Browse ACTL6A in pancancer
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00022 Actin |
||||||||||
| Function |
Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Required for maximal ATPase activity of SMARCA4/BRG1/BAF190A and for association of the SMARCA4/BRG1/BAF190A containing remodeling complex BAF with chromatin/nuclear matrix. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and is required for the proliferation of neural progenitors. During neural development a switch from a stem/progenitor to a post-mitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to post-mitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). Component of the NuA4 histone acetyltransferase (HAT) complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Putative core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000422 mitophagy GO:0001654 eye development GO:0003407 neural retina development GO:0006310 DNA recombination GO:0006338 chromatin remodeling GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0006626 protein targeting to mitochondrion GO:0006839 mitochondrial transport GO:0006914 autophagy GO:0007423 sensory organ development GO:0010506 regulation of autophagy GO:0010821 regulation of mitochondrion organization GO:0010822 positive regulation of mitochondrion organization GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0021510 spinal cord development GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0033157 regulation of intracellular protein transport GO:0043010 camera-type eye development GO:0043044 ATP-dependent chromatin remodeling GO:0043543 protein acylation GO:0043967 histone H4 acetylation GO:0043968 histone H2A acetylation GO:0051222 positive regulation of protein transport GO:0060041 retina development in camera-type eye GO:0061726 mitochondrion disassembly GO:0070585 protein localization to mitochondrion GO:0072655 establishment of protein localization to mitochondrion GO:0090316 positive regulation of intracellular protein transport GO:1903008 organelle disassembly GO:1903146 regulation of mitophagy GO:1903214 regulation of protein targeting to mitochondrion GO:1903533 regulation of protein targeting GO:1903747 regulation of establishment of protein localization to mitochondrion GO:1903749 positive regulation of establishment of protein localization to mitochondrion GO:1903829 positive regulation of cellular protein localization GO:1903955 positive regulation of protein targeting to mitochondrion GO:1904951 positive regulation of establishment of protein localization |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001158 enhancer sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0003682 chromatin binding GO:0003713 transcription coactivator activity GO:0031490 chromatin DNA binding GO:0031491 nucleosome binding GO:0031492 nucleosomal DNA binding GO:0035326 enhancer binding GO:0043566 structure-specific DNA binding |
| Cellular Component |
GO:0000123 histone acetyltransferase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016514 SWI/SNF complex GO:0031011 Ino80 complex GO:0031248 protein acetyltransferase complex GO:0033202 DNA helicase complex GO:0035267 NuA4 histone acetyltransferase complex GO:0043189 H4/H2A histone acetyltransferase complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0071564 npBAF complex GO:0090544 BAF-type complex GO:0097346 INO80-type complex GO:1902493 acetyltransferase complex GO:1902562 H4 histone acetyltransferase complex |
| KEGG | - |
| Reactome |
R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-5696394: DNA Damage Recognition in GG-NER R-HSA-73894: DNA Repair R-HSA-5688426: Deubiquitination R-HSA-5696399: Global Genome Nucleotide Excision Repair (GG-NER) R-HSA-3214847: HATs acetylate histones R-HSA-392499: Metabolism of proteins R-HSA-5696398: Nucleotide Excision Repair R-HSA-597592: Post-translational protein modification R-HSA-3214858: RMTs methylate histone arginines R-HSA-5689603: UCH proteinases |
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for ACTL6A. |
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
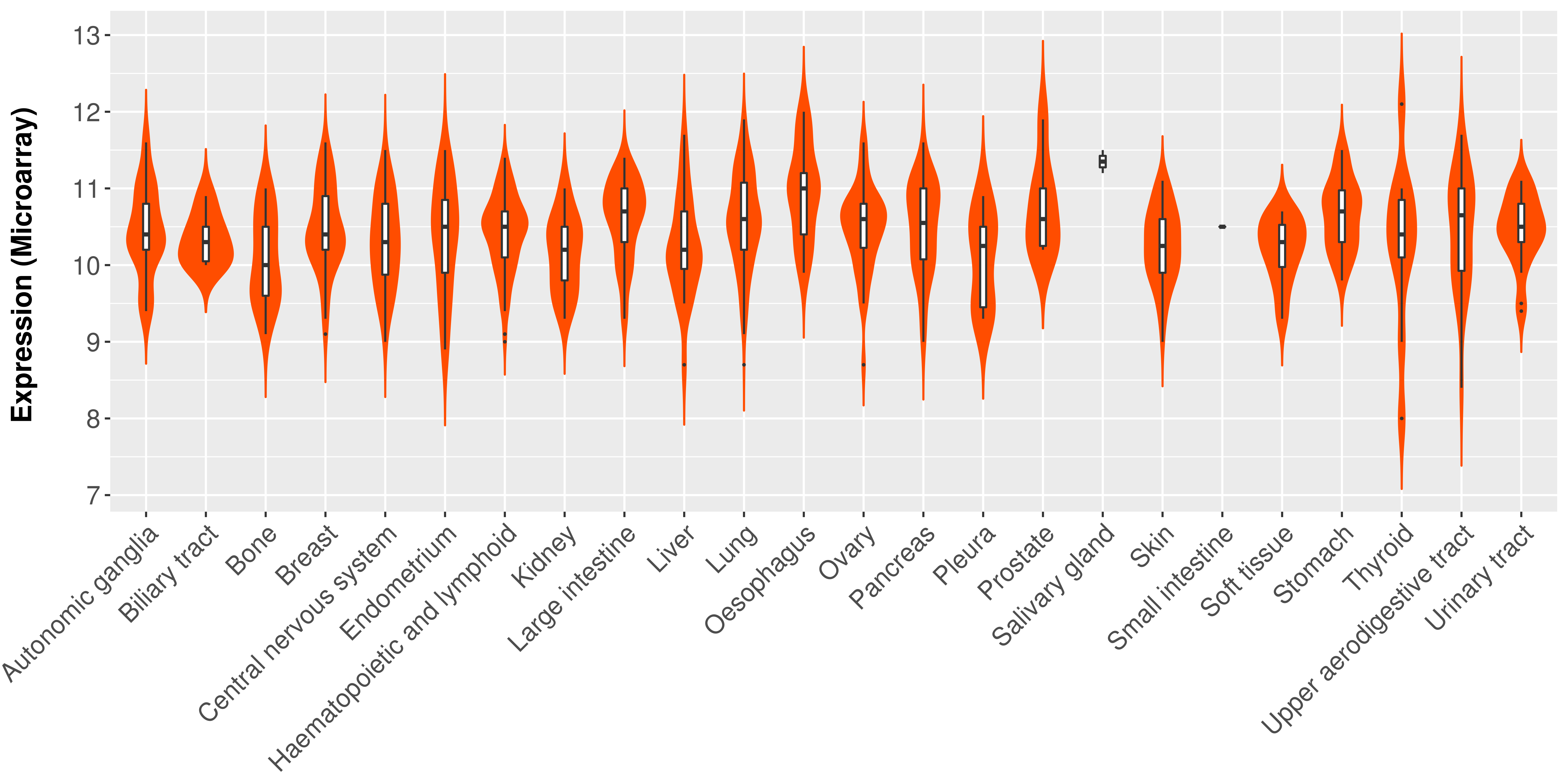
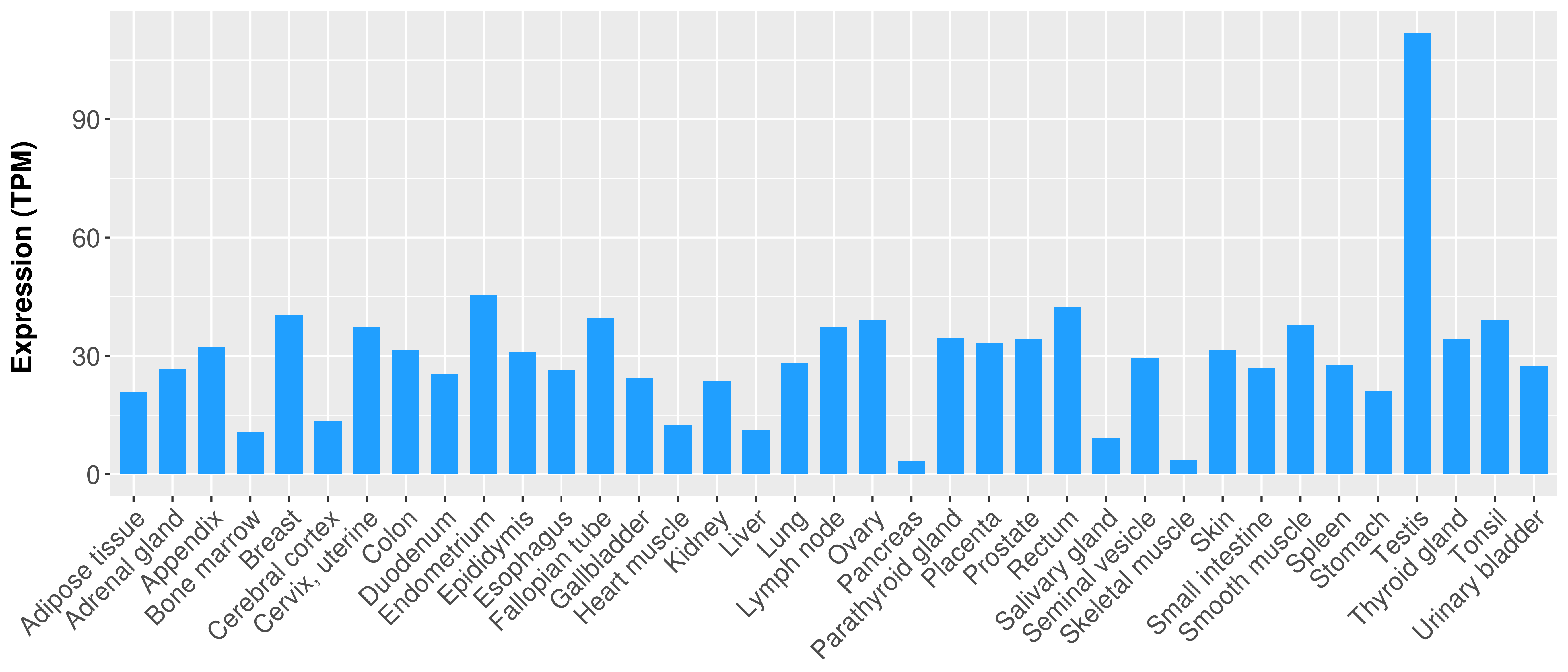
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
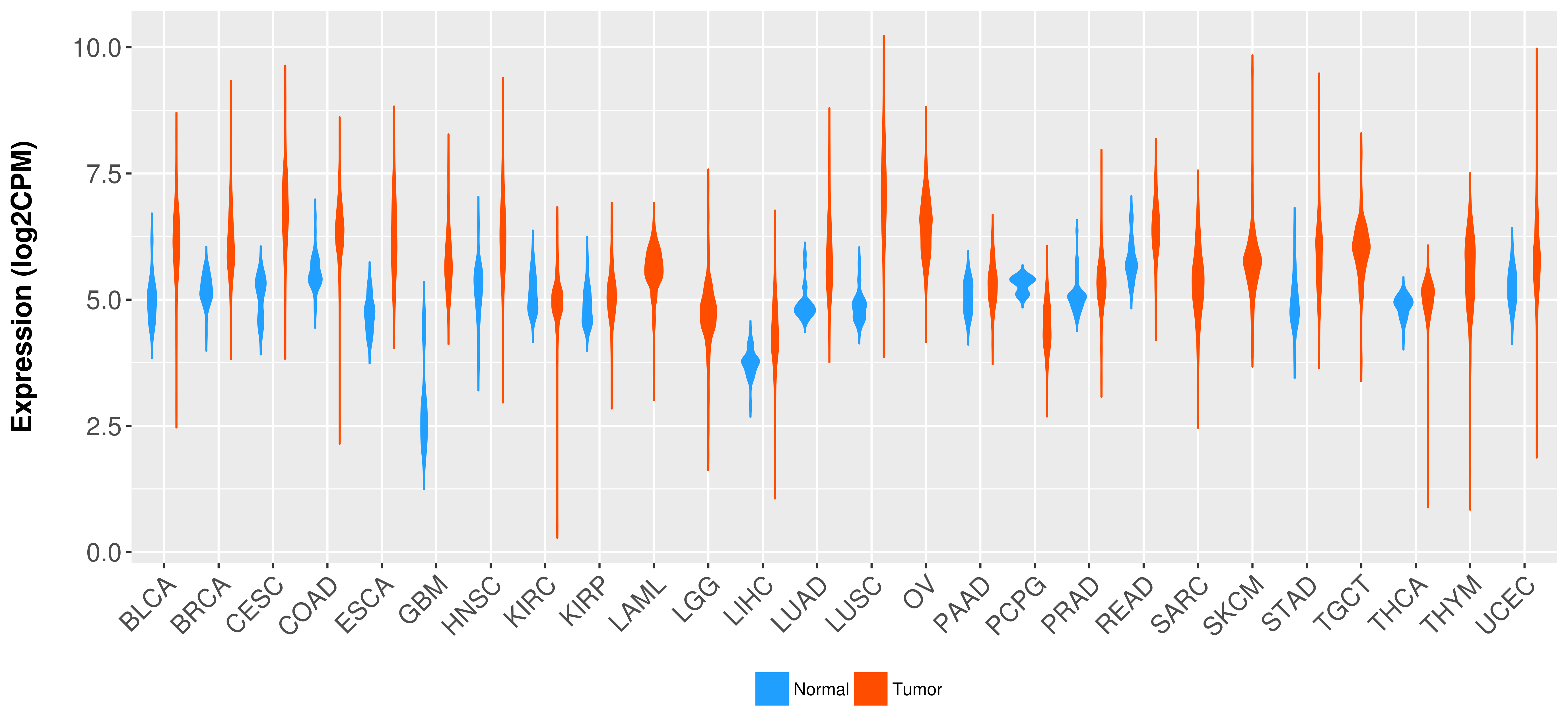
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
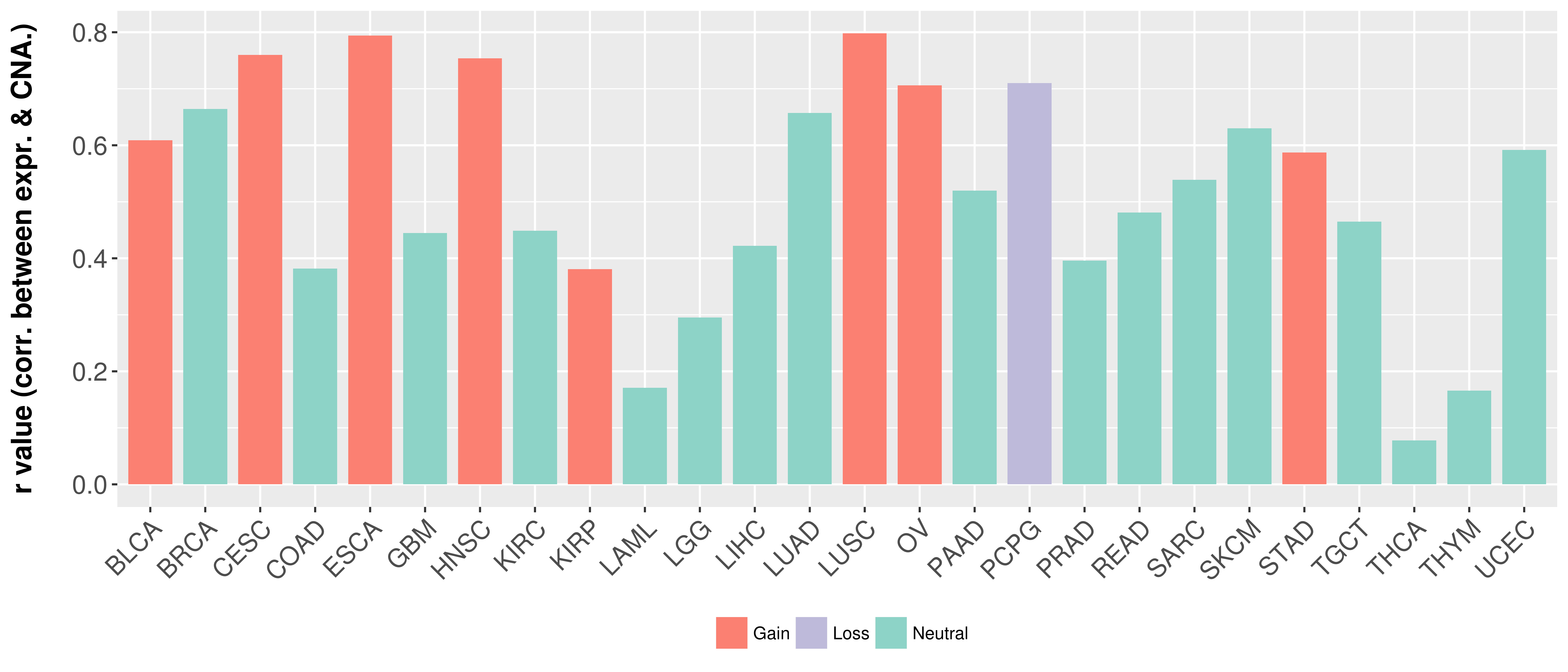
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
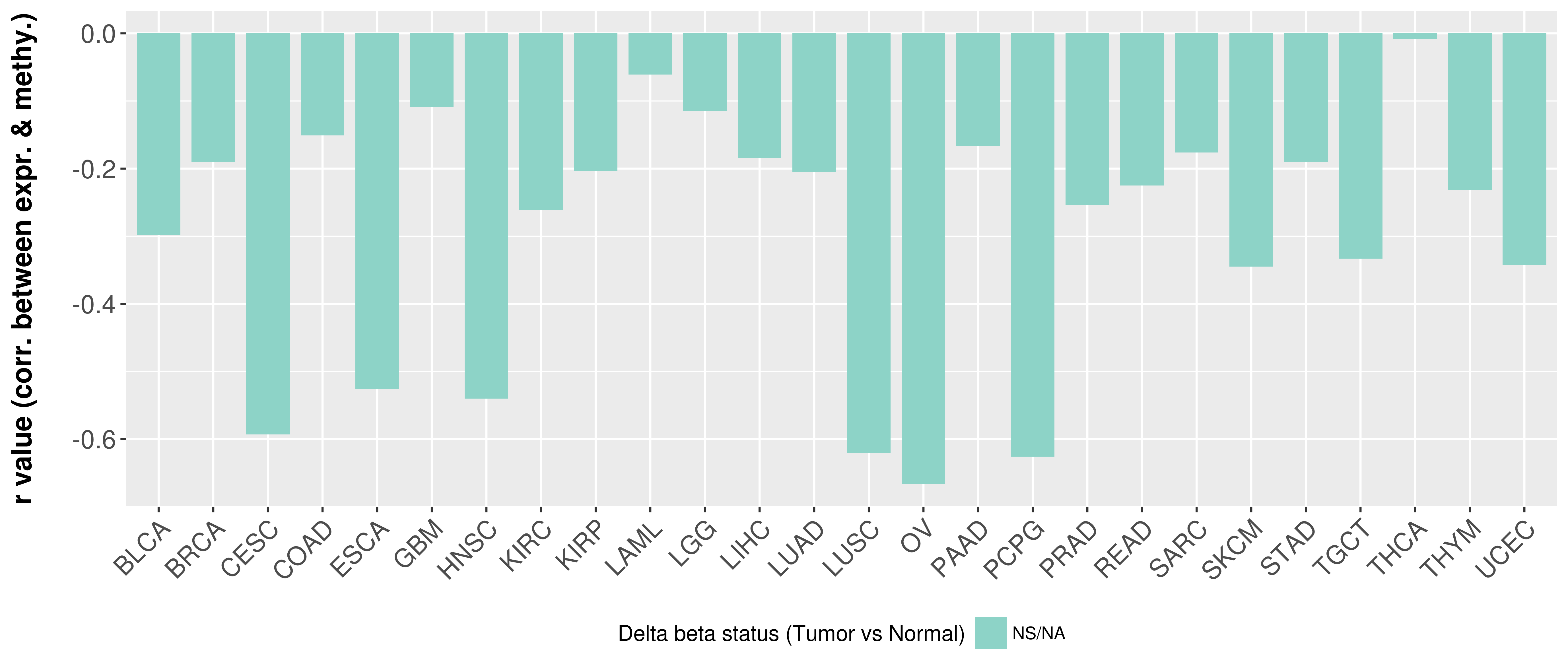
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
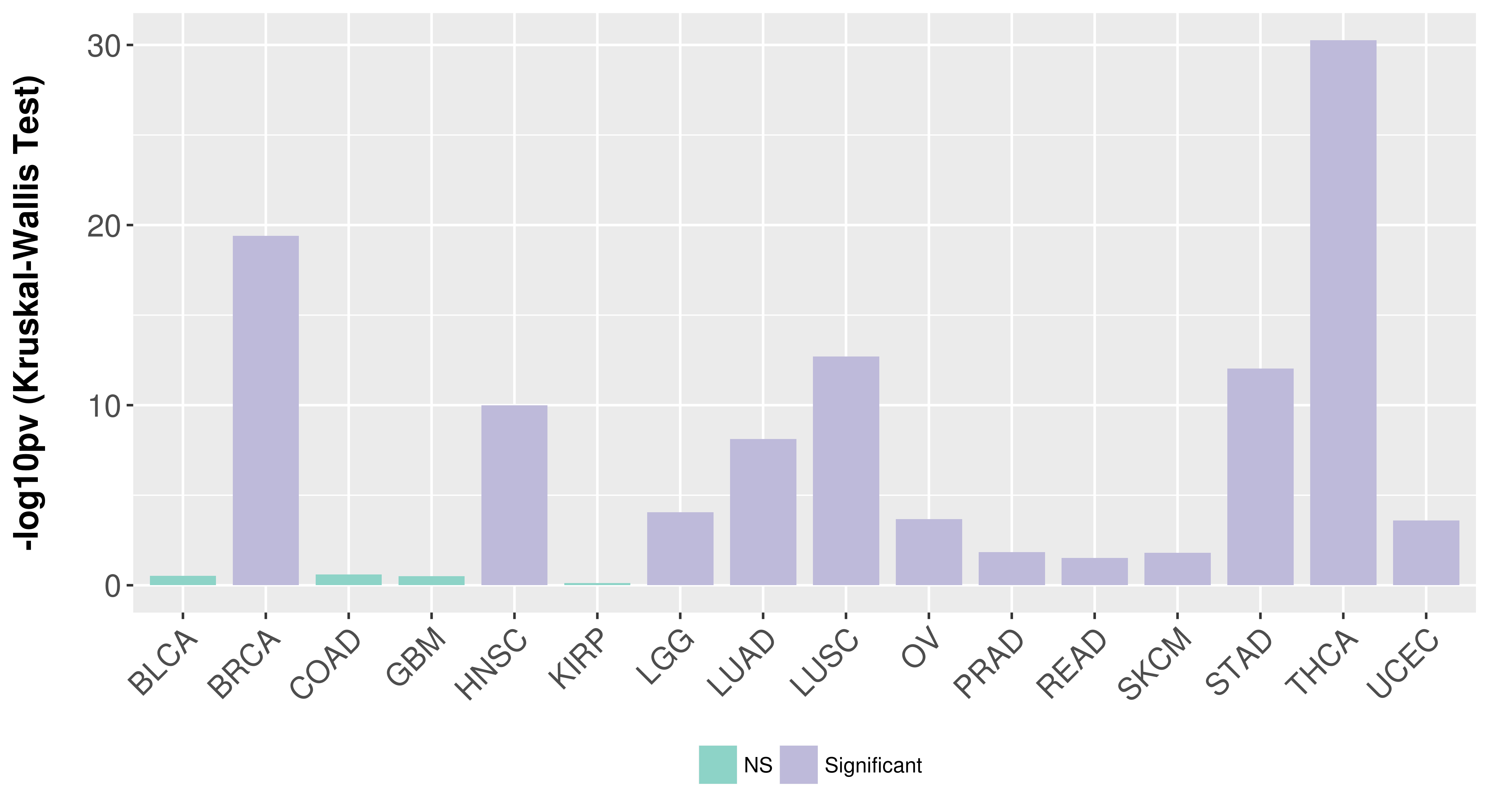
Association between expresson and subtype.
|
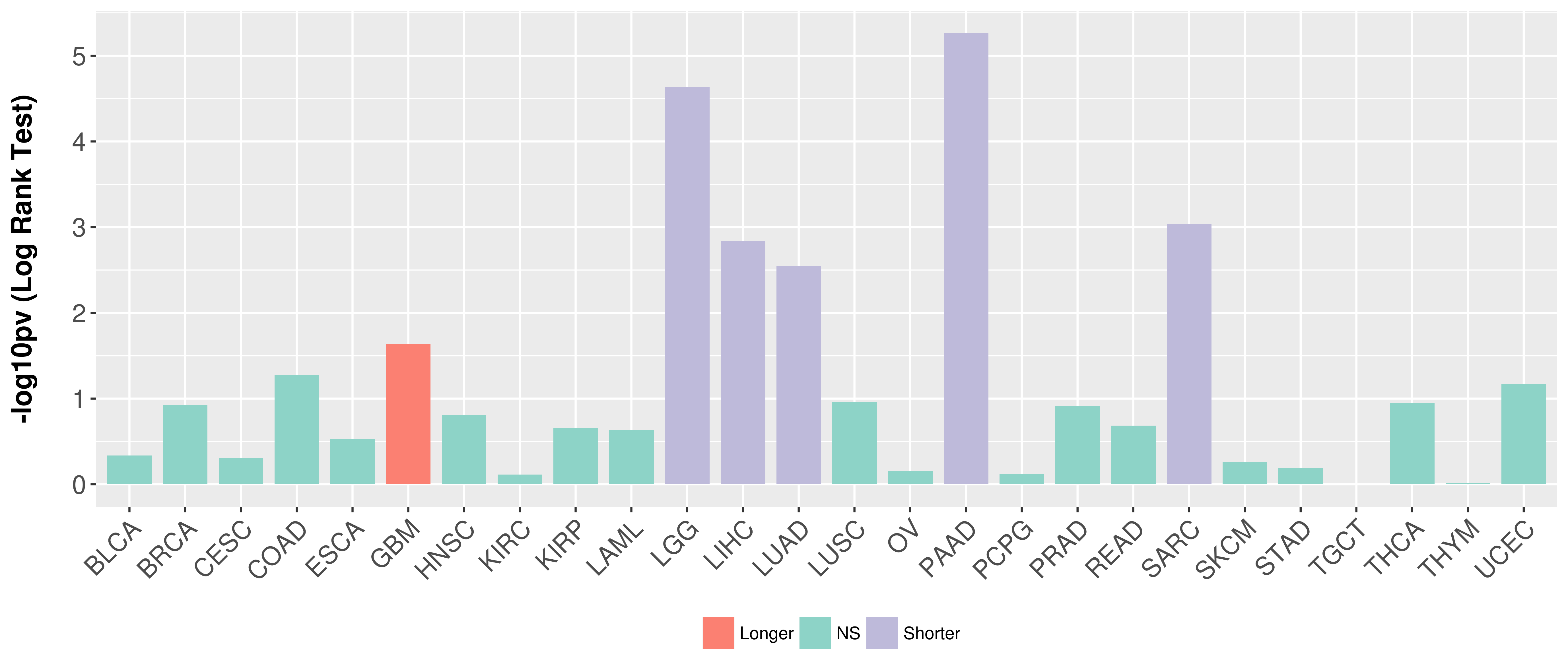
Overall survival analysis based on expression.
|
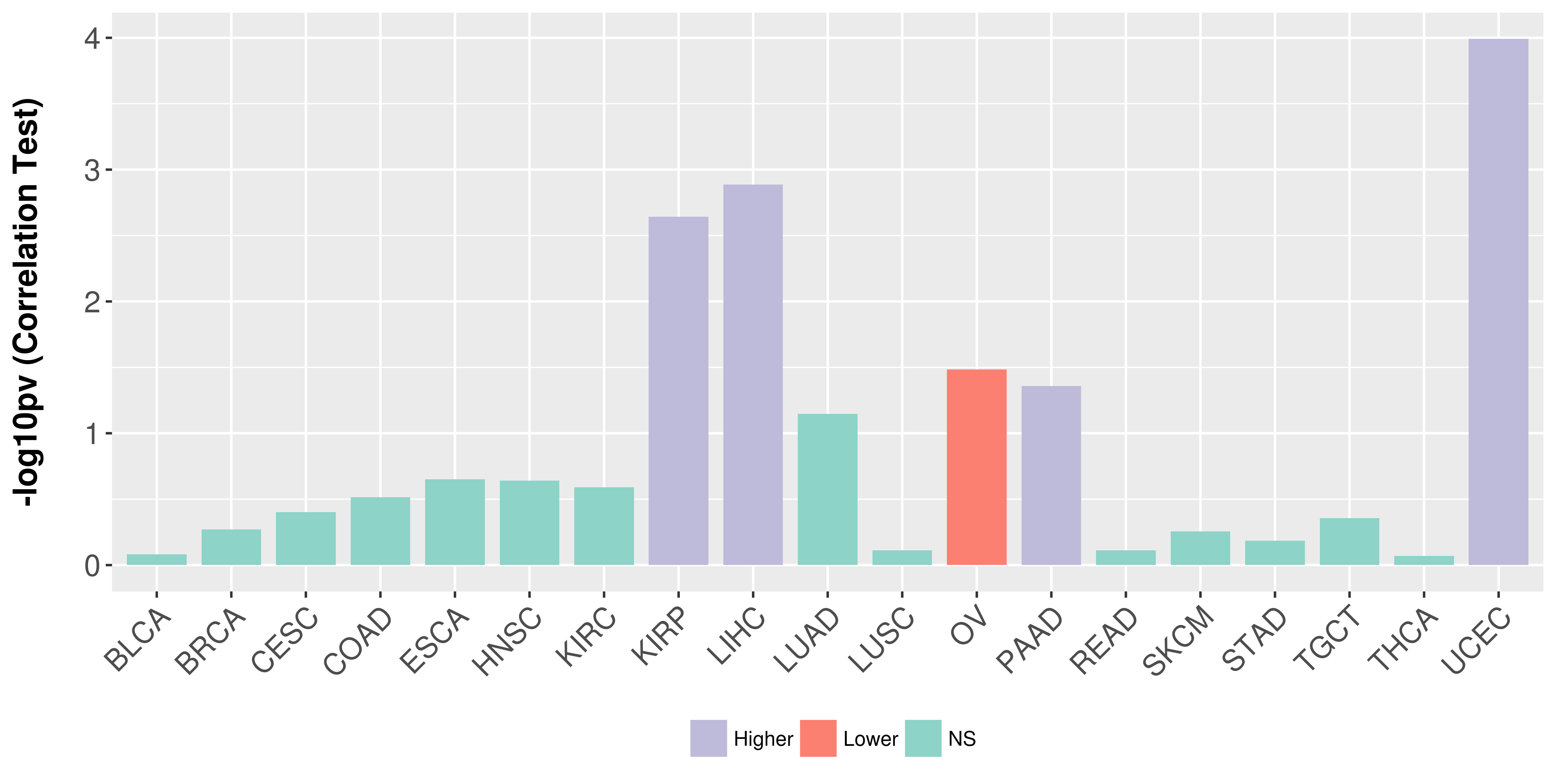
Association between expresson and stage.
|
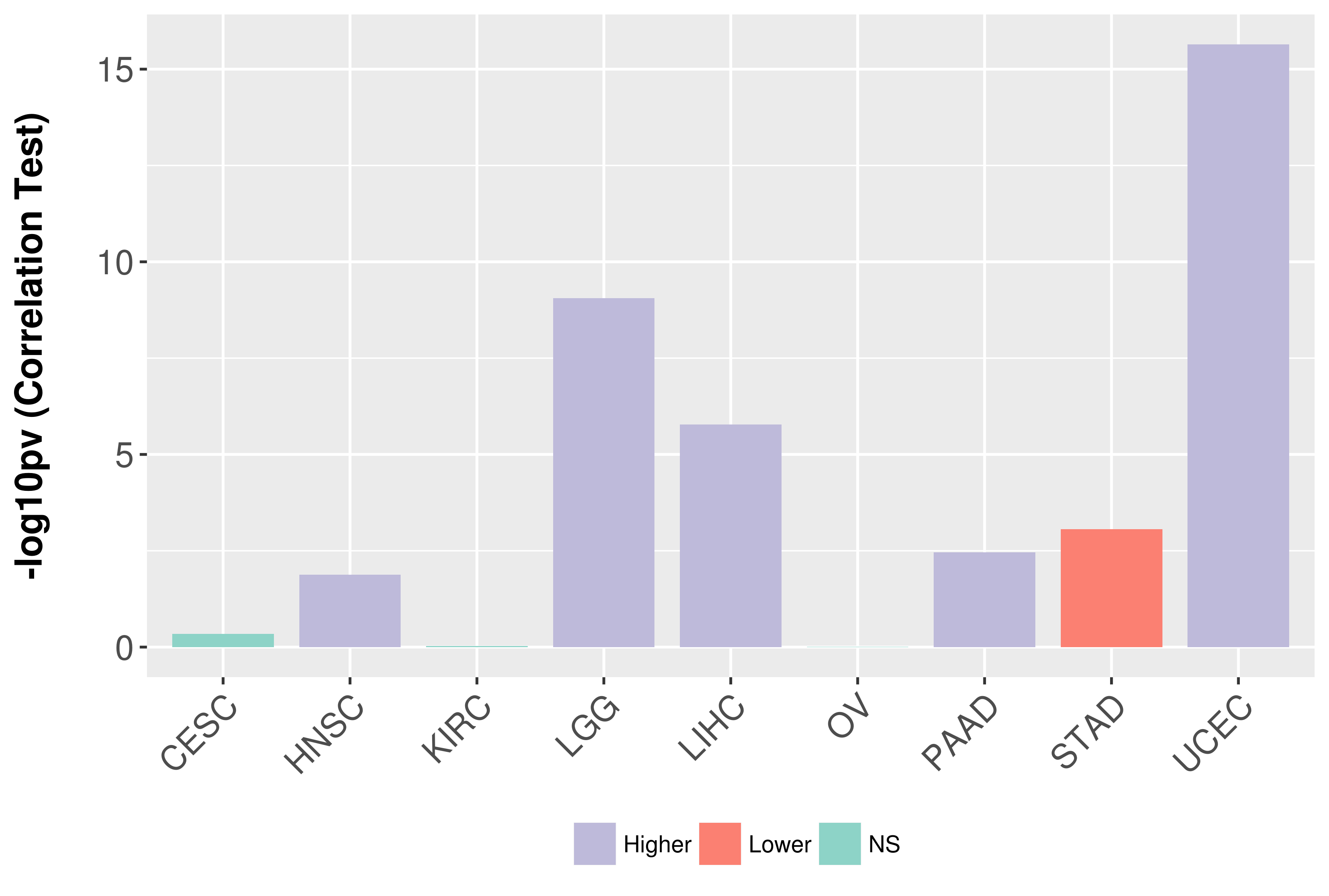
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ACTL6A. |
| Summary | |
|---|---|
| Symbol | ACTL6A |
| Name | actin like 6A |
| Aliases | BAF53A; INO80K; BAF complex 53 kDa subunit; BRG1-associated factor; actin-related protein 4; INO80 complex s ...... |
| Location | 3q26.33 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|