Browse ADNP in pancancer
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00046 Homeobox domain |
||||||||||
| Function |
Potential transcription factor. May mediate some of the neuroprotective peptide VIP-associated effects involving normal growth and cancer proliferation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001558 regulation of cell growth GO:0006140 regulation of nucleotide metabolic process GO:0006164 purine nucleotide biosynthetic process GO:0006182 cGMP biosynthetic process GO:0006471 protein ADP-ribosylation GO:0006486 protein glycosylation GO:0007409 axonogenesis GO:0007416 synapse assembly GO:0007611 learning or memory GO:0007613 memory GO:0007614 short-term memory GO:0008361 regulation of cell size GO:0009100 glycoprotein metabolic process GO:0009101 glycoprotein biosynthetic process GO:0009150 purine ribonucleotide metabolic process GO:0009152 purine ribonucleotide biosynthetic process GO:0009165 nucleotide biosynthetic process GO:0009187 cyclic nucleotide metabolic process GO:0009190 cyclic nucleotide biosynthetic process GO:0009260 ribonucleotide biosynthetic process GO:0009743 response to carbohydrate GO:0009991 response to extracellular stimulus GO:0010035 response to inorganic substance GO:0010559 regulation of glycoprotein biosynthetic process GO:0010720 positive regulation of cell development GO:0010769 regulation of cell morphogenesis involved in differentiation GO:0010770 positive regulation of cell morphogenesis involved in differentiation GO:0010835 regulation of protein ADP-ribosylation GO:0010975 regulation of neuron projection development GO:0010976 positive regulation of neuron projection development GO:0016049 cell growth GO:0018108 peptidyl-tyrosine phosphorylation GO:0018212 peptidyl-tyrosine modification GO:0022604 regulation of cell morphogenesis GO:0030307 positive regulation of cell growth GO:0030516 regulation of axon extension GO:0030799 regulation of cyclic nucleotide metabolic process GO:0030801 positive regulation of cyclic nucleotide metabolic process GO:0030802 regulation of cyclic nucleotide biosynthetic process GO:0030804 positive regulation of cyclic nucleotide biosynthetic process GO:0030808 regulation of nucleotide biosynthetic process GO:0030810 positive regulation of nucleotide biosynthetic process GO:0030823 regulation of cGMP metabolic process GO:0030825 positive regulation of cGMP metabolic process GO:0030826 regulation of cGMP biosynthetic process GO:0030828 positive regulation of cGMP biosynthetic process GO:0031346 positive regulation of cell projection organization GO:0031668 cellular response to extracellular stimulus GO:0032091 negative regulation of protein binding GO:0032147 activation of protein kinase activity GO:0032535 regulation of cellular component size GO:0033483 gas homeostasis GO:0033484 nitric oxide homeostasis GO:0033674 positive regulation of kinase activity GO:0042698 ovulation cycle GO:0043393 regulation of protein binding GO:0043413 macromolecule glycosylation GO:0043523 regulation of neuron apoptotic process GO:0043524 negative regulation of neuron apoptotic process GO:0044089 positive regulation of cellular component biogenesis GO:0044708 single-organism behavior GO:0044849 estrous cycle GO:0045666 positive regulation of neuron differentiation GO:0045773 positive regulation of axon extension GO:0045860 positive regulation of protein kinase activity GO:0045927 positive regulation of growth GO:0045981 positive regulation of nucleotide metabolic process GO:0046068 cGMP metabolic process GO:0046390 ribose phosphate biosynthetic process GO:0048511 rhythmic process GO:0048588 developmental cell growth GO:0048638 regulation of developmental growth GO:0048639 positive regulation of developmental growth GO:0048667 cell morphogenesis involved in neuron differentiation GO:0048675 axon extension GO:0050730 regulation of peptidyl-tyrosine phosphorylation GO:0050731 positive regulation of peptidyl-tyrosine phosphorylation GO:0050769 positive regulation of neurogenesis GO:0050770 regulation of axonogenesis GO:0050772 positive regulation of axonogenesis GO:0050803 regulation of synapse structure or activity GO:0050804 modulation of synaptic transmission GO:0050805 negative regulation of synaptic transmission GO:0050807 regulation of synapse organization GO:0050808 synapse organization GO:0050890 cognition GO:0051098 regulation of binding GO:0051100 negative regulation of binding GO:0051402 neuron apoptotic process GO:0051962 positive regulation of nervous system development GO:0051963 regulation of synapse assembly GO:0051965 positive regulation of synapse assembly GO:0052652 cyclic purine nucleotide metabolic process GO:0060049 regulation of protein glycosylation GO:0060560 developmental growth involved in morphogenesis GO:0061387 regulation of extent of cell growth GO:0061564 axon development GO:0070085 glycosylation GO:0070997 neuron death GO:0071496 cellular response to external stimulus GO:0072522 purine-containing compound biosynthetic process GO:0090066 regulation of anatomical structure size GO:1900371 regulation of purine nucleotide biosynthetic process GO:1900373 positive regulation of purine nucleotide biosynthetic process GO:1900542 regulation of purine nucleotide metabolic process GO:1900544 positive regulation of purine nucleotide metabolic process GO:1901214 regulation of neuron death GO:1901215 negative regulation of neuron death GO:1901293 nucleoside phosphate biosynthetic process GO:1903018 regulation of glycoprotein metabolic process GO:1990138 neuron projection extension |
| Molecular Function |
GO:0003682 chromatin binding GO:0005507 copper ion binding GO:0015631 tubulin binding GO:0033218 amide binding GO:0042277 peptide binding GO:0048487 beta-tubulin binding |
| Cellular Component |
GO:0030424 axon GO:0030425 dendrite GO:0043025 neuronal cell body GO:0044297 cell body |
| KEGG | - |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for ADNP. |
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
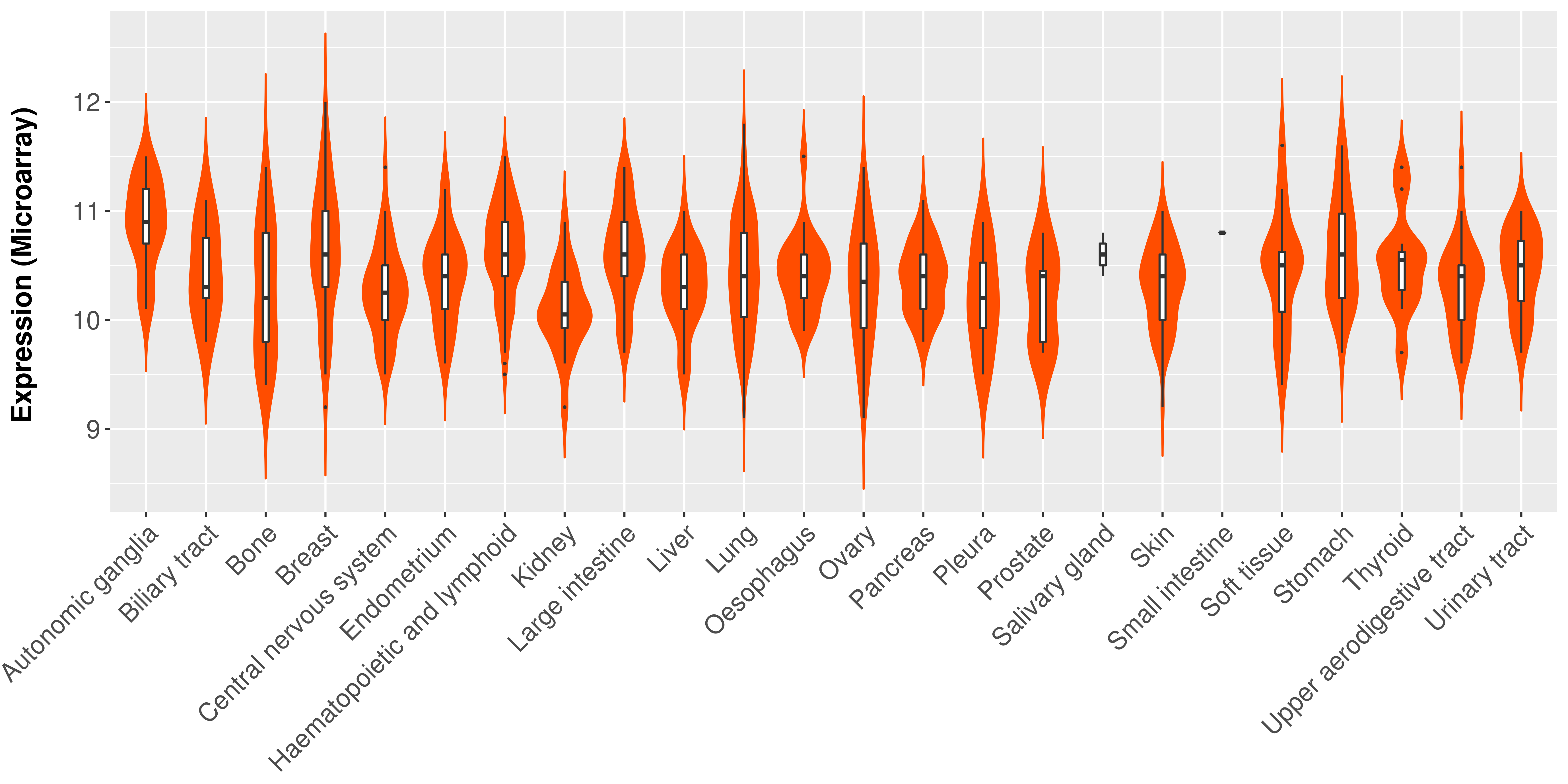
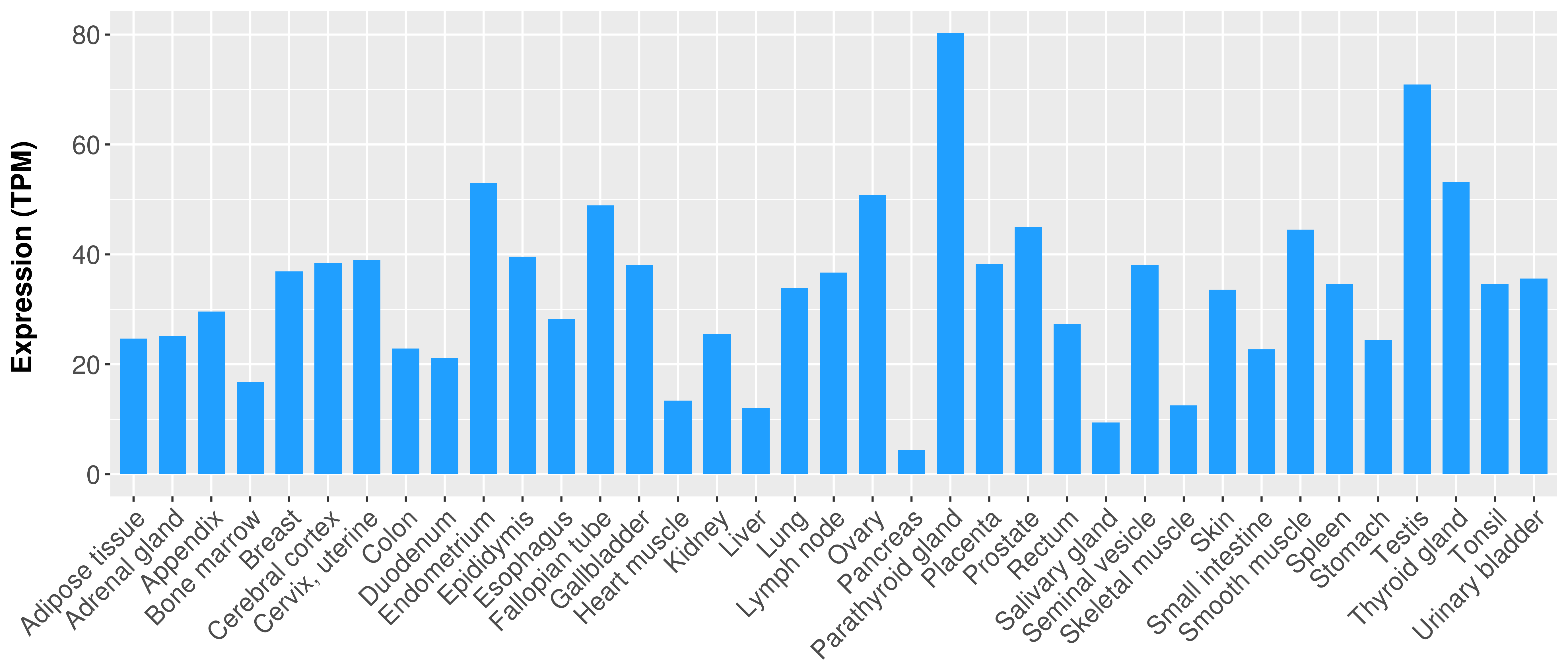
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
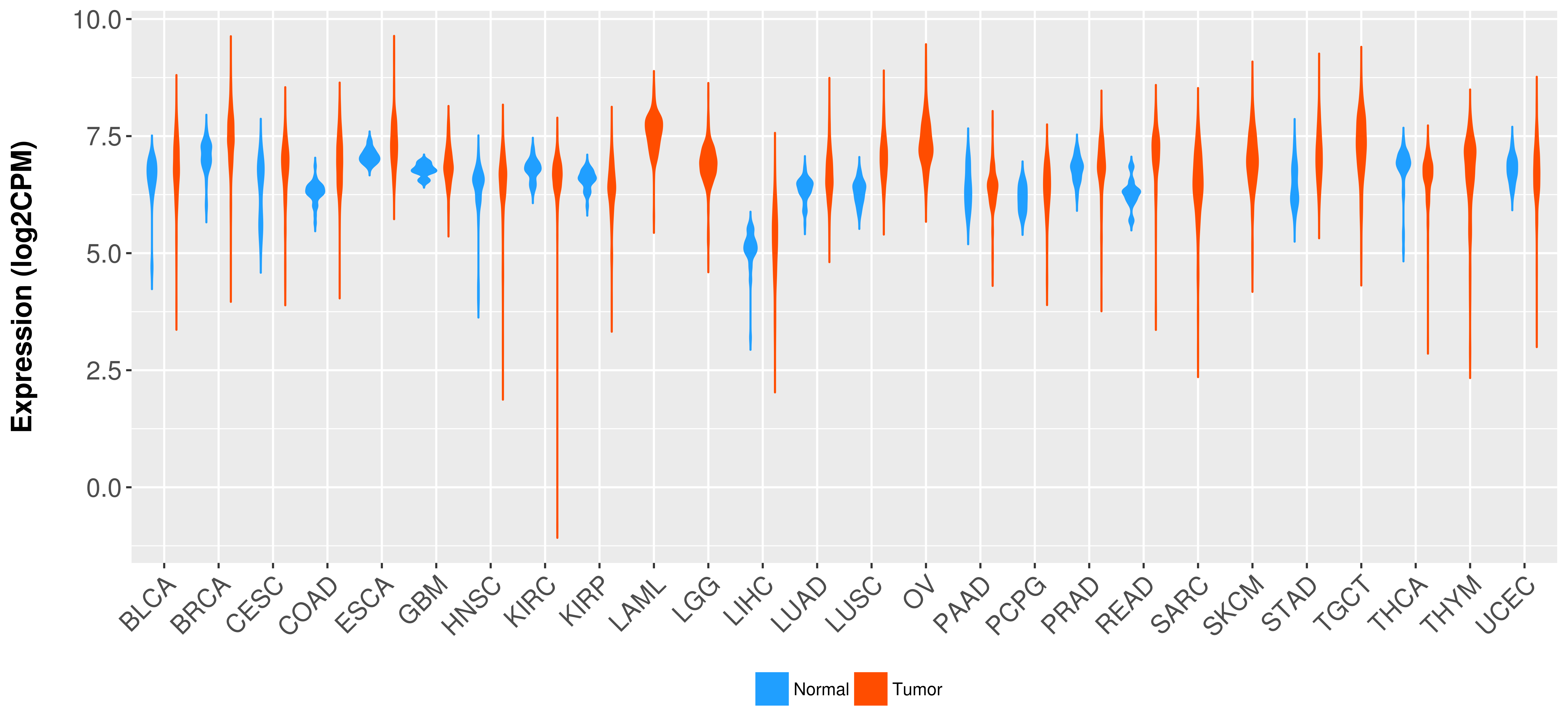
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
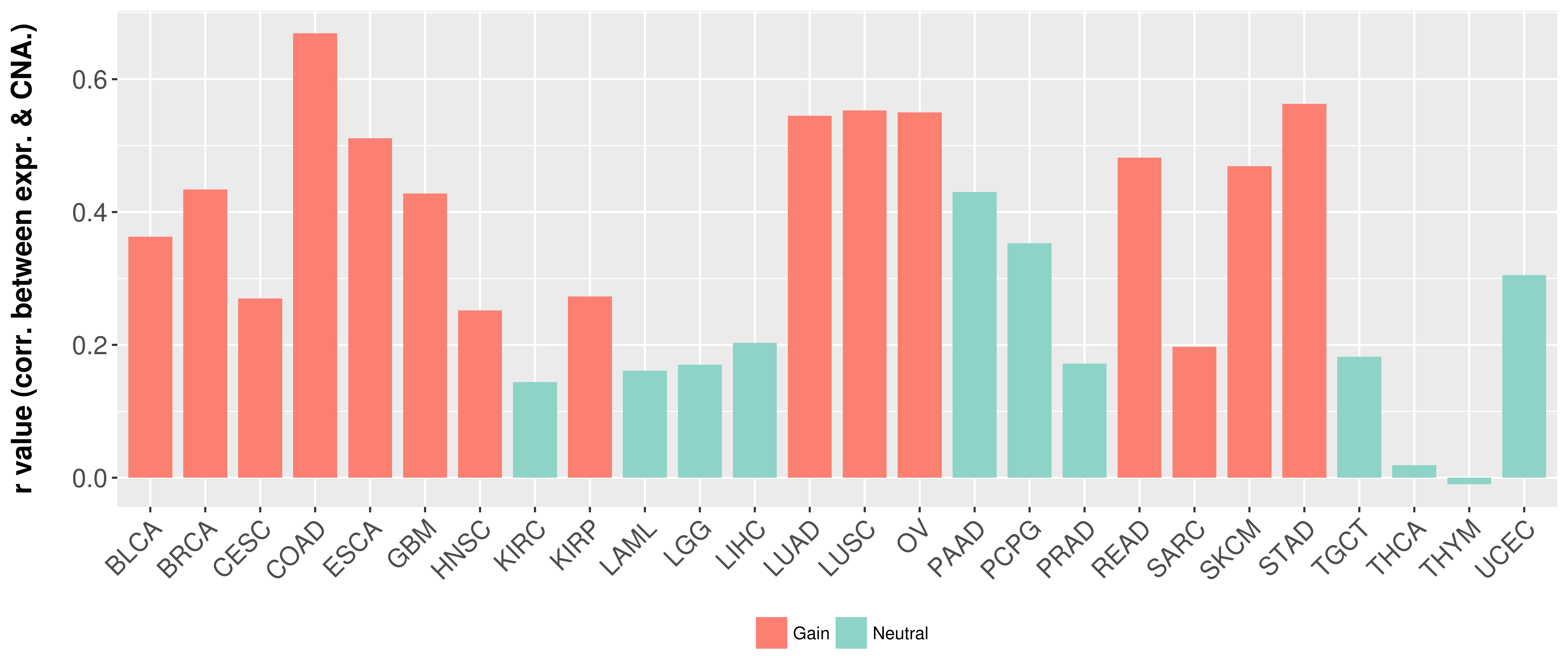
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
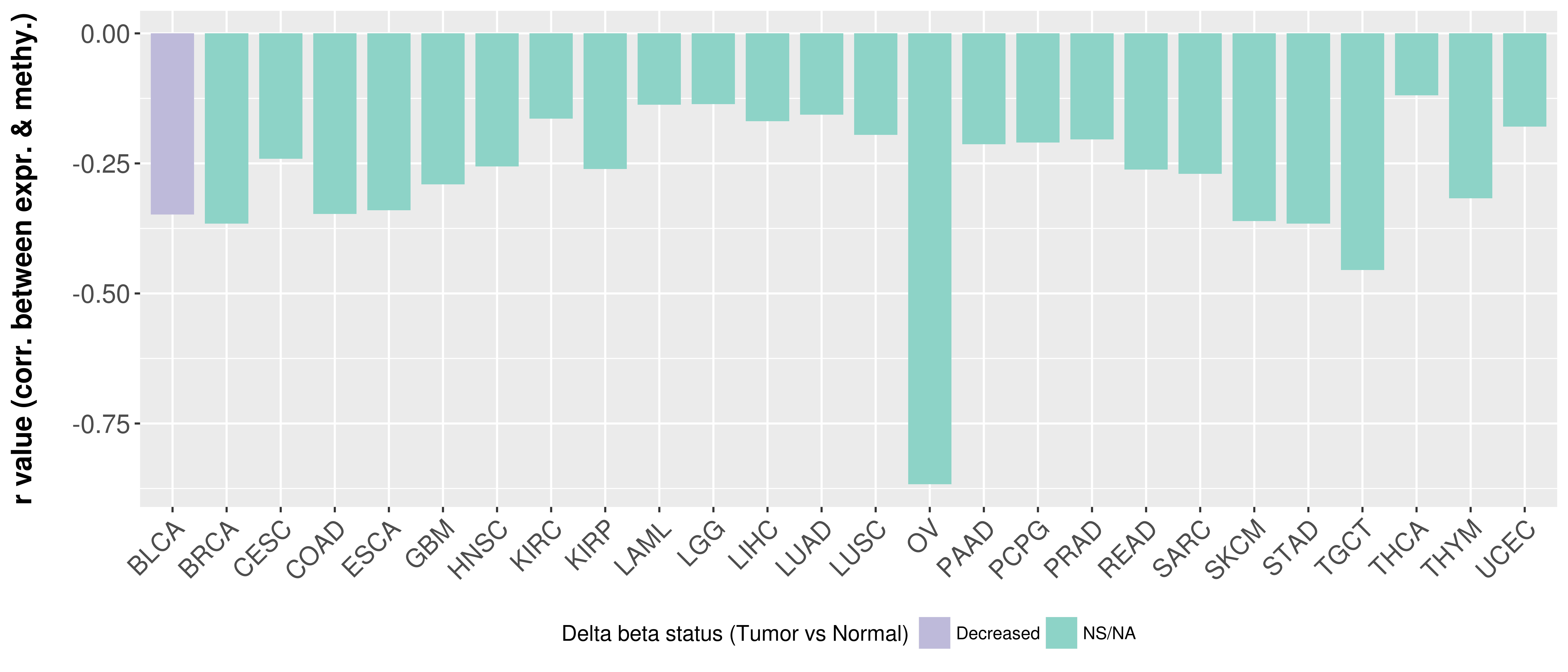
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
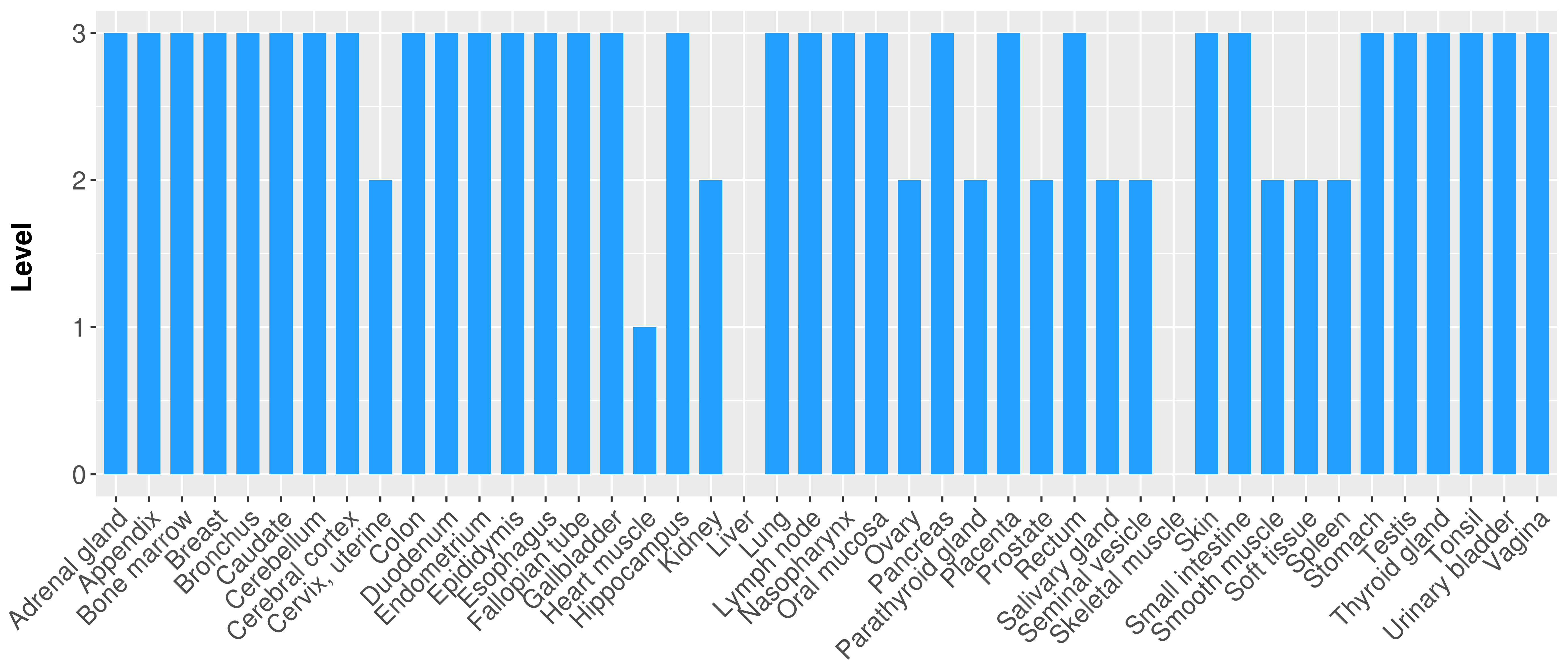
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
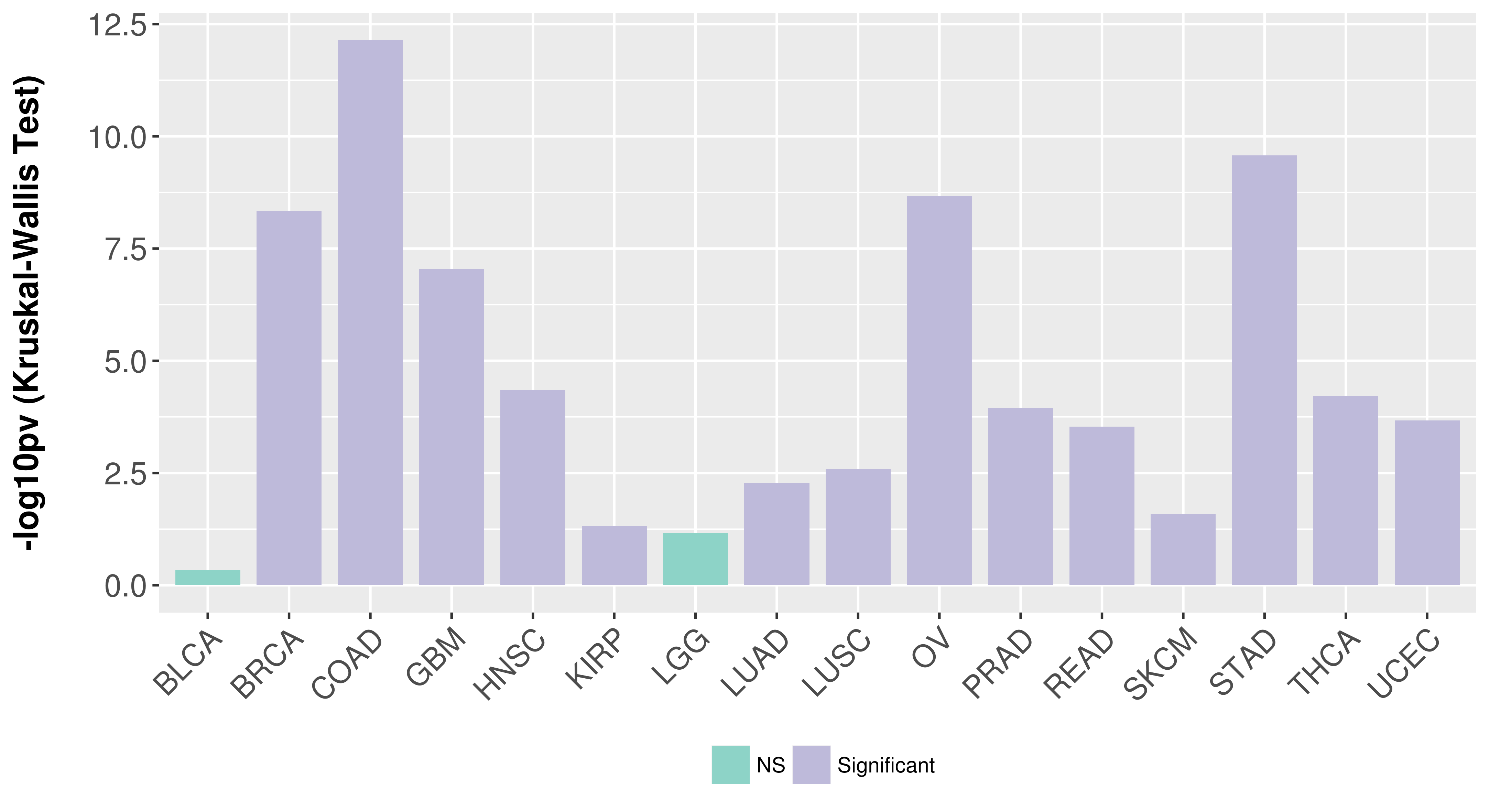
Association between expresson and subtype.
|
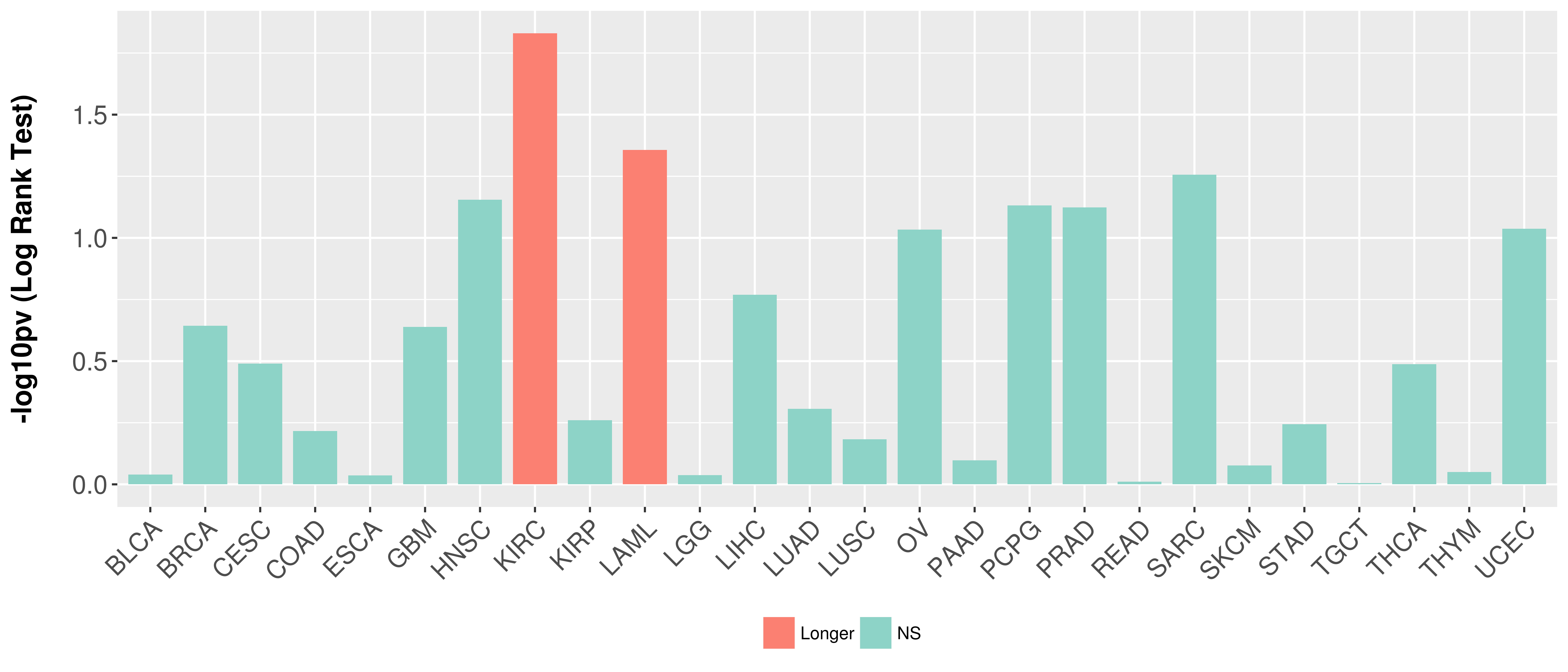
Overall survival analysis based on expression.
|
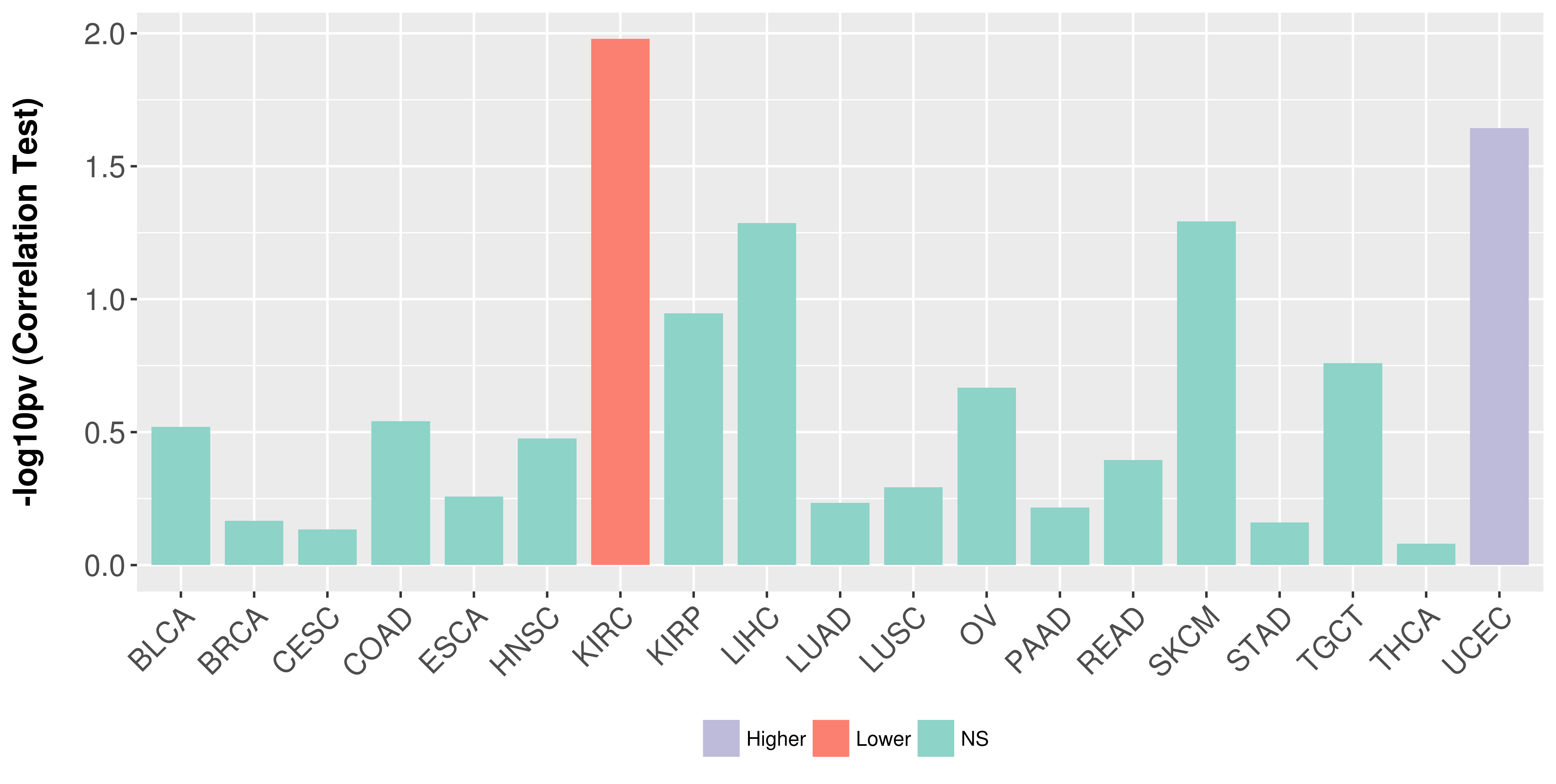
Association between expresson and stage.
|
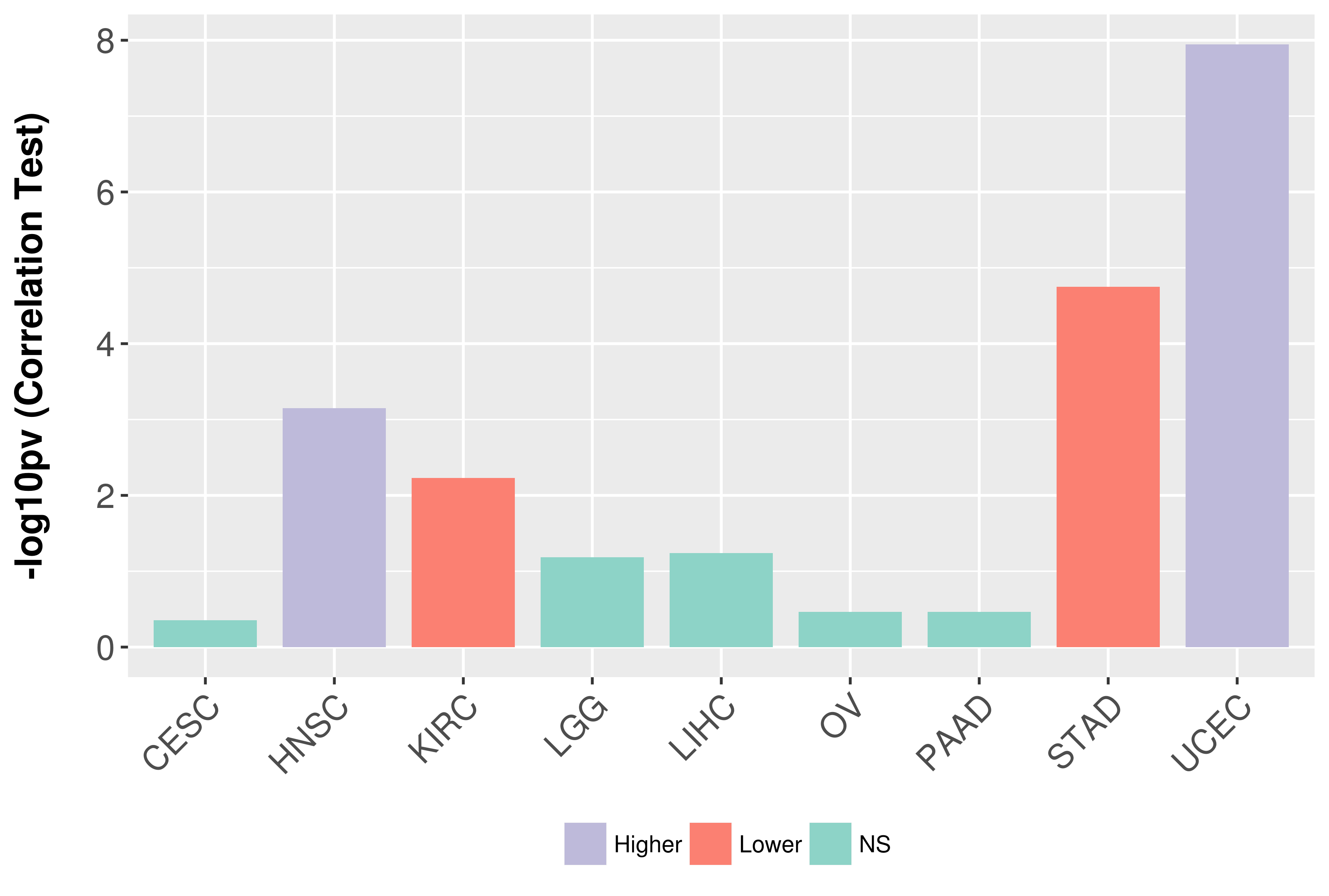
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for ADNP. |
| Summary | |
|---|---|
| Symbol | ADNP |
| Name | activity dependent neuroprotector homeobox |
| Aliases | KIAA0784; ADNP1; ADNP homeobox 1; activity-dependent neuroprotector; HVDAS; MRD28; activity-dependent neurop ...... |
| Location | 20q13.13 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|