This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-200c-3p | ACE2 | 2.07 | 0 | -2.45 | 0 | miRNATAP | -0.32 | 2.0E-5 | NA | |
| 2 | hsa-miR-200c-3p | ACTC1 | 2.07 | 0 | -1.25 | 0 | MirTarget | -0.45 | 0 | NA | |
| 3 | hsa-miR-200c-3p | ADAMTS3 | 2.07 | 0 | -1.85 | 0 | miRNATAP | -0.38 | 0 | NA | |
| 4 | hsa-miR-200c-3p | ARHGAP20 | 2.07 | 0 | -3.51 | 0 | MirTarget; miRNATAP | -0.69 | 0 | NA | |
| 5 | hsa-miR-200c-3p | ARHGAP28 | 2.07 | 0 | -1.18 | 0 | MirTarget | -0.43 | 0 | NA | |
| 6 | hsa-miR-200c-3p | ARHGAP6 | 2.07 | 0 | -1.65 | 0 | MirTarget | -0.46 | 0 | NA | |
| 7 | hsa-miR-200c-3p | BACH2 | 2.07 | 0 | -1.74 | 0 | mirMAP | -0.34 | 0 | NA | |
| 8 | hsa-miR-200c-3p | BNC2 | 2.07 | 0 | -0.78 | 0 | MirTarget | -0.46 | 0 | NA | |
| 9 | hsa-miR-200c-3p | C17orf51 | 2.07 | 0 | -1.38 | 0 | mirMAP | -0.36 | 0 | NA | |
| 10 | hsa-miR-200c-3p | CALCR | 2.07 | 0 | -2.1 | 0 | MirTarget | -0.36 | 0 | NA | |
| 11 | hsa-miR-200c-3p | CCDC82 | 2.07 | 0 | -1.57 | 0 | MirTarget | -0.3 | 0 | NA | |
| 12 | hsa-miR-200c-3p | CELF2 | 2.07 | 0 | -2.12 | 0 | mirMAP; miRNATAP | -0.45 | 0 | NA | |
| 13 | hsa-miR-200c-3p | CFL2 | 2.07 | 0 | -1.91 | 0 | MirTarget; miRNATAP | -0.38 | 0 | 23497265 | We characterized one of the target genes of miR-200c CFL2 and demonstrated that CFL2 is overexpressed in aggressive breast cancer cell lines and can be significantly down-regulated by exogenous miR-200c |
| 14 | hsa-miR-200c-3p | CHRDL1 | 2.07 | 0 | -5.4 | 0 | MirTarget | -0.92 | 0 | NA | |
| 15 | hsa-miR-200c-3p | CNTFR | 2.07 | 0 | -4.18 | 0 | miRNATAP | -0.65 | 0 | NA | |
| 16 | hsa-miR-200c-3p | CNTN1 | 2.07 | 0 | -2.3 | 0 | MirTarget; miRNATAP | -0.49 | 0 | NA | |
| 17 | hsa-miR-200c-3p | CNTN4 | 2.07 | 0 | -1.68 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 18 | hsa-miR-200c-3p | CREB5 | 2.07 | 0 | -2.35 | 0 | miRNATAP | -0.48 | 0 | NA | |
| 19 | hsa-miR-200c-3p | CRHBP | 2.07 | 0 | -3.67 | 0 | MirTarget | -0.75 | 0 | NA | |
| 20 | hsa-miR-200c-3p | CRYBG3 | 2.07 | 0 | -2.3 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 21 | hsa-miR-200c-3p | CSRNP3 | 2.07 | 0 | -3.02 | 0 | MirTarget; miRNATAP | -0.42 | 0 | NA | |
| 22 | hsa-miR-200c-3p | DDIT4L | 2.07 | 0 | -1.72 | 0 | MirTarget | -0.4 | 0 | NA | |
| 23 | hsa-miR-200c-3p | DIXDC1 | 2.07 | 0 | -1.64 | 0 | MirTarget | -0.32 | 0 | NA | |
| 24 | hsa-miR-200c-3p | DLC1 | 2.07 | 0 | -1.85 | 0 | miRNATAP | -0.45 | 0 | NA | |
| 25 | hsa-miR-200c-3p | DMD | 2.07 | 0 | -3.78 | 0 | miRNATAP | -0.59 | 0 | NA | |
| 26 | hsa-miR-200c-3p | DMRT2 | 2.07 | 0 | -3.09 | 0 | miRNATAP | -0.57 | 0 | NA | |
| 27 | hsa-miR-200c-3p | DST | 2.07 | 0 | -2.66 | 0 | mirMAP | -0.37 | 0 | NA | |
| 28 | hsa-miR-200c-3p | DUSP1 | 2.07 | 0 | -2.1 | 0 | MirTarget; miRNATAP | -0.46 | 0 | NA | |
| 29 | hsa-miR-200c-3p | DYNC1I1 | 2.07 | 0 | -1.58 | 0 | MirTarget | -0.31 | 0 | NA | |
| 30 | hsa-miR-200c-3p | FAT3 | 2.07 | 0 | -1.4 | 0 | MirTarget; miRNATAP | -0.55 | 0 | NA | |
| 31 | hsa-miR-200c-3p | FBLN5 | 2.07 | 0 | -2.09 | 0 | miRNAWalker2 validate; miRTarBase | -0.43 | 0 | NA | |
| 32 | hsa-miR-200c-3p | FGF2 | 2.07 | 0 | -3.62 | 0 | mirMAP | -0.62 | 0 | NA | |
| 33 | hsa-miR-200c-3p | FHL1 | 2.07 | 0 | -4.4 | 0 | MirTarget | -0.95 | 0 | NA | |
| 34 | hsa-miR-200c-3p | FIGN | 2.07 | 0 | -2.44 | 0 | MirTarget | -0.37 | 0 | NA | |
| 35 | hsa-miR-200c-3p | FLI1 | 2.07 | 0 | -1.26 | 0 | MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 36 | hsa-miR-200c-3p | FSTL1 | 2.07 | 0 | -1.28 | 0 | MirTarget | -0.45 | 0 | NA | |
| 37 | hsa-miR-200c-3p | FYN | 2.07 | 0 | -0.97 | 0 | miRNATAP | -0.31 | 0 | NA | |
| 38 | hsa-miR-200c-3p | FZD4 | 2.07 | 0 | -2.46 | 0 | mirMAP | -0.54 | 0 | NA | |
| 39 | hsa-miR-200c-3p | GABRB3 | 2.07 | 0 | -1.55 | 0 | mirMAP | -0.37 | 0 | NA | |
| 40 | hsa-miR-200c-3p | GEM | 2.07 | 0 | -1.31 | 0 | MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 41 | hsa-miR-200c-3p | GPM6A | 2.07 | 0 | -3.1 | 0 | miRNATAP | -0.52 | 0 | NA | |
| 42 | hsa-miR-200c-3p | GPR12 | 2.07 | 0 | -3.8 | 0 | mirMAP | -0.36 | 1.0E-5 | NA | |
| 43 | hsa-miR-200c-3p | GPR146 | 2.07 | 0 | -2.69 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 44 | hsa-miR-200c-3p | HLF | 2.07 | 0 | -3.73 | 0 | MirTarget; miRNATAP | -0.55 | 0 | NA | |
| 45 | hsa-miR-200c-3p | HTR2A | 2.07 | 0 | -3.25 | 0 | MirTarget | -0.55 | 0 | NA | |
| 46 | hsa-miR-200c-3p | IGSF10 | 2.07 | 0 | -3.95 | 0 | MirTarget | -0.72 | 0 | NA | |
| 47 | hsa-miR-200c-3p | IL6ST | 2.07 | 0 | -1.53 | 0 | mirMAP | -0.32 | 0 | NA | |
| 48 | hsa-miR-200c-3p | ITGA1 | 2.07 | 0 | -1.3 | 0 | MirTarget | -0.35 | 0 | NA | |
| 49 | hsa-miR-200c-3p | KCNJ2 | 2.07 | 0 | -1.76 | 0 | miRNATAP | -0.31 | 0 | NA | |
| 50 | hsa-miR-200c-3p | KIAA1462 | 2.07 | 0 | -0.89 | 0 | MirTarget | -0.36 | 0 | NA | |
| 51 | hsa-miR-200c-3p | KLF4 | 2.07 | 0 | -2.57 | 0 | MirTarget; miRNATAP | -0.48 | 0 | NA | |
| 52 | hsa-miR-200c-3p | KLF9 | 2.07 | 0 | -1.74 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.37 | 0 | NA | |
| 53 | hsa-miR-200c-3p | KLHL29 | 2.07 | 0 | -2.95 | 0 | MirTarget | -0.49 | 0 | NA | |
| 54 | hsa-miR-200c-3p | KLHL3 | 2.07 | 0 | -1.43 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 55 | hsa-miR-200c-3p | KLHL31 | 2.07 | 0 | -3.32 | 0 | MirTarget | -0.5 | 0 | NA | |
| 56 | hsa-miR-200c-3p | LAMC1 | 2.07 | 0 | -1.23 | 0 | MirTarget; miRNATAP | -0.33 | 0 | NA | |
| 57 | hsa-miR-200c-3p | LHFP | 2.07 | 0 | -2.16 | 0 | MirTarget; miRNATAP | -0.51 | 0 | NA | |
| 58 | hsa-miR-200c-3p | LIX1L | 2.07 | 0 | -1.12 | 0 | MirTarget | -0.32 | 0 | NA | |
| 59 | hsa-miR-200c-3p | LPAR1 | 2.07 | 0 | -1.21 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.35 | 0 | NA | |
| 60 | hsa-miR-200c-3p | LRP1B | 2.07 | 0 | -2.79 | 0 | MirTarget; miRNATAP | -0.48 | 0 | NA | |
| 61 | hsa-miR-200c-3p | LRP4 | 2.07 | 0 | -1.81 | 0 | MirTarget; miRNATAP | -0.35 | 0 | NA | |
| 62 | hsa-miR-200c-3p | MAF | 2.07 | 0 | -1.16 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 63 | hsa-miR-200c-3p | MBNL3 | 2.07 | 0 | -1.93 | 0 | MirTarget; mirMAP | -0.35 | 0 | NA | |
| 64 | hsa-miR-200c-3p | MFAP5 | 2.07 | 0 | -1.74 | 0 | miRNATAP | -0.5 | 0 | NA | |
| 65 | hsa-miR-200c-3p | MGAT3 | 2.07 | 0 | -1.92 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 66 | hsa-miR-200c-3p | MITF | 2.07 | 0 | -1.35 | 0 | miRNATAP | -0.38 | 0 | NA | |
| 67 | hsa-miR-200c-3p | MMD | 2.07 | 0 | -2.3 | 0 | miRNATAP | -0.5 | 0 | NA | |
| 68 | hsa-miR-200c-3p | MRVI1 | 2.07 | 0 | -1.29 | 0 | MirTarget | -0.35 | 0 | NA | |
| 69 | hsa-miR-200c-3p | MSRB3 | 2.07 | 0 | -2.05 | 0 | mirMAP | -0.51 | 0 | NA | |
| 70 | hsa-miR-200c-3p | NCAM1 | 2.07 | 0 | -2.68 | 0 | miRTarBase | -0.54 | 0 | NA | |
| 71 | hsa-miR-200c-3p | NDN | 2.07 | 0 | -1.9 | 0 | MirTarget; miRNATAP | -0.54 | 0 | NA | |
| 72 | hsa-miR-200c-3p | NEGR1 | 2.07 | 0 | -1.48 | 0 | miRNATAP | -0.49 | 0 | NA | |
| 73 | hsa-miR-200c-3p | NLGN4X | 2.07 | 0 | -2.19 | 0 | miRNATAP | -0.43 | 0 | NA | |
| 74 | hsa-miR-200c-3p | NOG | 2.07 | 0 | -0.84 | 4.0E-5 | MirTarget; miRNATAP | -0.35 | 0 | NA | |
| 75 | hsa-miR-200c-3p | NOVA1 | 2.07 | 0 | -2.65 | 0 | MirTarget; miRNATAP | -0.44 | 0 | NA | |
| 76 | hsa-miR-200c-3p | NR3C1 | 2.07 | 0 | -1.9 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 77 | hsa-miR-200c-3p | NR5A2 | 2.07 | 0 | -1.74 | 0 | miRNATAP | -0.42 | 0 | NA | |
| 78 | hsa-miR-200c-3p | NRG1 | 2.07 | 0 | -2.73 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 79 | hsa-miR-200c-3p | NTF3 | 2.07 | 0 | -2.35 | 0 | miRNAWalker2 validate; miRNATAP | -0.41 | 0 | NA | |
| 80 | hsa-miR-200c-3p | NTRK2 | 2.07 | 0 | -4.04 | 0 | miRNAWalker2 validate; miRTarBase | -0.54 | 0 | 23209748; 23074172 | miR 200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression;miR-200c also targets TrkB a mediator of resistance to anoikis |
| 81 | hsa-miR-200c-3p | OSR1 | 2.07 | 0 | -3.17 | 0 | miRNATAP | -0.44 | 0 | NA | |
| 82 | hsa-miR-200c-3p | PALM2-AKAP2 | 2.07 | 0 | -2.18 | 0 | miRNATAP | -0.45 | 0 | NA | |
| 83 | hsa-miR-200c-3p | PARD3B | 2.07 | 0 | -1.79 | 0 | MirTarget; miRNATAP | -0.35 | 0 | NA | |
| 84 | hsa-miR-200c-3p | PCDH19 | 2.07 | 0 | -2.02 | 0 | miRNATAP | -0.47 | 0 | NA | |
| 85 | hsa-miR-200c-3p | PCSK2 | 2.07 | 0 | -1.42 | 3.0E-5 | miRNATAP | -0.39 | 0 | NA | |
| 86 | hsa-miR-200c-3p | PDE7B | 2.07 | 0 | -1.82 | 0 | miRNATAP | -0.43 | 0 | NA | |
| 87 | hsa-miR-200c-3p | PHLDB1 | 2.07 | 0 | -1.05 | 0 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 88 | hsa-miR-200c-3p | PKIA | 2.07 | 0 | -1.54 | 0 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 89 | hsa-miR-200c-3p | PLXNA4 | 2.07 | 0 | -3.24 | 0 | miRNATAP | -0.7 | 0 | NA | |
| 90 | hsa-miR-200c-3p | PPARGC1A | 2.07 | 0 | -2.62 | 0 | mirMAP | -0.43 | 0 | NA | |
| 91 | hsa-miR-200c-3p | PRDM16 | 2.07 | 0 | -2.1 | 0 | MirTarget; miRNATAP | -0.47 | 0 | NA | |
| 92 | hsa-miR-200c-3p | PRKCA | 2.07 | 0 | -1.71 | 0 | miRNATAP | -0.38 | 0 | NA | |
| 93 | hsa-miR-200c-3p | PRKG1 | 2.07 | 0 | -1.38 | 0 | miRNATAP | -0.39 | 0 | NA | |
| 94 | hsa-miR-200c-3p | PTPN21 | 2.07 | 0 | -1.65 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 95 | hsa-miR-200c-3p | PTPRD | 2.07 | 0 | 0.15 | 0.42781 | miRNAWalker2 validate | -0.34 | 0 | NA | |
| 96 | hsa-miR-200c-3p | PTPRZ1 | 2.07 | 0 | -4.21 | 0 | MirTarget; miRNATAP | -0.51 | 0 | NA | |
| 97 | hsa-miR-200c-3p | PYGO1 | 2.07 | 0 | -1.89 | 0 | mirMAP | -0.38 | 0 | NA | |
| 98 | hsa-miR-200c-3p | RECK | 2.07 | 0 | -1.98 | 0 | miRNATAP | -0.48 | 0 | 27574450; 24647918 | MiR 200c promotes bladder cancer cell migration and invasion by directly targeting RECK; The luciferase reporter assay showed that RECK was a direct target of miR-200c;Finally we demonstrated that expression of miR-200c in H460 cells suppressed cell growth by targeting RECK followed by activation of the JNK signaling pathway and ER stress; Collectively these data show that miR-200c expression sensitizes H460 cells to RESV and this is likely due to RECK expression |
| 99 | hsa-miR-200c-3p | RELN | 2.07 | 0 | -4.67 | 0 | MirTarget | -0.83 | 0 | NA | |
| 100 | hsa-miR-200c-3p | RFTN2 | 2.07 | 0 | -1.5 | 0 | MirTarget | -0.45 | 0 | NA | |
| 101 | hsa-miR-200c-3p | RGL1 | 2.07 | 0 | -1.67 | 0 | MirTarget; miRNATAP | -0.37 | 0 | NA | |
| 102 | hsa-miR-200c-3p | SCN2A | 2.07 | 0 | -3.32 | 0 | MirTarget; miRNATAP | -0.42 | 0 | NA | |
| 103 | hsa-miR-200c-3p | SCN3A | 2.07 | 0 | -4.06 | 0 | mirMAP | -0.68 | 0 | NA | |
| 104 | hsa-miR-200c-3p | SCN5A | 2.07 | 0 | -2.93 | 0 | miRNATAP | -0.4 | 0 | NA | |
| 105 | hsa-miR-200c-3p | SEMA6D | 2.07 | 0 | -2.5 | 0 | miRNATAP | -0.47 | 0 | NA | |
| 106 | hsa-miR-200c-3p | SGCD | 2.07 | 0 | -1.03 | 0 | mirMAP | -0.43 | 0 | NA | |
| 107 | hsa-miR-200c-3p | SHE | 2.07 | 0 | -2.33 | 0 | mirMAP | -0.5 | 0 | NA | |
| 108 | hsa-miR-200c-3p | SHROOM4 | 2.07 | 0 | -1.47 | 0 | mirMAP | -0.35 | 0 | NA | |
| 109 | hsa-miR-200c-3p | SLC14A1 | 2.07 | 0 | -1.83 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 110 | hsa-miR-200c-3p | SLC16A12 | 2.07 | 0 | -3.4 | 0 | mirMAP; miRNATAP | -0.56 | 0 | NA | |
| 111 | hsa-miR-200c-3p | SLC38A4 | 2.07 | 0 | -1.23 | 0 | miRNATAP | -0.44 | 0 | NA | |
| 112 | hsa-miR-200c-3p | SLC4A4 | 2.07 | 0 | -3.22 | 0 | mirMAP | -0.63 | 0 | NA | |
| 113 | hsa-miR-200c-3p | SOX6 | 2.07 | 0 | -2.37 | 0 | mirMAP; miRNATAP | -0.41 | 0 | NA | |
| 114 | hsa-miR-200c-3p | STARD8 | 2.07 | 0 | -1.15 | 0 | mirMAP | -0.34 | 0 | NA | |
| 115 | hsa-miR-200c-3p | SV2B | 2.07 | 0 | -2.65 | 0 | mirMAP | -0.49 | 0 | NA | |
| 116 | hsa-miR-200c-3p | TBX18 | 2.07 | 0 | -1.63 | 0 | MirTarget | -0.46 | 0 | NA | |
| 117 | hsa-miR-200c-3p | TCF4 | 2.07 | 0 | -1.1 | 0 | mirMAP; miRNATAP | -0.37 | 0 | NA | |
| 118 | hsa-miR-200c-3p | TEAD1 | 2.07 | 0 | -1.4 | 0 | miRNATAP | -0.3 | 0 | NA | |
| 119 | hsa-miR-200c-3p | TFPI | 2.07 | 0 | -2.25 | 0 | MirTarget | -0.51 | 0 | NA | |
| 120 | hsa-miR-200c-3p | TIMP2 | 2.07 | 0 | -0.79 | 0 | miRTarBase | -0.35 | 0 | NA | |
| 121 | hsa-miR-200c-3p | TLN2 | 2.07 | 0 | -1.84 | 0 | miRNATAP | -0.4 | 0 | NA | |
| 122 | hsa-miR-200c-3p | TRHDE | 2.07 | 0 | -4.57 | 0 | MirTarget | -0.97 | 0 | NA | |
| 123 | hsa-miR-200c-3p | VAT1L | 2.07 | 0 | -1.43 | 0 | MirTarget; miRNATAP | -0.4 | 0 | NA | |
| 124 | hsa-miR-200c-3p | VGLL3 | 2.07 | 0 | -2.1 | 0 | mirMAP | -0.49 | 0 | NA | |
| 125 | hsa-miR-200c-3p | VLDLR | 2.07 | 0 | -1.77 | 0 | miRNATAP | -0.36 | 0 | NA | |
| 126 | hsa-miR-200c-3p | WASF3 | 2.07 | 0 | -2.53 | 0 | miRNATAP | -0.46 | 0 | NA | |
| 127 | hsa-miR-200c-3p | XG | 2.07 | 0 | -1.55 | 0 | MirTarget | -0.42 | 0 | NA | |
| 128 | hsa-miR-200c-3p | ZC3H12C | 2.07 | 0 | -1.87 | 0 | miRNATAP | -0.31 | 0 | NA | |
| 129 | hsa-miR-200c-3p | ZCCHC24 | 2.07 | 0 | -1.58 | 0 | MirTarget; miRNATAP | -0.47 | 0 | NA | |
| 130 | hsa-miR-200c-3p | ZEB1 | 2.07 | 0 | -1.2 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.42 | 0 | 24710933; 24615544; 21682933; 27717206; 23626803; 27666124; 23754305; 24424572; 24186205; 22407310 | miR-200c has been shown to regulate the epithelial-mesenchymal transition EMT by inhibiting ZEB1 and ZEB2 expression in breast cancer cells; This study further examined the role of miR-200c in the invasion and metastasis of breast cancer that goes beyond the regulation on ZEB1 and ZEB2 expression;MiR 200c suppresses TGF β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer; MiR-200c which was the most significantly downregulated miRNA in trastuzumab-resistant cells restored trastuzumab sensitivity and suppressed invasion of breast cancer cells by concurrently targeting ZNF217 a transcriptional activator of TGF-β and ZEB1 a known mediator of TGF-β signaling; Given the reported backward inhibition of miR-200c by ZEB1 ZNF217 also exerts a feedback suppression of miR-200c via TGF-β/ZEB1 signaling; Restoration of miR-200c silencing of ZEB1 or ZNF217 or blockade of TGF-β signaling increased trastuzumab sensitivity and suppressed invasiveness of breast cancer cells;Of this family miR-200c has garnered particular attention as a consequence of its ability to target ZEB1 and ZEB2 mediators of epithelial- mesenchymal transition;Epigenetic Silencing of miR 200c in Breast Cancer Is Associated with Aggressiveness and Is Modulated by ZEB1;Concomitant with the increase in miR-200b and miR-200c ZEB1 expression was decreased and cells appeared more epithelial in morphology and were sensitized to TAM and fulvestrant inhibition;miR 200c regulates crizotinib resistant ALK positive lung cancer cells by reversing epithelial mesenchymal transition via targeting ZEB1;Overexpression of miR-200c in SN12-PM6 and 786-0 cells was concurrent with downregulation of ZEB1 and upregulation of E-cadherin mRNA and protein; Thus our study demonstrated that miR-200c decreases the metastatic ability of renal carcinoma cells by upregulating E-cadherin through ZEB1 and that modulating the expression of miR-200c could influence Akt protein levels;Accordingly the enforced expression of miR-200c and mir-141 resulted in a significant upregulation in E-cadherin expression contrary to the significant downregulation in ZEB1 expression in 3 cell lines UTSCC-24A UTSCC-24B and UTSCC-6A cells;Furthermore miR-200c was found to be important in modulating ZEB1 upregulation by ERG;miR 200c inhibits invasion and migration in human colon cancer cells SW480/620 by targeting ZEB1; Among those dysregulated microRNAs miR-200c was speculated to inhibit metastasis by targeting ZEB1; Overexpression of miR-200c was concurrent with downregulation of ZEB1 mRNA and protein; Taken together our study demonstrated that miR-200c inhibits metastatic ability by targeting ZEB1 in colon cancer cells SW480/620 and suggested that modulation of miR-200c could serve as therapeutic tool for inhibiting metastasis in colorectal cancer |
| 131 | hsa-miR-200c-3p | ZEB2 | 2.07 | 0 | -1.6 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.47 | 0 | 24710933; 21682933; 26935975; 25052237; 18925646; 24885194 | miR-200c has been shown to regulate the epithelial-mesenchymal transition EMT by inhibiting ZEB1 and ZEB2 expression in breast cancer cells; This study further examined the role of miR-200c in the invasion and metastasis of breast cancer that goes beyond the regulation on ZEB1 and ZEB2 expression;Of this family miR-200c has garnered particular attention as a consequence of its ability to target ZEB1 and ZEB2 mediators of epithelial- mesenchymal transition;MicroRNA 200c inhibits the metastasis of non small cell lung cancer cells by targeting ZEB2 an epithelial mesenchymal transition regulator;miR 200c modulates ovarian cancer cell metastasis potential by targeting zinc finger E box binding homeobox 2 ZEB2 expression; Luciferase reporter assay confirmed the target of miR-200c as ZEB2; Furthermore miR-200c expression inhibited ovarian cancer cell ES-2 migration and invasion capacity by suppression of ZEB2 expression p < 0.01; Thus targeting of miR-200c or ZEB2 may serve as a potential therapeutic strategy for control of ovarian cancer;We established by quantitative RT-PCR that in CCCs in which miR-141 and miR-200c were down-regulated ZFHX1B a transcriptional repressor for CDH1/E-cadherin tended to be up-regulated; On the basis of these findings we suggest that down-regulation of miR-141 and miR-200c in CCCs might be involved in suppression of CDH1/E-cadherin transcription via up-regulation of ZFHX1B;Two gastric cancer cell lines were treated with IGF-I to induce EMT and levels of transcription factor ZEB2 and microRNA-200c miR-200c were measured; Furthermore both Akt/ERK inhibitors and knockdown of Akt/ERK gene reversed IGF-I-induced ZEB2 up-regulation and EMT through up-regulation of miR-200c suggesting the involvement of an Akt/ERK-miR-200c-ZEB2 axis in IGF-I-induced EMT |
| 132 | hsa-miR-200c-3p | ZFHX4 | 2.07 | 0 | -1.53 | 0 | miRNATAP | -0.51 | 0 | NA | |
| 133 | hsa-miR-200c-3p | ZFPM2 | 2.07 | 0 | -1.48 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.49 | 0 | NA | |
| 134 | hsa-miR-200c-3p | ZNF423 | 2.07 | 0 | -1.54 | 0 | MirTarget | -0.47 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF NEURON DIFFERENTIATION | 24 | 554 | 3.014e-13 | 1.402e-09 |
| 2 | NEUROGENESIS | 36 | 1402 | 1.221e-12 | 2.841e-09 |
| 3 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 26 | 750 | 4.441e-12 | 5.166e-09 |
| 4 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 28 | 872 | 3.654e-12 | 5.166e-09 |
| 5 | REGULATION OF CELL DIFFERENTIATION | 36 | 1492 | 7.444e-12 | 6.928e-09 |
| 6 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 38 | 1672 | 9.939e-12 | 7.708e-09 |
| 7 | REGULATION OF CELL DEVELOPMENT | 26 | 836 | 4.919e-11 | 3.27e-08 |
| 8 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 30 | 1142 | 8.041e-11 | 4.677e-08 |
| 9 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 19 | 437 | 1.064e-10 | 5.502e-08 |
| 10 | CELL DEVELOPMENT | 33 | 1426 | 2.071e-10 | 9.638e-08 |
| 11 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 16 | 306 | 2.444e-10 | 1.034e-07 |
| 12 | HEAD DEVELOPMENT | 22 | 709 | 1.915e-09 | 7.427e-07 |
| 13 | POSITIVE REGULATION OF CELL DEVELOPMENT | 18 | 472 | 2.759e-09 | 9.875e-07 |
| 14 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 23 | 823 | 5.738e-09 | 1.907e-06 |
| 15 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 30 | 1395 | 9.071e-09 | 2.814e-06 |
| 16 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 16 | 408 | 1.52e-08 | 4.421e-06 |
| 17 | FOREBRAIN DEVELOPMENT | 15 | 357 | 1.776e-08 | 4.86e-06 |
| 18 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 12 | 232 | 5.413e-08 | 1.399e-05 |
| 19 | NEURON DIFFERENTIATION | 22 | 874 | 8.174e-08 | 2.002e-05 |
| 20 | CELLULAR COMPONENT MORPHOGENESIS | 22 | 900 | 1.359e-07 | 3.012e-05 |
| 21 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 13 | 303 | 1.316e-07 | 3.012e-05 |
| 22 | REGULATION OF CELL PROJECTION ORGANIZATION | 17 | 558 | 2.033e-07 | 4.3e-05 |
| 23 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 23 | 1021 | 2.91e-07 | 5.888e-05 |
| 24 | LOCOMOTION | 24 | 1114 | 3.481e-07 | 6.749e-05 |
| 25 | NEURON PROJECTION MORPHOGENESIS | 14 | 402 | 5.377e-07 | 0.0001001 |
| 26 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 31 | 1784 | 6.081e-07 | 0.0001088 |
| 27 | NEURON DEVELOPMENT | 18 | 687 | 7.88e-07 | 0.0001309 |
| 28 | NEURON PROJECTION DEVELOPMENT | 16 | 545 | 7.784e-07 | 0.0001309 |
| 29 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 25 | 1275 | 1.091e-06 | 0.0001751 |
| 30 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 22 | 1036 | 1.46e-06 | 0.0002147 |
| 31 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 22 | 1036 | 1.46e-06 | 0.0002147 |
| 32 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 9 | 162 | 1.477e-06 | 0.0002147 |
| 33 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 30 | 1805 | 2.423e-06 | 0.0003417 |
| 34 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 28 | 1618 | 2.616e-06 | 0.0003581 |
| 35 | OLIGODENDROCYTE DIFFERENTIATION | 6 | 60 | 3.008e-06 | 0.0003998 |
| 36 | POSITIVE REGULATION OF GENE EXPRESSION | 29 | 1733 | 3.248e-06 | 0.0004051 |
| 37 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 21 | 1004 | 3.308e-06 | 0.0004051 |
| 38 | TELENCEPHALON DEVELOPMENT | 10 | 228 | 3.235e-06 | 0.0004051 |
| 39 | HINDBRAIN DEVELOPMENT | 8 | 137 | 3.984e-06 | 0.0004754 |
| 40 | POSITIVE REGULATION OF DENDRITE DEVELOPMENT | 6 | 64 | 4.406e-06 | 0.0005126 |
| 41 | BIOLOGICAL ADHESION | 21 | 1032 | 5.069e-06 | 0.0005753 |
| 42 | REGULATION OF PHOSPHOLIPASE C ACTIVITY | 5 | 39 | 6.006e-06 | 0.0006654 |
| 43 | CIRCULATORY SYSTEM PROCESS | 12 | 366 | 6.703e-06 | 0.0007153 |
| 44 | REGULATION OF MUSCLE SYSTEM PROCESS | 9 | 195 | 6.764e-06 | 0.0007153 |
| 45 | REGULATION OF SYSTEM PROCESS | 14 | 507 | 8.018e-06 | 0.000829 |
| 46 | CELL PROJECTION ORGANIZATION | 19 | 902 | 9.259e-06 | 0.0009366 |
| 47 | BEHAVIOR | 14 | 516 | 9.778e-06 | 0.0009681 |
| 48 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 9 | 213 | 1.375e-05 | 0.001333 |
| 49 | REGULATION OF DENDRITE DEVELOPMENT | 7 | 120 | 1.642e-05 | 0.001559 |
| 50 | MUSCLE ORGAN DEVELOPMENT | 10 | 277 | 1.781e-05 | 0.001658 |
| 51 | REGULATION OF CELL MORPHOGENESIS | 14 | 552 | 2.074e-05 | 0.001892 |
| 52 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 21 | 1135 | 2.138e-05 | 0.001909 |
| 53 | REGULATION OF PROTEIN MODIFICATION PROCESS | 27 | 1710 | 2.174e-05 | 0.001909 |
| 54 | CELL PART MORPHOGENESIS | 15 | 633 | 2.289e-05 | 0.001972 |
| 55 | GLIOGENESIS | 8 | 175 | 2.391e-05 | 0.002023 |
| 56 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 9 | 229 | 2.441e-05 | 0.002028 |
| 57 | POSITIVE REGULATION OF PHOSPHOLIPASE ACTIVITY | 5 | 53 | 2.777e-05 | 0.002267 |
| 58 | ACTIVATION OF PHOSPHOLIPASE C ACTIVITY | 4 | 27 | 3e-05 | 0.002407 |
| 59 | GLIAL CELL DIFFERENTIATION | 7 | 136 | 3.687e-05 | 0.002908 |
| 60 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 11 | 368 | 3.848e-05 | 0.002984 |
| 61 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 13 | 513 | 4.188e-05 | 0.003195 |
| 62 | MESENCHYME DEVELOPMENT | 8 | 190 | 4.301e-05 | 0.003228 |
| 63 | SYSTEM PROCESS | 27 | 1785 | 4.613e-05 | 0.003407 |
| 64 | COGNITION | 9 | 251 | 4.996e-05 | 0.003632 |
| 65 | REGULATION OF CELL PROLIFERATION | 24 | 1496 | 5.258e-05 | 0.003764 |
| 66 | SYNAPSE ORGANIZATION | 7 | 145 | 5.548e-05 | 0.003912 |
| 67 | METENCEPHALON DEVELOPMENT | 6 | 100 | 5.754e-05 | 0.003996 |
| 68 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 16 | 771 | 5.959e-05 | 0.004077 |
| 69 | REGULATION OF MUSCLE ADAPTATION | 5 | 63 | 6.447e-05 | 0.004347 |
| 70 | TISSUE DEVELOPMENT | 24 | 1518 | 6.618e-05 | 0.004399 |
| 71 | REGULATION OF HYDROLASE ACTIVITY | 22 | 1327 | 6.984e-05 | 0.004451 |
| 72 | TAXIS | 12 | 464 | 6.895e-05 | 0.004451 |
| 73 | REGULATION OF PHOSPHOLIPASE ACTIVITY | 5 | 64 | 6.956e-05 | 0.004451 |
| 74 | POSITIVE REGULATION OF LIPASE ACTIVITY | 5 | 66 | 8.066e-05 | 0.005072 |
| 75 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 20 | 1152 | 8.317e-05 | 0.00516 |
| 76 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 10 | 337 | 9.291e-05 | 0.005628 |
| 77 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 16 | 801 | 9.314e-05 | 0.005628 |
| 78 | NEURON MIGRATION | 6 | 110 | 9.789e-05 | 0.005839 |
| 79 | SYNAPSE ASSEMBLY | 5 | 69 | 9.982e-05 | 0.005879 |
| 80 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 15 | 724 | 0.0001047 | 0.006088 |
| 81 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 9 | 278 | 0.0001092 | 0.006274 |
| 82 | SENSORY ORGAN DEVELOPMENT | 12 | 493 | 0.0001221 | 0.00693 |
| 83 | REGULATION OF N METHYL D ASPARTATE SELECTIVE GLUTAMATE RECEPTOR ACTIVITY | 3 | 15 | 0.0001262 | 0.007072 |
| 84 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 4 | 39 | 0.0001321 | 0.007318 |
| 85 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 18 | 1008 | 0.0001375 | 0.007528 |
| 86 | CELL MOTILITY | 16 | 835 | 0.0001503 | 0.008039 |
| 87 | LOCALIZATION OF CELL | 16 | 835 | 0.0001503 | 0.008039 |
| 88 | MUSCLE STRUCTURE DEVELOPMENT | 11 | 432 | 0.0001599 | 0.008416 |
| 89 | CELL SUBSTRATE JUNCTION ASSEMBLY | 4 | 41 | 0.000161 | 0.008416 |
| 90 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 23 | 1518 | 0.0001806 | 0.009338 |
| 91 | DENDRITE DEVELOPMENT | 5 | 79 | 0.0001898 | 0.009703 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL BODY | 17 | 494 | 3.556e-08 | 2.077e-05 |
| 2 | SOMATODENDRITIC COMPARTMENT | 18 | 650 | 3.535e-07 | 0.0001032 |
| 3 | NEURON PROJECTION | 21 | 942 | 1.21e-06 | 0.0002355 |
| 4 | DENDRITE | 14 | 451 | 2.091e-06 | 0.0003052 |
| 5 | NEURON PART | 23 | 1265 | 1.087e-05 | 0.001104 |
| 6 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 27 | 1649 | 1.134e-05 | 0.001104 |
| 7 | MEMBRANE REGION | 21 | 1134 | 2.11e-05 | 0.00176 |
| 8 | MEMBRANE MICRODOMAIN | 10 | 288 | 2.487e-05 | 0.001815 |
| 9 | ANCHORED COMPONENT OF MEMBRANE | 7 | 152 | 7.476e-05 | 0.004851 |
| 10 | VOLTAGE GATED SODIUM CHANNEL COMPLEX | 3 | 14 | 0.0001014 | 0.005923 |
| 11 | CELL PROJECTION | 26 | 1786 | 0.000123 | 0.006528 |
| 12 | CELL SURFACE | 15 | 757 | 0.0001705 | 0.008297 |
| 13 | SODIUM CHANNEL COMPLEX | 3 | 17 | 0.0001867 | 0.008387 |
| 14 | POSTSYNAPSE | 10 | 378 | 0.0002359 | 0.009464 |
| 15 | CELL JUNCTION | 19 | 1151 | 0.0002431 | 0.009464 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04510_Focal_adhesion | 6 | 200 | 0.002309 | 0.1861 | |
| 2 | hsa04512_ECM.receptor_interaction | 4 | 85 | 0.002565 | 0.1861 | |
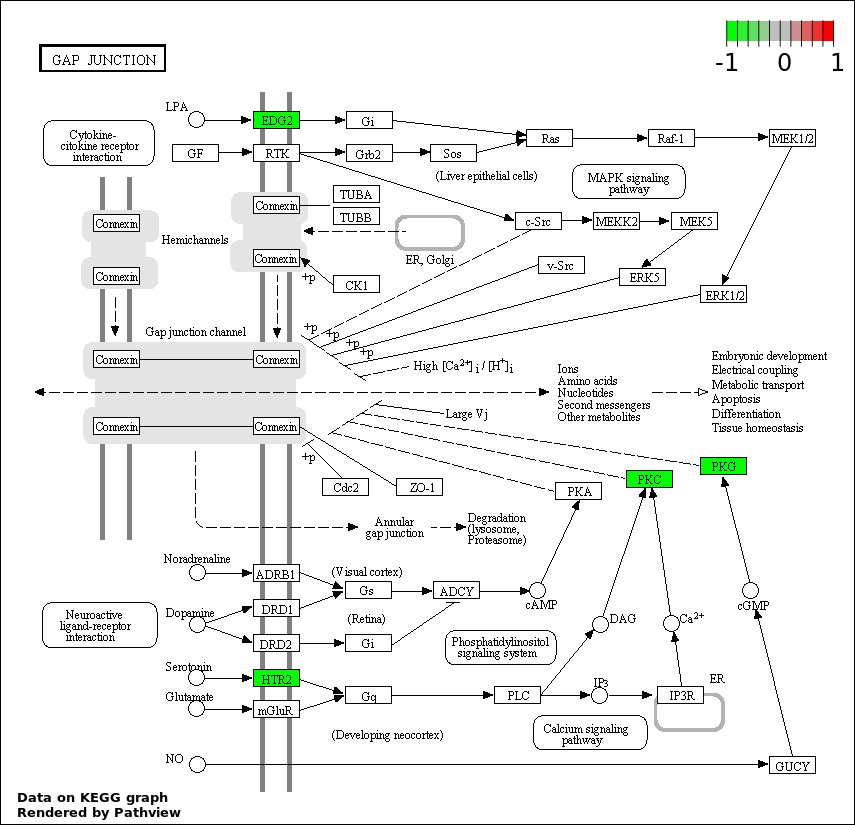
| 3 | hsa04540_Gap_junction | 4 | 90 | 0.003155 | 0.1861 | |
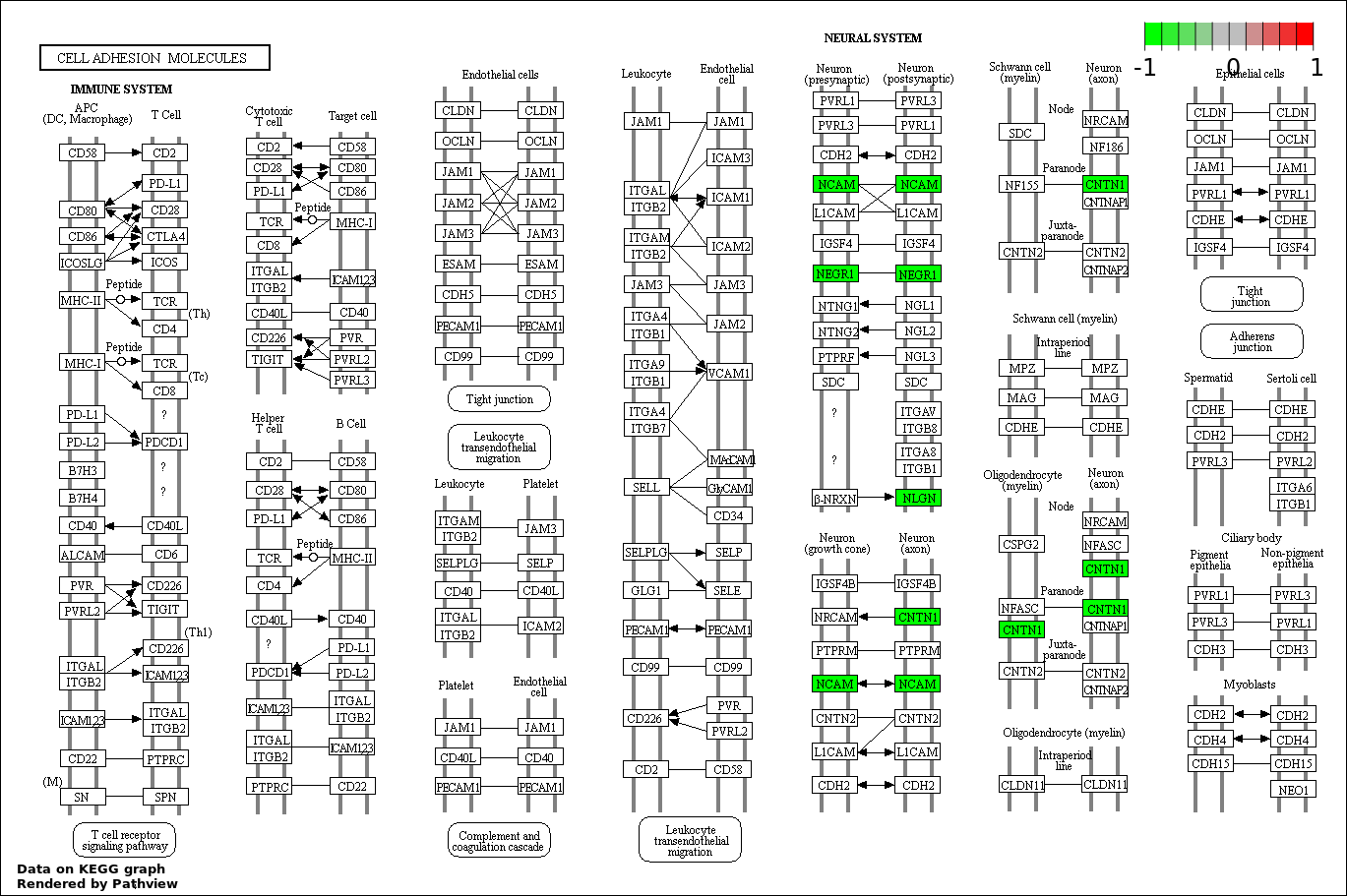
| 4 | hsa04514_Cell_adhesion_molecules_.CAMs. | 4 | 136 | 0.01331 | 0.589 | |
| 5 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 3 | 95 | 0.02604 | 0.7765 | |
| 6 | hsa04916_Melanogenesis | 3 | 101 | 0.03046 | 0.7765 | |
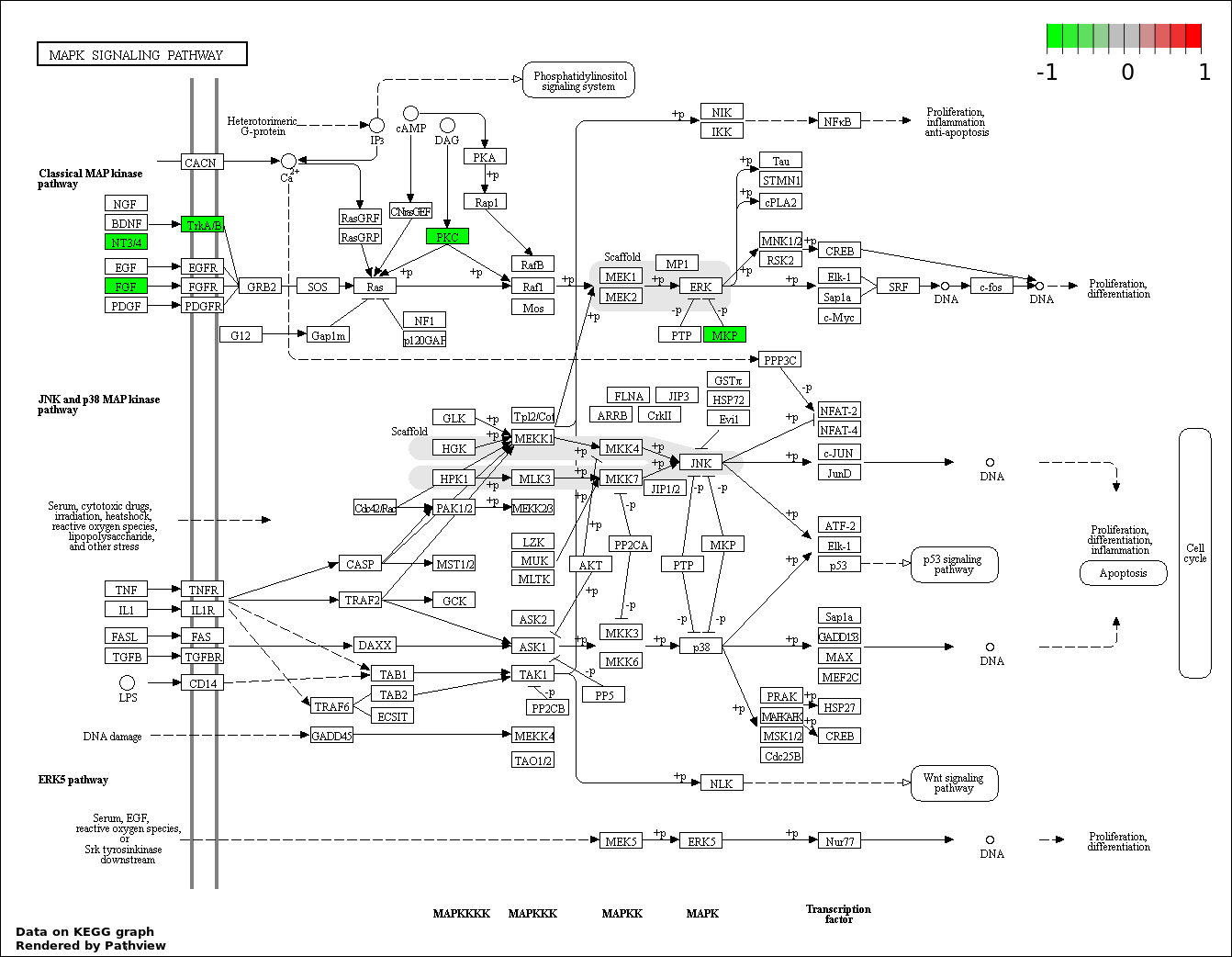
| 7 | hsa04010_MAPK_signaling_pathway | 5 | 268 | 0.0345 | 0.7765 | |
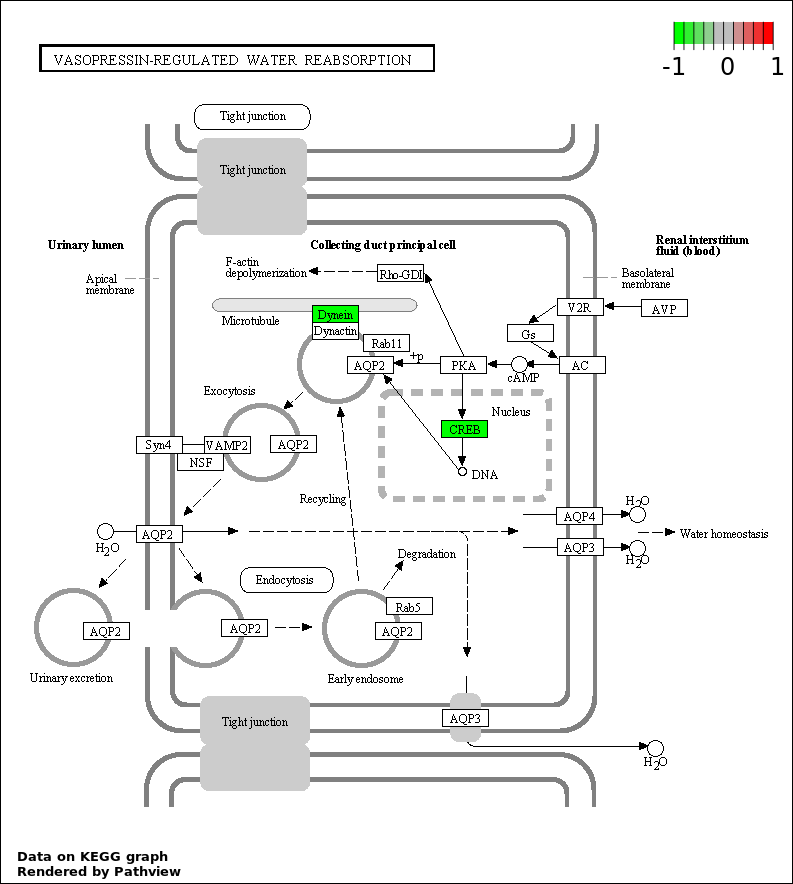
| 8 | hsa04962_Vasopressin.regulated_water_reabsorption | 2 | 44 | 0.03509 | 0.7765 | |
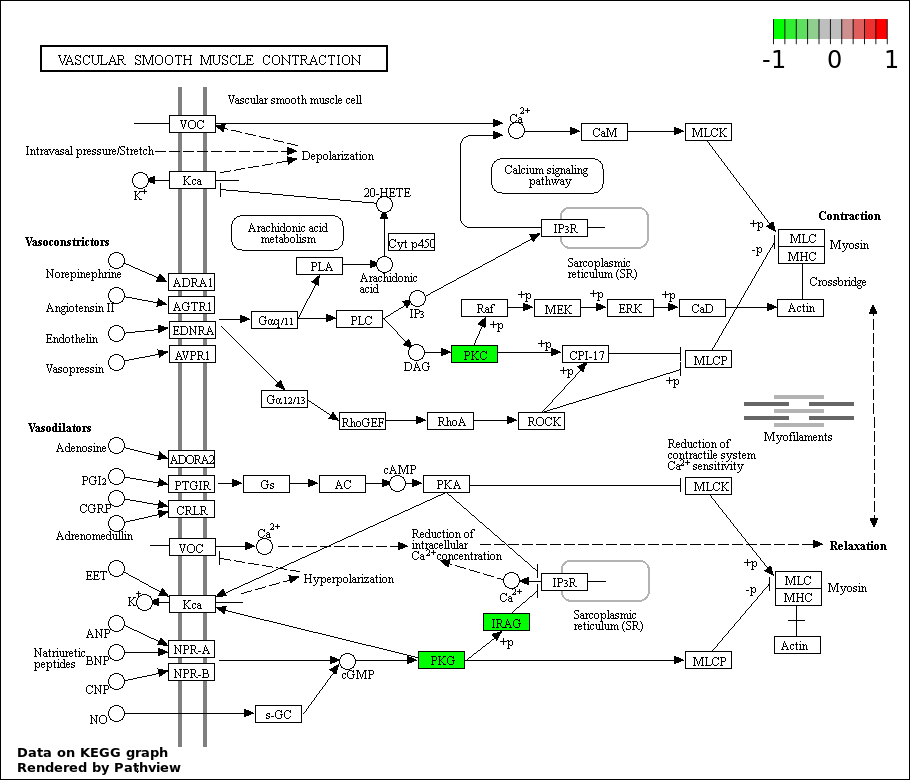
| 9 | hsa04270_Vascular_smooth_muscle_contraction | 3 | 116 | 0.04314 | 0.8483 | |
| 10 | hsa04380_Osteoclast_differentiation | 3 | 128 | 0.05489 | 0.9168 | |
| 11 | hsa04360_Axon_guidance | 3 | 130 | 0.05698 | 0.9168 | |
| 12 | hsa04730_Long.term_depression | 2 | 70 | 0.08015 | 1 | |
| 13 | hsa04520_Adherens_junction | 2 | 73 | 0.08611 | 1 | |
| 14 | hsa04971_Gastric_acid_secretion | 2 | 74 | 0.08813 | 1 | |
| 15 | hsa04664_Fc_epsilon_RI_signaling_pathway | 2 | 79 | 0.09842 | 1 | |
| 16 | hsa04012_ErbB_signaling_pathway | 2 | 87 | 0.1155 | 1 | |
| 17 | hsa04970_Salivary_secretion | 2 | 89 | 0.1199 | 1 | |
| 18 | hsa04972_Pancreatic_secretion | 2 | 101 | 0.1471 | 1 | |
| 19 | hsa04810_Regulation_of_actin_cytoskeleton | 3 | 214 | 0.1735 | 1 | |
| 20 | hsa04722_Neurotrophin_signaling_pathway | 2 | 127 | 0.2092 | 1 | |
| 21 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 2 | 136 | 0.2314 | 1 | |
| 22 | hsa04310_Wnt_signaling_pathway | 2 | 151 | 0.2686 | 1 | |
| 23 | hsa04630_Jak.STAT_signaling_pathway | 2 | 155 | 0.2785 | 1 | |
| 24 | hsa04020_Calcium_signaling_pathway | 2 | 177 | 0.3328 | 1 |