Browse AICDA in pancancer
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF08210 APOBEC-like N-terminal domain |
||||||||||
| Function |
Single-stranded DNA-specific cytidine deaminase. Involved in somatic hypermutation (SHM), gene conversion, and class-switch recombination (CSR) in B-lymphocytes by deaminating C to U during transcription of Ig-variable (V) and Ig-switch (S) region DNA. Required for several crucial steps of B-cell terminal differentiation necessary for efficient antibody responses (PubMed:18722174, PubMed:21385873, PubMed:21518874, PubMed:27716525). May also play a role in the epigenetic regulation of gene expression by participating in DNA demethylation (PubMed:21496894). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0002200 somatic diversification of immune receptors GO:0002204 somatic recombination of immunoglobulin genes involved in immune response GO:0002208 somatic diversification of immunoglobulins involved in immune response GO:0002237 response to molecule of bacterial origin GO:0002250 adaptive immune response GO:0002263 cell activation involved in immune response GO:0002285 lymphocyte activation involved in immune response GO:0002312 B cell activation involved in immune response GO:0002366 leukocyte activation involved in immune response GO:0002377 immunoglobulin production GO:0002381 immunoglobulin production involved in immunoglobulin mediated immune response GO:0002440 production of molecular mediator of immune response GO:0002443 leukocyte mediated immunity GO:0002449 lymphocyte mediated immunity GO:0002460 adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains GO:0002521 leukocyte differentiation GO:0002562 somatic diversification of immune receptors via germline recombination within a single locus GO:0002566 somatic diversification of immune receptors via somatic mutation GO:0006213 pyrimidine nucleoside metabolic process GO:0006216 cytidine catabolic process GO:0006304 DNA modification GO:0006310 DNA recombination GO:0006342 chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006397 mRNA processing GO:0009116 nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009164 nucleoside catabolic process GO:0009972 cytidine deamination GO:0016064 immunoglobulin mediated immune response GO:0016444 somatic cell DNA recombination GO:0016445 somatic diversification of immunoglobulins GO:0016446 somatic hypermutation of immunoglobulin genes GO:0016447 somatic recombination of immunoglobulin gene segments GO:0016458 gene silencing GO:0019439 aromatic compound catabolic process GO:0019724 B cell mediated immunity GO:0030098 lymphocyte differentiation GO:0030183 B cell differentiation GO:0031935 regulation of chromatin silencing GO:0031936 negative regulation of chromatin silencing GO:0032496 response to lipopolysaccharide GO:0034655 nucleobase-containing compound catabolic process GO:0035510 DNA dealkylation GO:0040029 regulation of gene expression, epigenetic GO:0042113 B cell activation GO:0042454 ribonucleoside catabolic process GO:0044270 cellular nitrogen compound catabolic process GO:0044728 DNA methylation or demethylation GO:0045190 isotype switching GO:0045814 negative regulation of gene expression, epigenetic GO:0045815 positive regulation of gene expression, epigenetic GO:0046087 cytidine metabolic process GO:0046131 pyrimidine ribonucleoside metabolic process GO:0046133 pyrimidine ribonucleoside catabolic process GO:0046135 pyrimidine nucleoside catabolic process GO:0046700 heterocycle catabolic process GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0060968 regulation of gene silencing GO:0060969 negative regulation of gene silencing GO:0070988 demethylation GO:0071216 cellular response to biotic stimulus GO:0071219 cellular response to molecule of bacterial origin GO:0071222 cellular response to lipopolysaccharide GO:0071396 cellular response to lipid GO:0072527 pyrimidine-containing compound metabolic process GO:0072529 pyrimidine-containing compound catabolic process GO:0080111 DNA demethylation GO:0090308 regulation of methylation-dependent chromatin silencing GO:0090310 negative regulation of methylation-dependent chromatin silencing GO:1901136 carbohydrate derivative catabolic process GO:1901361 organic cyclic compound catabolic process GO:1901565 organonitrogen compound catabolic process GO:1901657 glycosyl compound metabolic process GO:1901658 glycosyl compound catabolic process GO:1902275 regulation of chromatin organization GO:1905268 negative regulation of chromatin organization |
| Molecular Function |
GO:0004126 cytidine deaminase activity GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016814 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines GO:0019239 deaminase activity GO:0031625 ubiquitin protein ligase binding GO:0044389 ubiquitin-like protein ligase binding |
| Cellular Component |
GO:0000178 exosome (RNase complex) GO:1905354 exoribonuclease complex |
| KEGG |
hsa04672 Intestinal immune network for IgA production |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for AICDA. |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
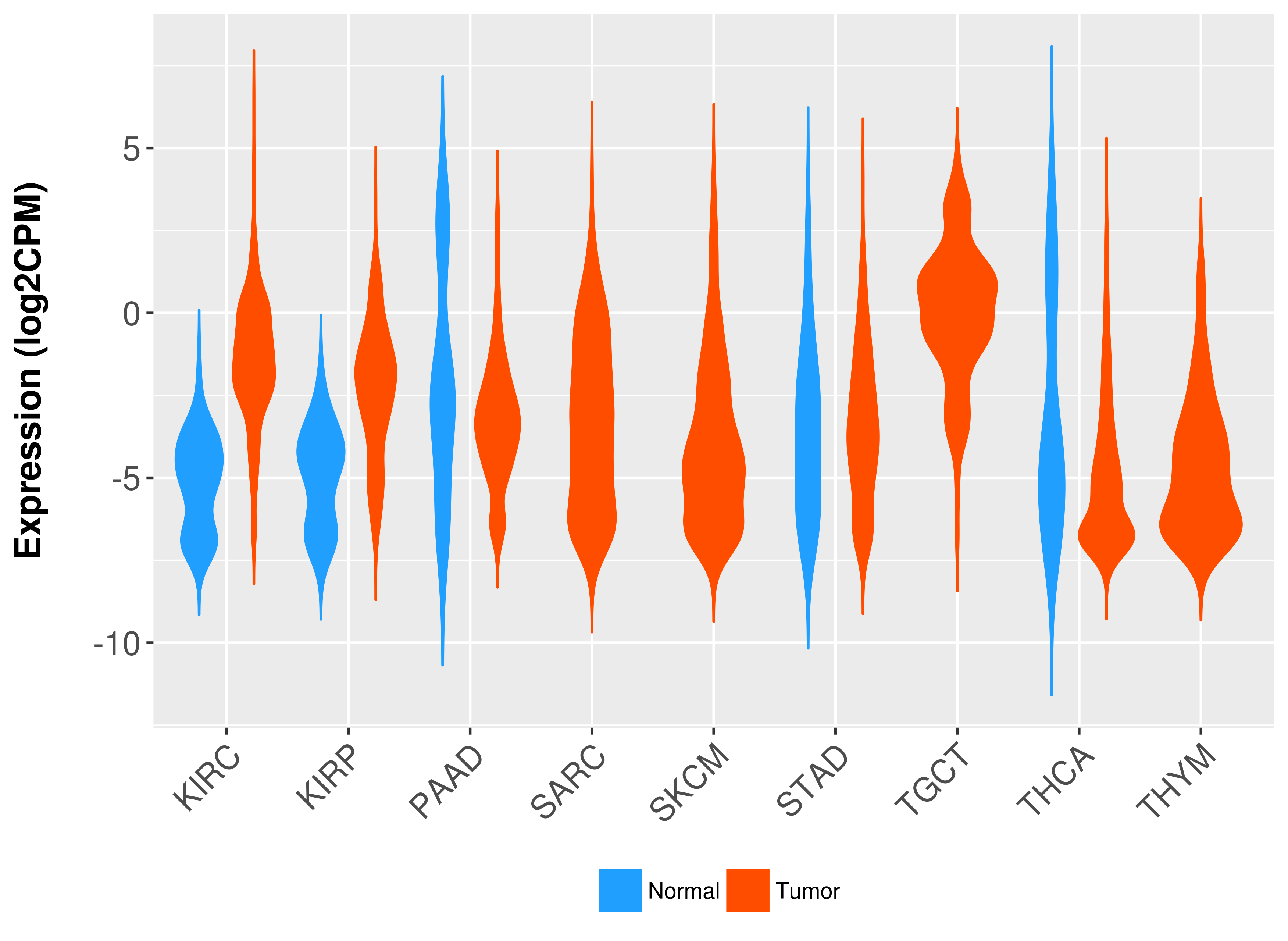
Differential expression analysis for cancers with more than 10 normal samples
|
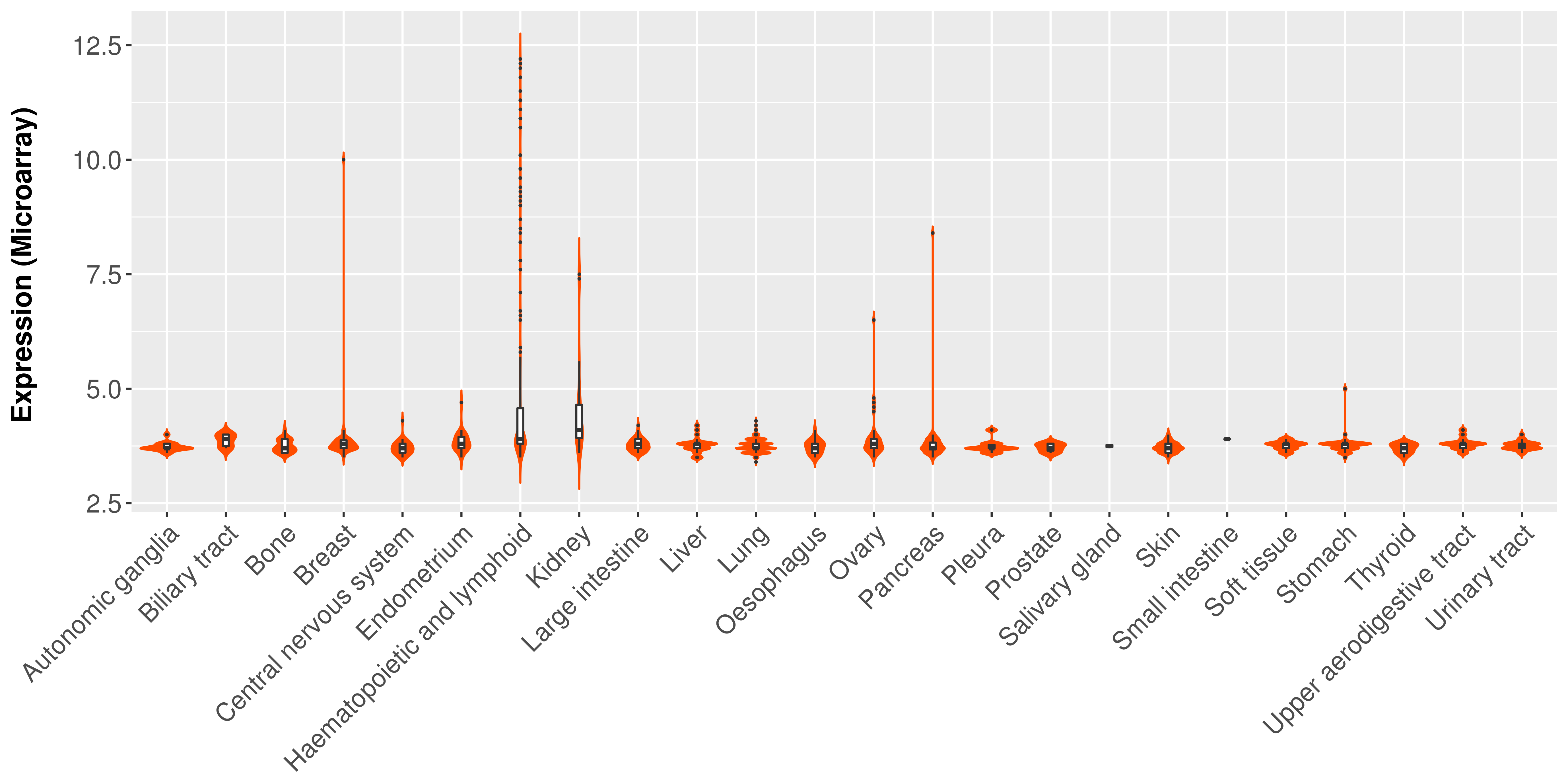
|
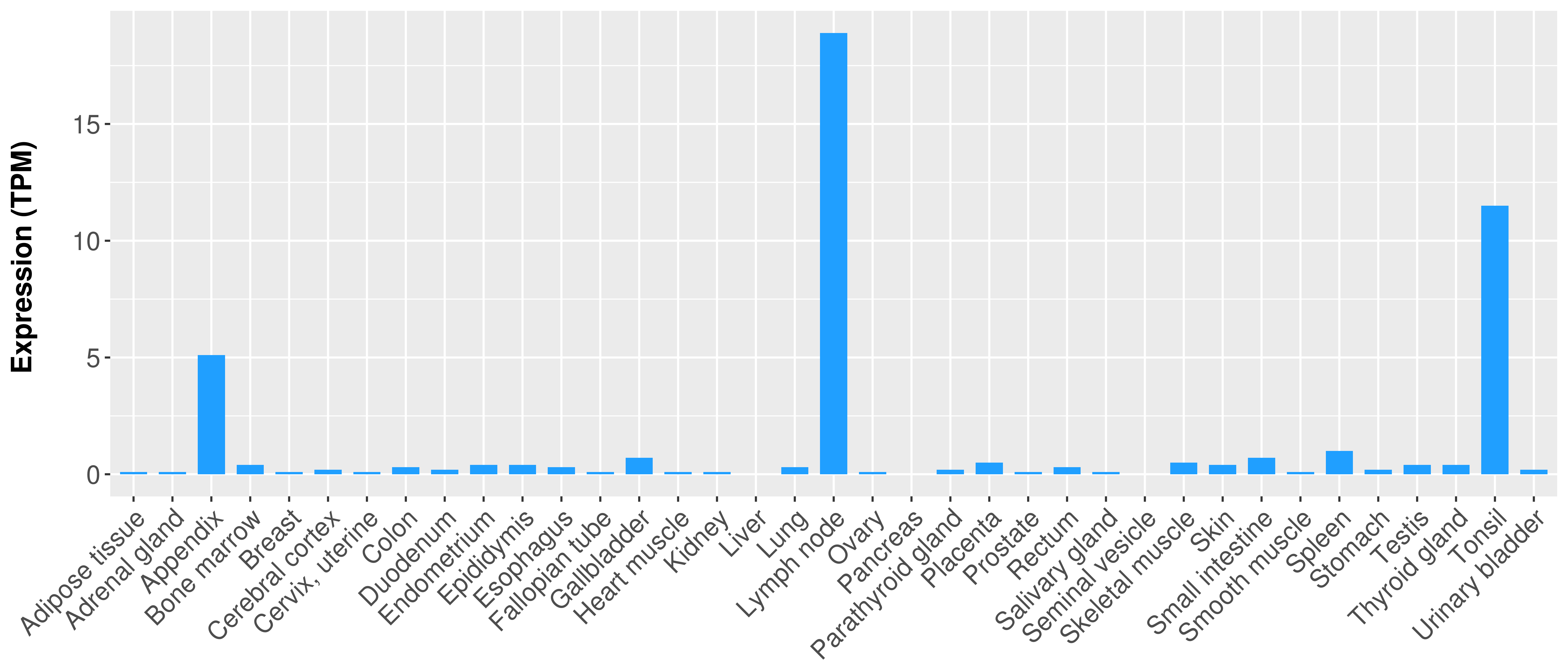
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
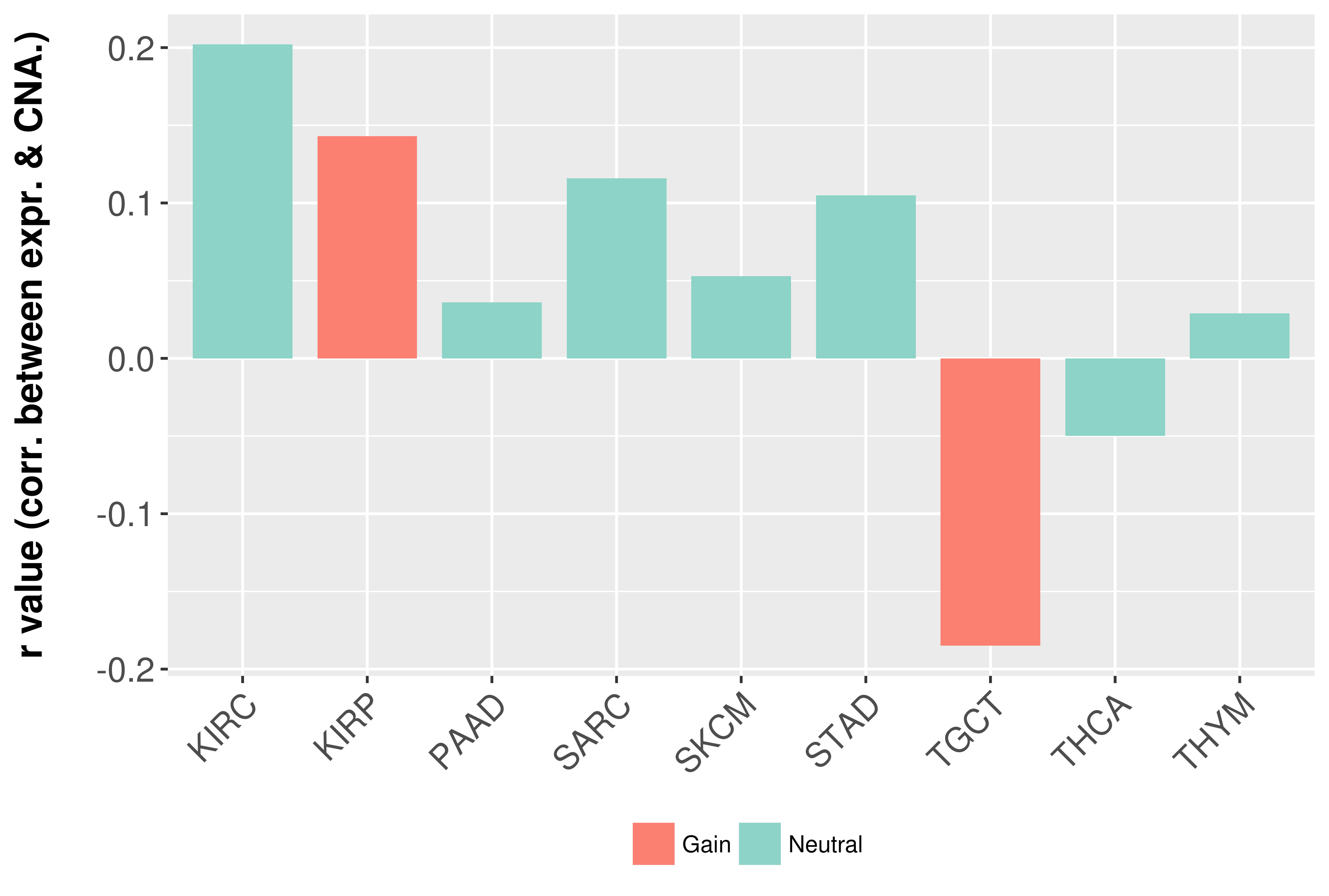
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
| There is no record. |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
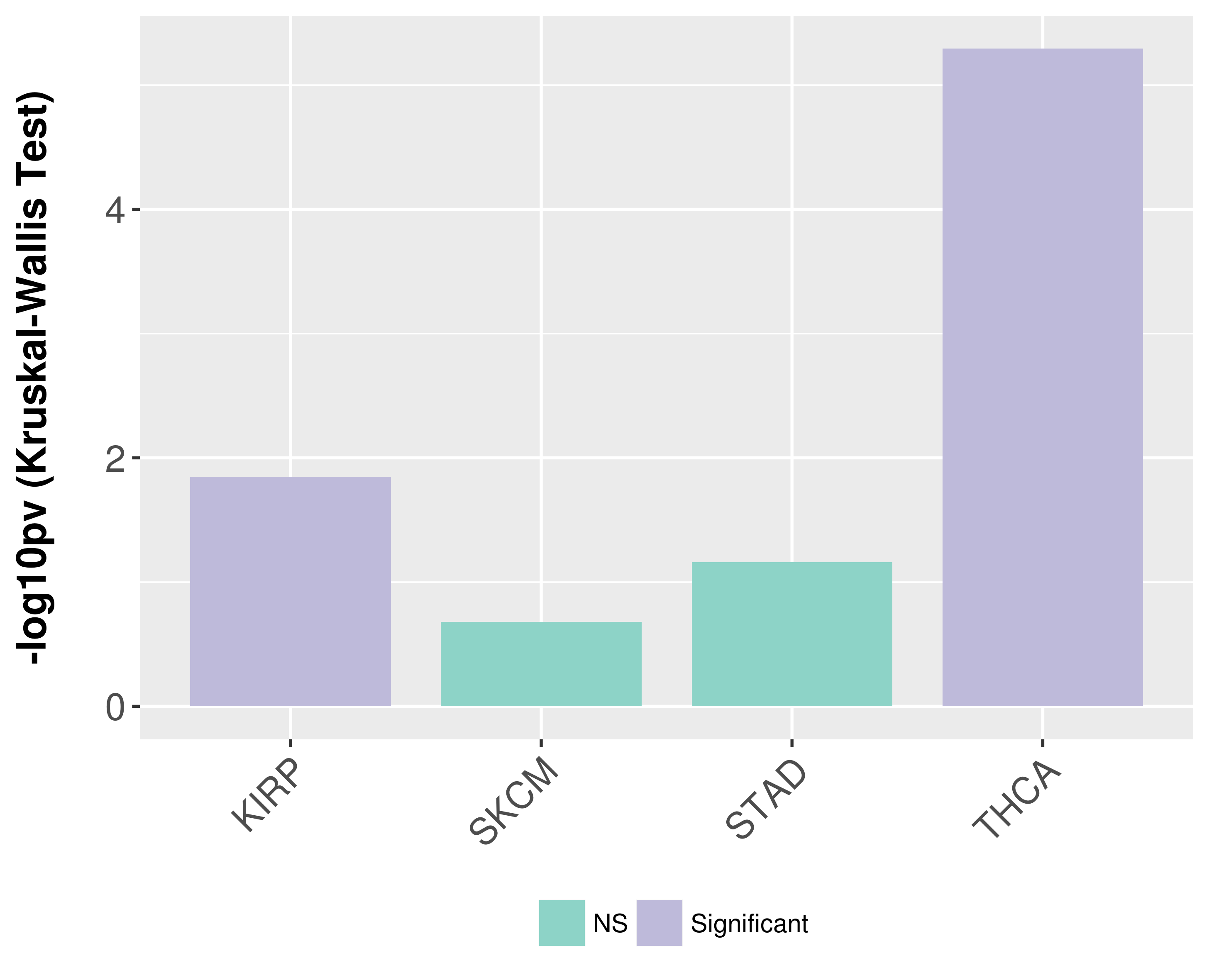
Association between expresson and subtype.
|

Overall survival analysis based on expression.
|
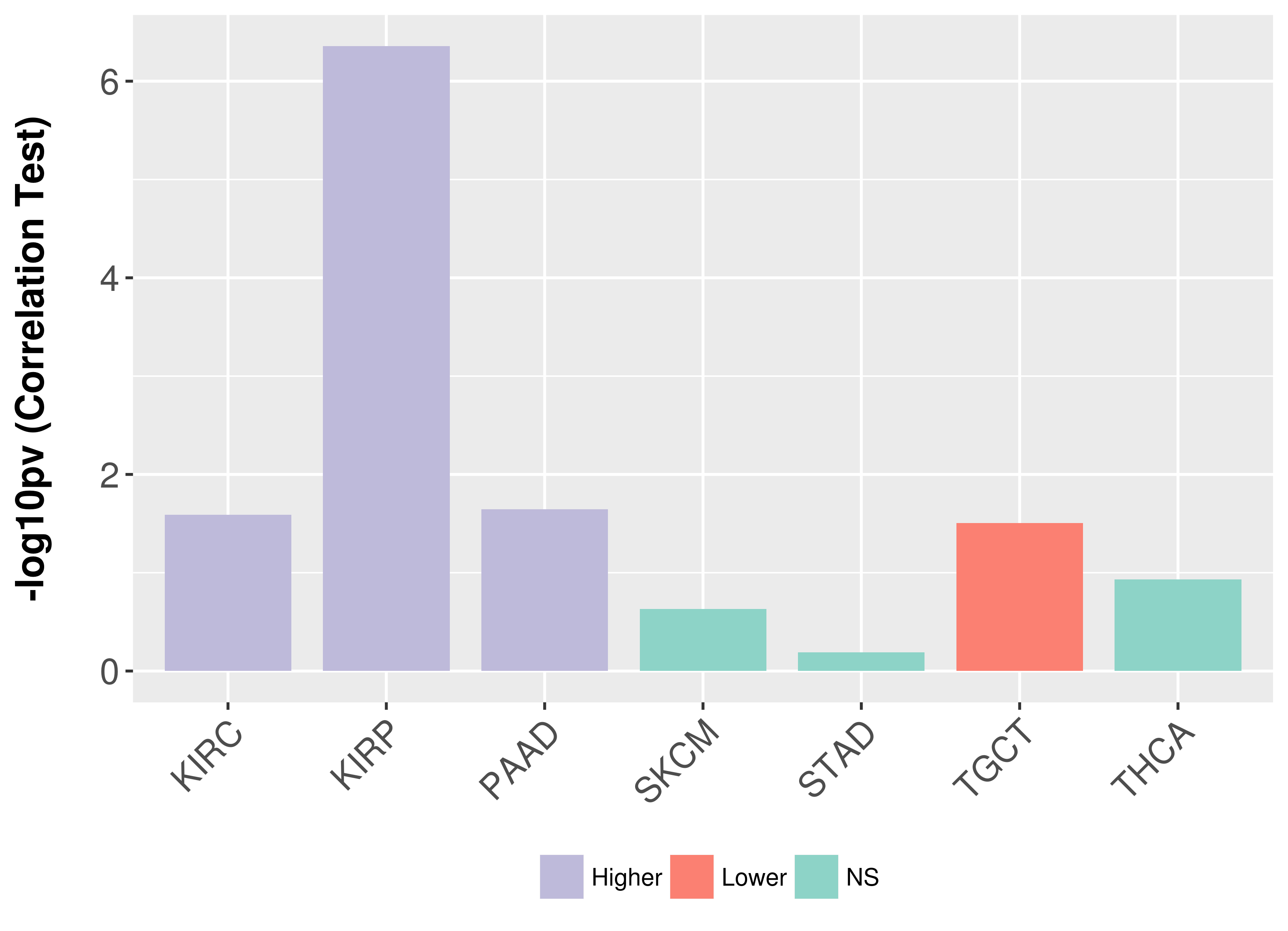
Association between expresson and stage.
|
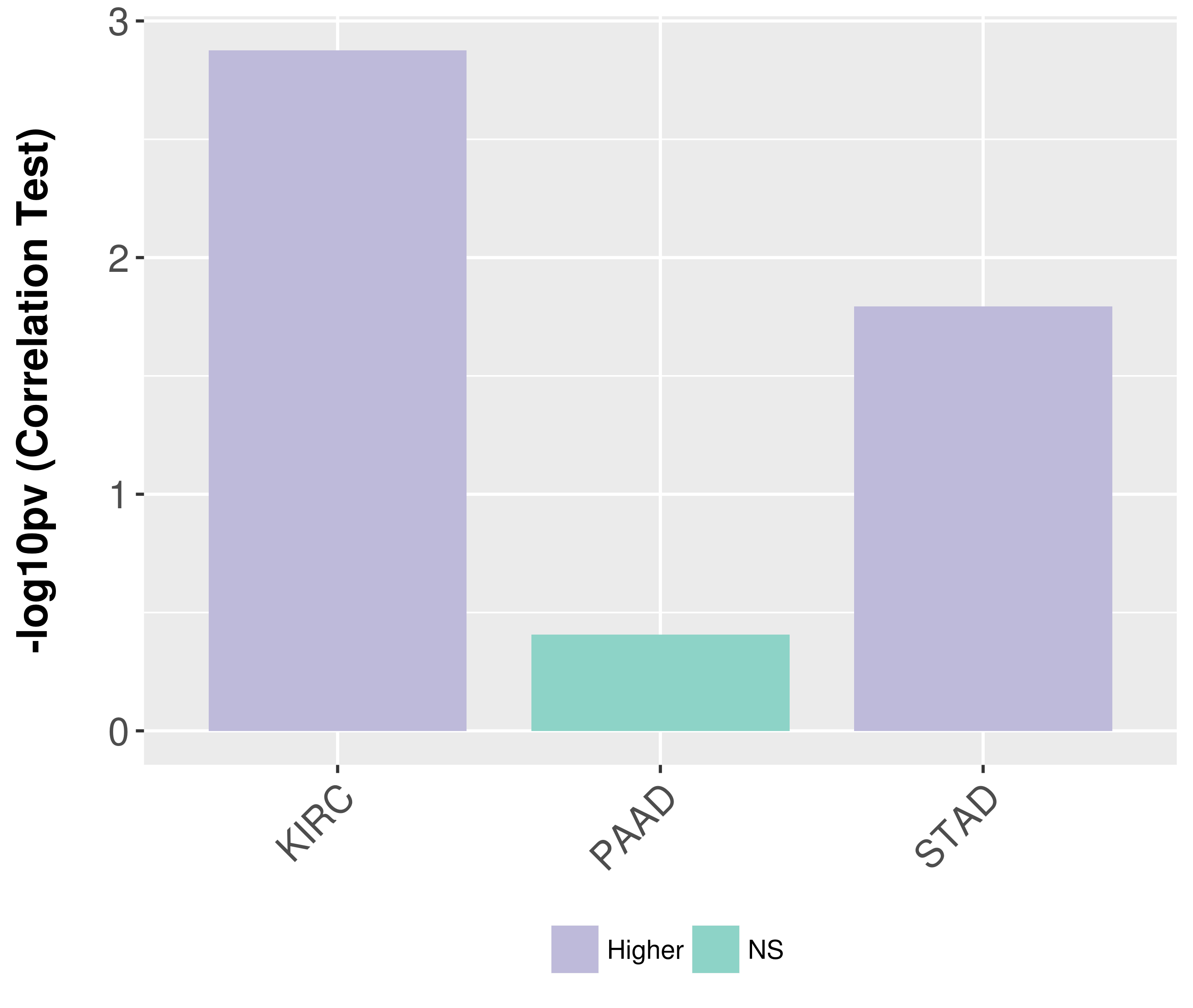
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for AICDA. |
| Summary | |
|---|---|
| Symbol | AICDA |
| Name | activation induced cytidine deaminase |
| Aliases | HIGM2; CDA2; AID; epididymis secretory protein Li 284; integrated into Burkitt's lymphoma cell line Ramos; S ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for AICDA. |