p | lung squamous cell cancer | CRK | "Crk is a predicted putative target gene for miR-12 ......" | 18602365 |
| hsa-miR-126-5p | pancreatic cancer | CRK | "miR-126 is also known to target other crucial onco ......" | 22845403 |
| hsa-miR-126-5p | bladder cancer | PIK3R2 | "MiR 126 regulates proliferation and invasion in th ......" | 27578985 |
| hsa-miR-126-5p | breast cancer | PIK3R2 | "Endothelial specific intron derived miR 126 is dow ......" | 21249429 |
| hsa-miR-126-5p | esophageal cancer | PIK3R2 | "Restoration of miR-126 in EC109 cells induced a re ......" | 26191164 |
| hsa-miR-126-5p | liver cancer | PIK3R2 | "MiR 126 3p suppresses tumor metastasis and angioge ......" | 25240815 |
| hsa-miR-126-5p | thyroid cancer | PIK3R2 | "MicroRNA 126 suppresses proliferation of undiffere ......" | 26384552 |
| hsa-miR-126-5p | colon cancer | CXCR4 | "Expression of miR 126 suppresses migration and inv ......" | 23744532 |
| hsa-miR-126-5p | colon cancer | CXCR4 | "Moreover we verified that miR-126 negatively regul ......" | 27517626 |
| hsa-miR-126-5p | colorectal cancer | CXCR4 | "MicroRNA-126 miR-126 has been reported to be a tum ......" | 24532280 |
| hsa-miR-126-5p | colorectal cancer | CXCR4 | "MicroRNA 126 functions as a tumor suppressor in co ......" | 24189753 |
| hsa-miR-126-5p | colorectal cancer | EGF | "MicroRNA 126 and epidermal growth factor like doma ......" | 23922111 |
| hsa-miR-126-5p | colorectal cancer | EGF | "Intra tumoural vessel area estimated by expression ......" | 25592646 |
| hsa-miR-126-5p | esophageal cancer | EGF | "Ectopic expression of miR-126 or silencing of ADAM ......" | 25512445 |
| hsa-miR-126-5p | colon cancer | IRS1 | "It has been shown that IRS1 SLC75A and TOM1 were t ......" | 23981989 |
| hsa-miR-126-5p | colorectal cancer | IRS1 | "Down regulation of miR 126 is associated with colo ......" | 24312276 |
| hsa-miR-126-5p | endometrial cancer | IRS1 | "Molecular mechanism investigation established that ......" | 26893720 |
| hsa-miR-126-5p | colon cancer | MVD | "We analysed the prognostic value of two angiogenes ......" | 25199818 |
| hsa-miR-126-5p | gastric cancer | MVD | "Down-regulation of miR-126 was found to inversely ......" | 25428912 |
| hsa-miR-126-5p | lung cancer | MVD | "We demonstrated significantly higher MVD and decre ......" | 23029111 |
| hsa-miR-126-5p | liver cancer | SOX2 | "miR 126 inhibits cell proliferation and induces ce ......" | 25585946 |
| hsa-miR-126-5p | prostate cancer | SOX2 | "The expression levels of SOX2 NANOG Oct4 miR-126 a ......" | 25462564 |
| hsa-miR-126-5p | sarcoma | SOX2 | "miR 126 functions as a tumor suppressor in osteosa ......" | 24384842 |
| hsa-miR-126-5p | colorectal cancer | INSR | "In this study we identified the potential effects ......" | 24312276 |
| hsa-miR-126-5p | endometrial cancer | INSR | "MicroRNA 126 inhibits the migration and invasion o ......" | 26893720 |
| hsa-miR-126-5p | liver cancer | LRP6 | "MiR 126 3p suppresses tumor metastasis and angioge ......" | 25240815 |
| hsa-miR-126-5p | thyroid cancer | LRP6 | "miR 126 inhibits papillary thyroid carcinoma growt ......" | 26239517 |
| hsa-miR-126-5p | prostate cancer | VCAM1 | "Osteoblast derived WNT induced secreted protein 1 ......" | 25277191 |
| hsa-miR-126-5p | sarcoma | VCAM1 | "We also observed that naringin enhancing miR-126 e ......" | 24975661 |
| hsa-miR-126-5p | cervical and endocervical cancer | ADM | "Repression of miR 126 and upregulation of adrenome ......" | 24037526 |
| hsa-miR-126-5p | liver cancer | AFP | "Interestingly triple combination of markers miR-12 ......" | 26756996 |
| hsa-miR-126-5p | gastric cancer | AKR1B1 | "Ectopic expression of miR-126 increased sensitivit ......" | 27622325 |
| hsa-miR-126-5p | acute myeloid leukemia | ARHGEF1 | "Therefore we hypothesized that miR-126 contributes ......" | 26055302 |
| hsa-miR-126-5p | cervical and endocervical cancer | BLM | "The viability of Siha cervical cancer cells was fu ......" | 24377569 |
| hsa-miR-126-5p | gastric cancer | CADM1 | "MicroRNA 126 regulates migration and invasion of g ......" | 26464628 |
| hsa-miR-126-5p | thyroid cancer | CD79A | "miR-126 expression of undifferentiated thyroid car ......" | 26384552 |
| hsa-miR-126-5p | colon cancer | CDC37 | "MicroRNA 126 inhibits colon cancer cell proliferat ......" | 27517626 |
| hsa-miR-126-5p | pancreatic cancer | CDH1 | "Reexpression of miR-126 and siRNA-based knockdown ......" | 22064652 |
| hsa-miR-126-5p | gastric cancer | CRKL | "CRKL promotes cell proliferation in gastric cancer ......" | 24055140 |
| hsa-miR-126-5p | kidney renal cell cancer | CRS | "A factor derived from the z-score resulting from t ......" | 24428907 |
| hsa-miR-126-5p | lung squamous cell cancer | DICER1 | "We have investigated the prognostic impact of Dice ......" | 25525410 |
| hsa-miR-126-5p | esophageal cancer | DNMT1 | "Intriguingly DNMT1 was suppressed by overexpressio ......" | 25512445 |
| hsa-miR-126-5p | lung squamous cell cancer | DROSHA | "We have investigated the prognostic impact of Dice ......" | 25525410 |
| hsa-miR-126-5p | gastric cancer | EZH2 | "MicroRNA 126 increases chemosensitivity in drug re ......" | 27622325 |
| hsa-miR-126-5p | colorectal cancer | FAP | "We analyzed the expressions of miR-126 and miR-20b ......" | 24994098 |
| hsa-miR-126-5p | gastric cancer | GNB2 | "Regulator of G protein signaling 3 targeted by miR ......" | 27754994 |
| hsa-miR-126-5p | colorectal cancer | IARS | "Down regulation of miR 126 is associated with colo ......" | 24312276 |
| hsa-miR-126-5p | pancreatic cancer | IDUA | "Along with higher expression of miR-21 which has b ......" | 22064652 |
| hsa-miR-126-5p | pancreatic cancer | KRAS | "miR-126 is also known to target other crucial onco ......" | 22845403 |
| hsa-miR-126-5p | gastric cancer | LAT | "MicroRNA 126 inhibits cell proliferation in gastri ......" | 26054677 |
| hsa-miR-126-5p | lung squamous cell cancer | MAX | "Moreover forced expression of miRNAs contributed t ......" | 20097187 |
| hsa-miR-126-5p | lung squamous cell cancer | MET | "The overexpression of mir-34b and mir-126 decrease ......" | 21702040 |
| hsa-miR-126-5p | gastric cancer | MTOR | "In addition the restoration of miR-126 expression ......" | 25428912 |
| hsa-miR-126-5p | prostate cancer | NANOG | "The expression levels of SOX2 NANOG Oct4 miR-126 a ......" | 25462564 |
| hsa-miR-126-5p | bladder cancer | NMI | "We first profiled the expression of miRNAs and mRN ......" | 24823697 |
| hsa-miR-126-5p | ovarian cancer | PAK4 | "microRNA 126 suppresses PAK4 expression in ovarian ......" | 26137045 |
| hsa-miR-126-5p | colon cancer | PIK3CD | "The noncoding RNA miR 126 suppresses the growth of ......" | 18663744 |
| hsa-miR-126-5p | bladder cancer | RFXANK | "MiR 126 regulates proliferation and invasion in th ......" | 27578985 |
| hsa-miR-126-5p | gastric cancer | RGS3 | "Interestingly our pathways analysis and the follow ......" | 27754994 |
| hsa-miR-126-5p | colon cancer | RHOA | "Moreover we verified that miR-126 negatively regul ......" | 27517626 |
| hsa-miR-126-5p | kidney renal cell cancer | ROCK1 | "MicroRNA 126 inhibits tumor cell invasion and meta ......" | 27108693 |
| hsa-miR-126-5p | lung squamous cell cancer | SCLC1 | "Since miR-126 is under-expressed in the majority o ......" | 21439283 |
| hsa-miR-126-5p | prostate cancer | SDC1 | "Syndecan 1 responsive microRNA 126 and 149 regulat ......" | 25462564 |
| hsa-miR-126-5p | sarcoma | SIRT1 | "MicroRNA 126 inhibits osteosarcoma cells prolifera ......" | 23877372 |
| hsa-miR-126-5p | lung squamous cell cancer | SLC7A5 | "miR 126 inhibits proliferation of small cell lung ......" | 21439283 |
| hsa-miR-126-5p | cervical and endocervical cancer | TBATA | "This study investigated the temporal and spatial e ......" | 24037526 |
| hsa-miR-126-5p | colon cancer | TOM1 | "It has been shown that IRS1 SLC75A and TOM1 were t ......" | 23981989 |
| hsa-miR-126-5p | colon cancer | TUBA1B | "In the current study using microRNA arrays we foun ......" | 18663744 |
| hsa-miR-126-5p | prostate cancer | WISP1 | "Osteoblast-derived WISP-1 inhibited miR-126 expres ......" | 25277191 |
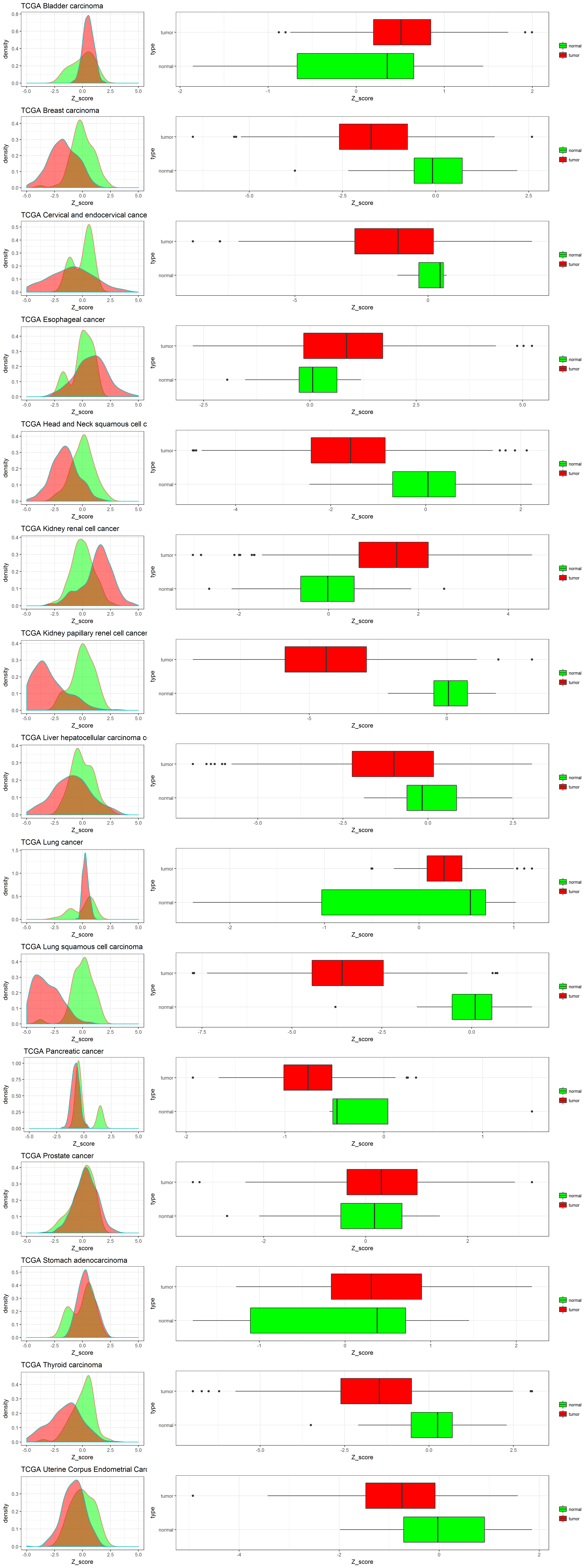
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-126-5p | MIB1 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUSC; PAAD; SARC; THCA; STAD | MirTarget; mirMAP | TCGA BLCA -0.107; TCGA COAD -0.096; TCGA ESCA -0.176; TCGA HNSC -0.082; TCGA LGG -0.054; TCGA LUSC -0.185; TCGA PAAD -0.177; TCGA SARC -0.139; TCGA THCA -0.114; TCGA STAD -0.19 |
hsa-miR-126-5p | MAP3K2 | 9 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.133; TCGA COAD -0.15; TCGA ESCA -0.186; TCGA LUAD -0.073; TCGA LUSC -0.144; TCGA PAAD -0.145; TCGA SARC -0.201; TCGA THCA -0.112; TCGA STAD -0.181 |
hsa-miR-126-5p | CALU | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.142; TCGA BRCA -0.056; TCGA COAD -0.162; TCGA ESCA -0.18; TCGA HNSC -0.134; TCGA KIRP -0.094; TCGA LIHC -0.128; TCGA LUAD -0.081; TCGA LUSC -0.265; TCGA PAAD -0.319; TCGA THCA -0.135; TCGA STAD -0.09 |
hsa-miR-126-5p | COL11A1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; THCA | MirTarget; mirMAP | TCGA BLCA -0.604; TCGA BRCA -1.033; TCGA COAD -0.431; TCGA ESCA -2.364; TCGA HNSC -1.342; TCGA LUAD -0.661; TCGA LUSC -2.048; TCGA PAAD -1.782; TCGA THCA -2.11 |
hsa-miR-126-5p | MITF | 10 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD | MirTarget; mirMAP | TCGA BLCA -0.467; TCGA COAD -0.392; TCGA ESCA -0.295; TCGA KIRC -0.121; TCGA KIRP -0.294; TCGA LIHC -0.241; TCGA PAAD -0.324; TCGA PRAD -0.104; TCGA SARC -0.378; TCGA STAD -0.49 |
hsa-miR-126-5p | ETV1 | 10 cancers: BLCA; COAD; ESCA; KIRP; LGG; LIHC; PAAD; PRAD; THCA; STAD | MirTarget; mirMAP | TCGA BLCA -0.311; TCGA COAD -0.284; TCGA ESCA -0.369; TCGA KIRP -0.353; TCGA LGG -0.302; TCGA LIHC -0.252; TCGA PAAD -0.218; TCGA PRAD -0.218; TCGA THCA -0.351; TCGA STAD -0.451 |
hsa-miR-126-5p | TRPS1 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUSC; PRAD; STAD; UCEC | MirTarget; mirMAP; miRNATAP | TCGA BLCA -0.316; TCGA BRCA -0.126; TCGA ESCA -0.29; TCGA KIRC -0.126; TCGA KIRP -0.089; TCGA LUSC -0.269; TCGA PRAD -0.135; TCGA STAD -0.444; TCGA UCEC -0.265 |
hsa-miR-126-5p | VCAN | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA BLCA -0.376; TCGA BRCA -0.111; TCGA COAD -0.325; TCGA ESCA -0.652; TCGA HNSC -0.35; TCGA KIRP -0.915; TCGA LGG -0.276; TCGA LUAD -0.194; TCGA LUSC -0.396; TCGA PAAD -0.459; TCGA THCA -0.939 |
hsa-miR-126-5p | SEMA3C | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.225; TCGA COAD -0.262; TCGA ESCA -0.558; TCGA HNSC -0.332; TCGA KIRC -0.438; TCGA LUSC -0.209; TCGA PAAD -0.394; TCGA PRAD -0.298; TCGA SARC -0.204; TCGA THCA -0.775; TCGA STAD -0.518 |
hsa-miR-126-5p | KATNAL1 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.431; TCGA COAD -0.153; TCGA ESCA -0.477; TCGA HNSC -0.119; TCGA KIRP -0.089; TCGA PRAD -0.117; TCGA SARC -0.187; TCGA THCA -0.132; TCGA STAD -0.521 |
hsa-miR-126-5p | TMEM106B | 9 cancers: BLCA; COAD; KIRC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.086; TCGA COAD -0.233; TCGA KIRC -0.113; TCGA LUAD -0.089; TCGA LUSC -0.091; TCGA PRAD -0.12; TCGA SARC -0.124; TCGA THCA -0.131; TCGA STAD -0.259 |
hsa-miR-126-5p | ASPN | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.642; TCGA BRCA -0.282; TCGA COAD -0.516; TCGA ESCA -0.655; TCGA HNSC -0.458; TCGA LUSC -0.277; TCGA PAAD -0.287; TCGA SARC -0.372; TCGA THCA -0.247; TCGA STAD -0.499 |
hsa-miR-126-5p | COL12A1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA BLCA -0.255; TCGA BRCA -0.214; TCGA COAD -0.263; TCGA ESCA -0.643; TCGA HNSC -0.507; TCGA LUAD -0.228; TCGA LUSC -0.488; TCGA PAAD -0.647; TCGA THCA -0.206 |
hsa-miR-126-5p | TNFSF4 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA BLCA -0.36; TCGA BRCA -0.212; TCGA COAD -0.372; TCGA ESCA -0.509; TCGA HNSC -0.305; TCGA LIHC -0.211; TCGA LUAD -0.162; TCGA LUSC -0.278; TCGA PAAD -0.347; TCGA THCA -0.591 |
hsa-miR-126-5p | NFYB | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.086; TCGA CESC -0.065; TCGA COAD -0.073; TCGA ESCA -0.177; TCGA KIRC -0.106; TCGA LGG -0.058; TCGA PRAD -0.064; TCGA SARC -0.091; TCGA STAD -0.17 |
hsa-miR-126-5p | INHBA | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.272; TCGA BRCA -0.365; TCGA COAD -0.356; TCGA ESCA -0.852; TCGA HNSC -0.89; TCGA LUAD -0.254; TCGA PAAD -0.899; TCGA SARC -0.607; TCGA THCA -0.79 |
hsa-miR-126-5p | SSPN | 10 cancers: BLCA; COAD; ESCA; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.346; TCGA COAD -0.282; TCGA ESCA -0.591; TCGA KIRP -0.086; TCGA LGG -0.094; TCGA LUAD -0.119; TCGA PRAD -0.207; TCGA SARC -0.243; TCGA THCA -0.224; TCGA STAD -0.558 |
hsa-miR-126-5p | NRK | 9 cancers: BLCA; COAD; ESCA; KIRC; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.355; TCGA COAD -0.665; TCGA ESCA -1.212; TCGA KIRC -1.332; TCGA LUSC -0.362; TCGA PRAD -0.288; TCGA SARC -0.765; TCGA THCA -0.759; TCGA STAD -0.686 |
hsa-miR-126-5p | SULF1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.518; TCGA BRCA -0.139; TCGA COAD -0.4; TCGA ESCA -0.883; TCGA HNSC -0.33; TCGA LGG -0.271; TCGA LUAD -0.228; TCGA LUSC -0.659; TCGA PAAD -0.508; TCGA SARC -0.472; TCGA THCA -0.424 |
hsa-miR-126-5p | PHLDB2 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.381; TCGA COAD -0.458; TCGA ESCA -0.637; TCGA HNSC -0.274; TCGA KIRP -0.083; TCGA PRAD -0.115; TCGA SARC -0.295; TCGA THCA -0.539; TCGA STAD -0.575 |
hsa-miR-126-5p | SCD5 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.463; TCGA CESC -0.293; TCGA COAD -0.311; TCGA ESCA -0.442; TCGA HNSC -0.222; TCGA LGG -0.122; TCGA PRAD -0.179; TCGA SARC -0.236; TCGA STAD -0.206 |
hsa-miR-126-5p | TPBG | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.146; TCGA BRCA -0.188; TCGA ESCA -0.575; TCGA HNSC -0.207; TCGA KIRP -0.339; TCGA LUAD -0.126; TCGA LUSC -0.564; TCGA PAAD -0.373; TCGA PRAD -0.169; TCGA THCA -0.308 |
hsa-miR-126-5p | BEND6 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.276; TCGA BRCA -0.098; TCGA COAD -0.397; TCGA ESCA -0.74; TCGA HNSC -0.324; TCGA LUSC -0.493; TCGA PRAD -0.164; TCGA THCA -0.956; TCGA STAD -0.418 |
hsa-miR-126-5p | LSM11 | 11 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.291; TCGA COAD -0.092; TCGA ESCA -0.149; TCGA KIRP -0.141; TCGA LIHC -0.111; TCGA LUAD -0.053; TCGA LUSC -0.127; TCGA PRAD -0.108; TCGA SARC -0.402; TCGA THCA -0.114; TCGA STAD -0.289 |
hsa-miR-126-5p | PDLIM5 | 9 cancers: BLCA; COAD; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP; miRNATAP | TCGA BLCA -0.206; TCGA COAD -0.081; TCGA LGG -0.177; TCGA LUAD -0.094; TCGA LUSC -0.095; TCGA PAAD -0.26; TCGA SARC -0.435; TCGA THCA -0.084; TCGA STAD -0.138 |
hsa-miR-126-5p | PIGX | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.07; TCGA BRCA -0.103; TCGA ESCA -0.269; TCGA HNSC -0.095; TCGA KIRC -0.087; TCGA LIHC -0.178; TCGA LUAD -0.055; TCGA LUSC -0.368; TCGA PAAD -0.12; TCGA SARC -0.117; TCGA STAD -0.122 |
hsa-miR-126-5p | RAB31 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.246; TCGA BRCA -0.178; TCGA COAD -0.292; TCGA ESCA -0.31; TCGA HNSC -0.198; TCGA KIRP -0.245; TCGA PAAD -0.212; TCGA THCA -0.244; TCGA STAD -0.287 |
hsa-miR-126-5p | PCDH7 | 11 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.662; TCGA COAD -0.357; TCGA ESCA -0.343; TCGA KIRC -0.369; TCGA LUAD -0.154; TCGA LUSC -0.265; TCGA PAAD -0.476; TCGA PRAD -0.309; TCGA SARC -0.656; TCGA STAD -0.489; TCGA UCEC -0.325 |
hsa-miR-126-5p | PAWR | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.181; TCGA BRCA -0.077; TCGA COAD -0.098; TCGA ESCA -0.186; TCGA HNSC -0.152; TCGA KIRC -0.194; TCGA LUAD -0.066; TCGA LUSC -0.253; TCGA PAAD -0.275; TCGA SARC -0.718; TCGA THCA -0.18 |
hsa-miR-126-5p | ARSJ | 9 cancers: BLCA; ESCA; HNSC; KIRP; LUAD; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.293; TCGA ESCA -0.416; TCGA HNSC -0.239; TCGA KIRP -0.312; TCGA LUAD -0.145; TCGA PAAD -0.234; TCGA PRAD -0.3; TCGA THCA -0.924; TCGA STAD -0.181 |
hsa-miR-126-5p | IPO9 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; THCA | mirMAP | TCGA BLCA -0.061; TCGA BRCA -0.056; TCGA ESCA -0.103; TCGA HNSC -0.074; TCGA LIHC -0.141; TCGA LUAD -0.064; TCGA LUSC -0.184; TCGA SARC -0.118; TCGA THCA -0.08 |
hsa-miR-126-5p | PPP1CB | 11 cancers: BLCA; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.181; TCGA COAD -0.066; TCGA ESCA -0.13; TCGA LIHC -0.072; TCGA LUAD -0.066; TCGA LUSC -0.113; TCGA PRAD -0.076; TCGA SARC -0.228; TCGA THCA -0.166; TCGA STAD -0.28; TCGA UCEC -0.087 |
hsa-miR-126-5p | UBE2QL1 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.215; TCGA BRCA -0.135; TCGA COAD -0.292; TCGA ESCA -0.78; TCGA LUAD -0.235; TCGA LUSC -0.791; TCGA PRAD -0.226; TCGA SARC -0.827; TCGA THCA -0.29; TCGA STAD -0.466 |
hsa-miR-126-5p | TNC | 9 cancers: BLCA; ESCA; HNSC; KIRP; LUSC; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.358; TCGA ESCA -0.983; TCGA HNSC -0.284; TCGA KIRP -0.48; TCGA LUSC -0.445; TCGA PRAD -0.205; TCGA SARC -0.619; TCGA THCA -1.18; TCGA STAD -0.434 |
hsa-miR-126-5p | MLF1 | 9 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; STAD | MirTarget; mirMAP | TCGA BRCA -0.117; TCGA CESC -0.301; TCGA ESCA -0.877; TCGA KIRC -0.337; TCGA KIRP -0.082; TCGA LUAD -0.094; TCGA LUSC -0.158; TCGA PAAD -0.188; TCGA STAD -0.333 |
hsa-miR-126-5p | LIN9 | 9 cancers: BRCA; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | MirTarget; mirMAP; miRNATAP | TCGA BRCA -0.065; TCGA LGG -0.061; TCGA LIHC -0.213; TCGA LUAD -0.075; TCGA LUSC -0.312; TCGA PAAD -0.159; TCGA PRAD -0.081; TCGA SARC -0.236; TCGA UCEC -0.14 |
hsa-miR-126-5p | ANLN | 9 cancers: BRCA; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.336; TCGA KIRP -0.185; TCGA LIHC -0.789; TCGA LUAD -0.159; TCGA LUSC -0.977; TCGA PAAD -0.528; TCGA SARC -0.202; TCGA THCA -0.512; TCGA UCEC -0.235 |
hsa-miR-126-5p | SIX4 | 11 cancers: BRCA; COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | mirMAP | TCGA BRCA -0.165; TCGA COAD -0.384; TCGA ESCA -0.546; TCGA KIRC -1.126; TCGA LGG -0.134; TCGA LIHC -0.548; TCGA LUAD -0.196; TCGA LUSC -0.49; TCGA PAAD -0.613; TCGA THCA -0.229; TCGA UCEC -0.193 |
hsa-miR-126-5p | CEP70 | 9 cancers: BRCA; KIRC; KIRP; LGG; LIHC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.078; TCGA KIRC -0.293; TCGA KIRP -0.249; TCGA LGG -0.12; TCGA LIHC -0.109; TCGA PAAD -0.108; TCGA SARC -0.239; TCGA STAD -0.156; TCGA UCEC -0.091 |
hsa-miR-126-5p | GMPS | 9 cancers: BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BRCA -0.055; TCGA ESCA -0.15; TCGA HNSC -0.118; TCGA LIHC -0.145; TCGA LUAD -0.113; TCGA LUSC -0.422; TCGA PAAD -0.248; TCGA SARC -0.065; TCGA THCA -0.072 |
hsa-miR-126-5p | LRRC42 | 10 cancers: BRCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRNATAP | TCGA BRCA -0.14; TCGA ESCA -0.101; TCGA HNSC -0.087; TCGA LGG -0.094; TCGA LIHC -0.221; TCGA LUAD -0.056; TCGA LUSC -0.186; TCGA PAAD -0.21; TCGA THCA -0.057; TCGA UCEC -0.16 |
hsa-miR-126-5p | AXIN1 | 9 cancers: BRCA; HNSC; KIRP; LGG; LUSC; PAAD; SARC; THCA; UCEC | miRNATAP | TCGA BRCA -0.233; TCGA HNSC -0.086; TCGA KIRP -0.202; TCGA LGG -0.103; TCGA LUSC -0.124; TCGA PAAD -0.124; TCGA SARC -0.052; TCGA THCA -0.185; TCGA UCEC -0.115 |
hsa-miR-126-5p | DDR1 | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; THCA | miRNATAP | TCGA BRCA -0.184; TCGA ESCA -0.199; TCGA HNSC -0.132; TCGA KIRC -0.138; TCGA KIRP -0.276; TCGA LGG -0.12; TCGA LIHC -0.376; TCGA LUSC -0.371; TCGA PAAD -0.336; TCGA SARC -0.406; TCGA THCA -0.17 |
hsa-miR-126-5p | PEX13 | 10 cancers: CESC; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.136; TCGA HNSC -0.075; TCGA KIRC -0.07; TCGA LUAD -0.055; TCGA LUSC -0.176; TCGA PAAD -0.235; TCGA PRAD -0.056; TCGA SARC -0.101; TCGA STAD -0.097; TCGA UCEC -0.083 |
hsa-miR-126-5p | DHX40 | 9 cancers: CESC; COAD; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA CESC -0.092; TCGA COAD -0.162; TCGA KIRP -0.193; TCGA LGG -0.054; TCGA LIHC -0.116; TCGA LUAD -0.098; TCGA LUSC -0.134; TCGA SARC -0.058; TCGA STAD -0.1 |
hsa-miR-126-5p | PRKCI | 10 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; THCA | mirMAP | TCGA CESC -0.158; TCGA COAD -0.144; TCGA HNSC -0.071; TCGA KIRC -0.162; TCGA KIRP -0.138; TCGA LIHC -0.157; TCGA LUSC -0.246; TCGA PAAD -0.429; TCGA SARC -0.06; TCGA THCA -0.059 |
hsa-miR-126-5p | UBXN2A | 9 cancers: CESC; COAD; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA CESC -0.089; TCGA COAD -0.126; TCGA LGG -0.059; TCGA LIHC -0.098; TCGA LUAD -0.071; TCGA LUSC -0.202; TCGA PAAD -0.073; TCGA THCA -0.064; TCGA STAD -0.105 |
hsa-miR-126-5p | GSK3B | 9 cancers: COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA | mirMAP; miRNATAP | TCGA COAD -0.086; TCGA ESCA -0.135; TCGA KIRP -0.086; TCGA LIHC -0.06; TCGA LUAD -0.054; TCGA LUSC -0.155; TCGA PAAD -0.116; TCGA SARC -0.061; TCGA THCA -0.163 |
hsa-miR-126-5p | TRIM9 | 9 cancers: COAD; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; SARC; STAD | mirMAP | TCGA COAD -0.321; TCGA ESCA -0.511; TCGA HNSC -0.236; TCGA KIRP -0.652; TCGA LGG -0.073; TCGA LUSC -0.575; TCGA PRAD -0.188; TCGA SARC -0.598; TCGA STAD -0.378 |
hsa-miR-126-5p | SLC25A36 | 9 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; SARC; STAD | mirMAP | TCGA COAD -0.089; TCGA ESCA -0.187; TCGA HNSC -0.11; TCGA KIRC -0.119; TCGA KIRP -0.103; TCGA LGG -0.075; TCGA LUSC -0.131; TCGA SARC -0.059; TCGA STAD -0.268 |
hsa-miR-126-5p | FAM60A | 10 cancers: COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; THCA | mirMAP | TCGA COAD -0.148; TCGA HNSC -0.082; TCGA KIRC -0.28; TCGA KIRP -0.087; TCGA LGG -0.226; TCGA LIHC -0.183; TCGA LUSC -0.283; TCGA PAAD -0.242; TCGA SARC -0.156; TCGA THCA -0.23 |
hsa-miR-126-5p | FZD6 | 10 cancers: COAD; ESCA; HNSC; KIRC; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP; miRNATAP | TCGA COAD -0.203; TCGA ESCA -0.607; TCGA HNSC -0.187; TCGA KIRC -0.167; TCGA LUSC -0.239; TCGA PAAD -0.233; TCGA SARC -0.283; TCGA THCA -0.224; TCGA STAD -0.123; TCGA UCEC -0.143 |
hsa-miR-126-5p | IAH1 | 9 cancers: ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA ESCA -0.194; TCGA HNSC -0.116; TCGA KIRP -0.082; TCGA LIHC -0.073; TCGA LUAD -0.053; TCGA LUSC -0.093; TCGA PAAD -0.112; TCGA SARC -0.096; TCGA STAD -0.073 |
hsa-miR-126-5p | DCBLD2 | 9 cancers: ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA ESCA -0.697; TCGA HNSC -0.143; TCGA KIRC -0.216; TCGA KIRP -0.516; TCGA PAAD -0.329; TCGA PRAD -0.13; TCGA THCA -0.467; TCGA STAD -0.388; TCGA UCEC -0.139 |
hsa-miR-126-5p | SPATS2L | 9 cancers: HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | MirTarget | TCGA HNSC -0.076; TCGA KIRP -0.34; TCGA LIHC -0.138; TCGA LUAD -0.106; TCGA LUSC -0.164; TCGA PAAD -0.386; TCGA PRAD -0.102; TCGA SARC -0.121; TCGA THCA -0.392 |
hsa-miR-126-5p | TFDP2 | 10 cancers: HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA HNSC -0.133; TCGA KIRC -0.251; TCGA LGG -0.068; TCGA LIHC -0.084; TCGA LUAD -0.092; TCGA LUSC -0.355; TCGA PAAD -0.217; TCGA THCA -0.159; TCGA STAD -0.341; TCGA UCEC -0.1 |
Enriched cancer pathways of putative targets