Browse APOBEC1 in pancancer
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF08210 APOBEC-like N-terminal domain |
||||||||||
| Function |
Catalytic component of the apolipoprotein B mRNA editing enzyme complex which is responsible for the postranscriptional editing of a CAA codon for Gln to a UAA codon for stop in the APOB mRNA. Also involved in CGA (Arg) to UGA (Stop) editing in the NF1 mRNA. May also play a role in the epigenetic regulation of gene expression by participating in DNA demethylation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0006213 pyrimidine nucleoside metabolic process GO:0006216 cytidine catabolic process GO:0006304 DNA modification GO:0006342 chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006397 mRNA processing GO:0006970 response to osmotic stress GO:0009116 nucleoside metabolic process GO:0009119 ribonucleoside metabolic process GO:0009164 nucleoside catabolic process GO:0009314 response to radiation GO:0009451 RNA modification GO:0009615 response to virus GO:0009972 cytidine deamination GO:0010035 response to inorganic substance GO:0010038 response to metal ion GO:0010043 response to zinc ion GO:0010212 response to ionizing radiation GO:0010332 response to gamma radiation GO:0010608 posttranscriptional regulation of gene expression GO:0016458 gene silencing GO:0016553 base conversion or substitution editing GO:0016554 cytidine to uridine editing GO:0016556 mRNA modification GO:0019439 aromatic compound catabolic process GO:0031935 regulation of chromatin silencing GO:0031936 negative regulation of chromatin silencing GO:0032868 response to insulin GO:0032869 cellular response to insulin stimulus GO:0034655 nucleobase-containing compound catabolic process GO:0035510 DNA dealkylation GO:0040029 regulation of gene expression, epigenetic GO:0042157 lipoprotein metabolic process GO:0042158 lipoprotein biosynthetic process GO:0042454 ribonucleoside catabolic process GO:0042493 response to drug GO:0042953 lipoprotein transport GO:0043434 response to peptide hormone GO:0043487 regulation of RNA stability GO:0043488 regulation of mRNA stability GO:0043489 RNA stabilization GO:0044270 cellular nitrogen compound catabolic process GO:0044728 DNA methylation or demethylation GO:0044872 lipoprotein localization GO:0045006 DNA deamination GO:0045471 response to ethanol GO:0045814 negative regulation of gene expression, epigenetic GO:0045815 positive regulation of gene expression, epigenetic GO:0046087 cytidine metabolic process GO:0046131 pyrimidine ribonucleoside metabolic process GO:0046133 pyrimidine ribonucleoside catabolic process GO:0046135 pyrimidine nucleoside catabolic process GO:0046700 heterocycle catabolic process GO:0048255 mRNA stabilization GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051592 response to calcium ion GO:0051607 defense response to virus GO:0060968 regulation of gene silencing GO:0060969 negative regulation of gene silencing GO:0070383 DNA cytosine deamination GO:0070988 demethylation GO:0071375 cellular response to peptide hormone stimulus GO:0071417 cellular response to organonitrogen compound GO:0072527 pyrimidine-containing compound metabolic process GO:0072529 pyrimidine-containing compound catabolic process GO:0080111 DNA demethylation GO:0090308 regulation of methylation-dependent chromatin silencing GO:0090310 negative regulation of methylation-dependent chromatin silencing GO:0090365 regulation of mRNA modification GO:0090366 positive regulation of mRNA modification GO:0097305 response to alcohol GO:0098542 defense response to other organism GO:1901136 carbohydrate derivative catabolic process GO:1901361 organic cyclic compound catabolic process GO:1901565 organonitrogen compound catabolic process GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1901657 glycosyl compound metabolic process GO:1901658 glycosyl compound catabolic process GO:1902275 regulation of chromatin organization GO:1903311 regulation of mRNA metabolic process GO:1903313 positive regulation of mRNA metabolic process GO:1905268 negative regulation of chromatin organization GO:1990267 response to transition metal nanoparticle |
| Molecular Function |
GO:0003729 mRNA binding GO:0004126 cytidine deaminase activity GO:0004131 cytosine deaminase activity GO:0008047 enzyme activator activity GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016814 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines GO:0017091 AU-rich element binding GO:0019239 deaminase activity GO:0043021 ribonucleoprotein complex binding |
| Cellular Component | - |
| KEGG | - |
| Reactome |
R-HSA-75094: Formation of the Editosome R-HSA-74160: Gene Expression R-HSA-75072: mRNA Editing R-HSA-72200: mRNA Editing |
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
| There is no PTM data |
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
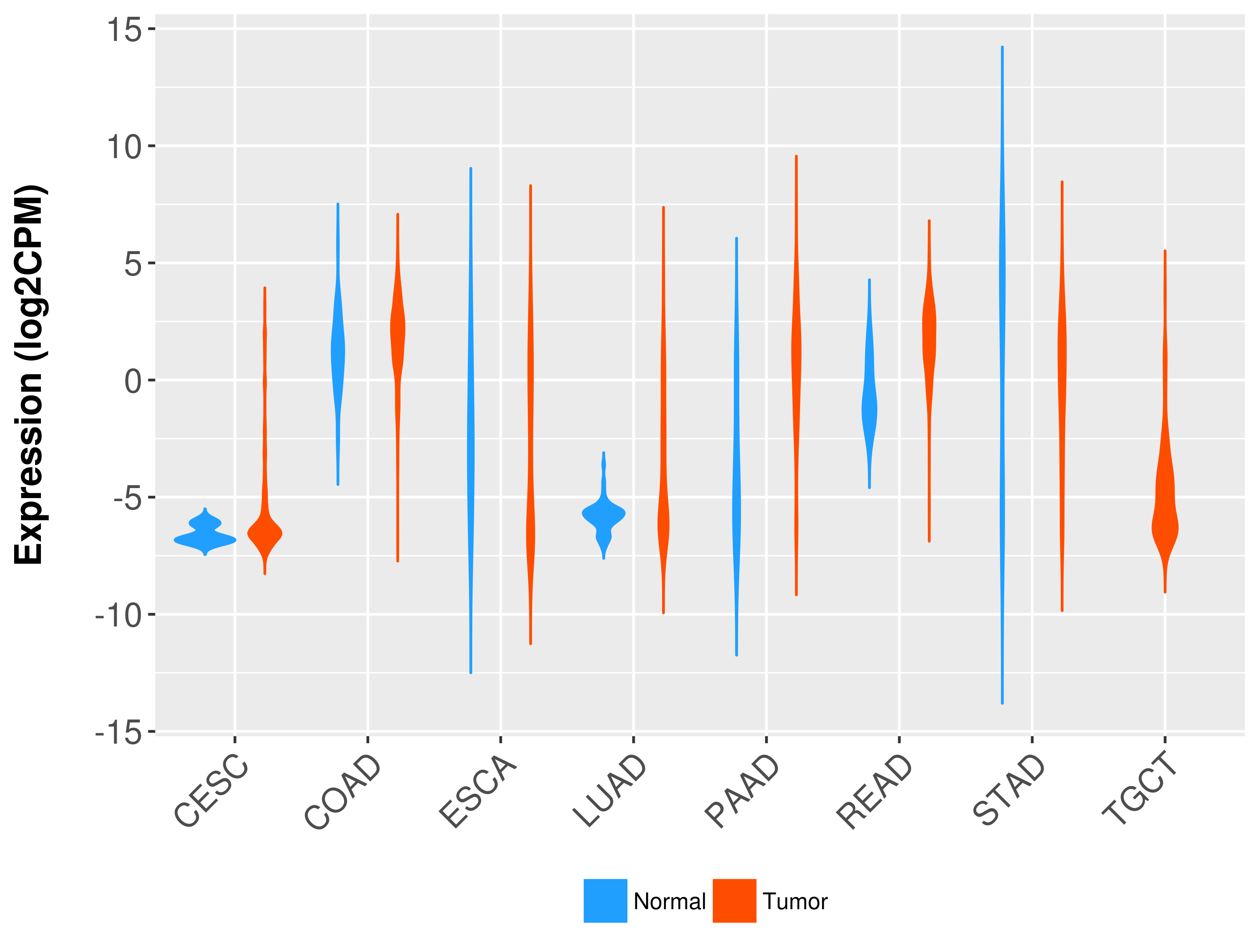
Differential expression analysis for cancers with more than 10 normal samples
|
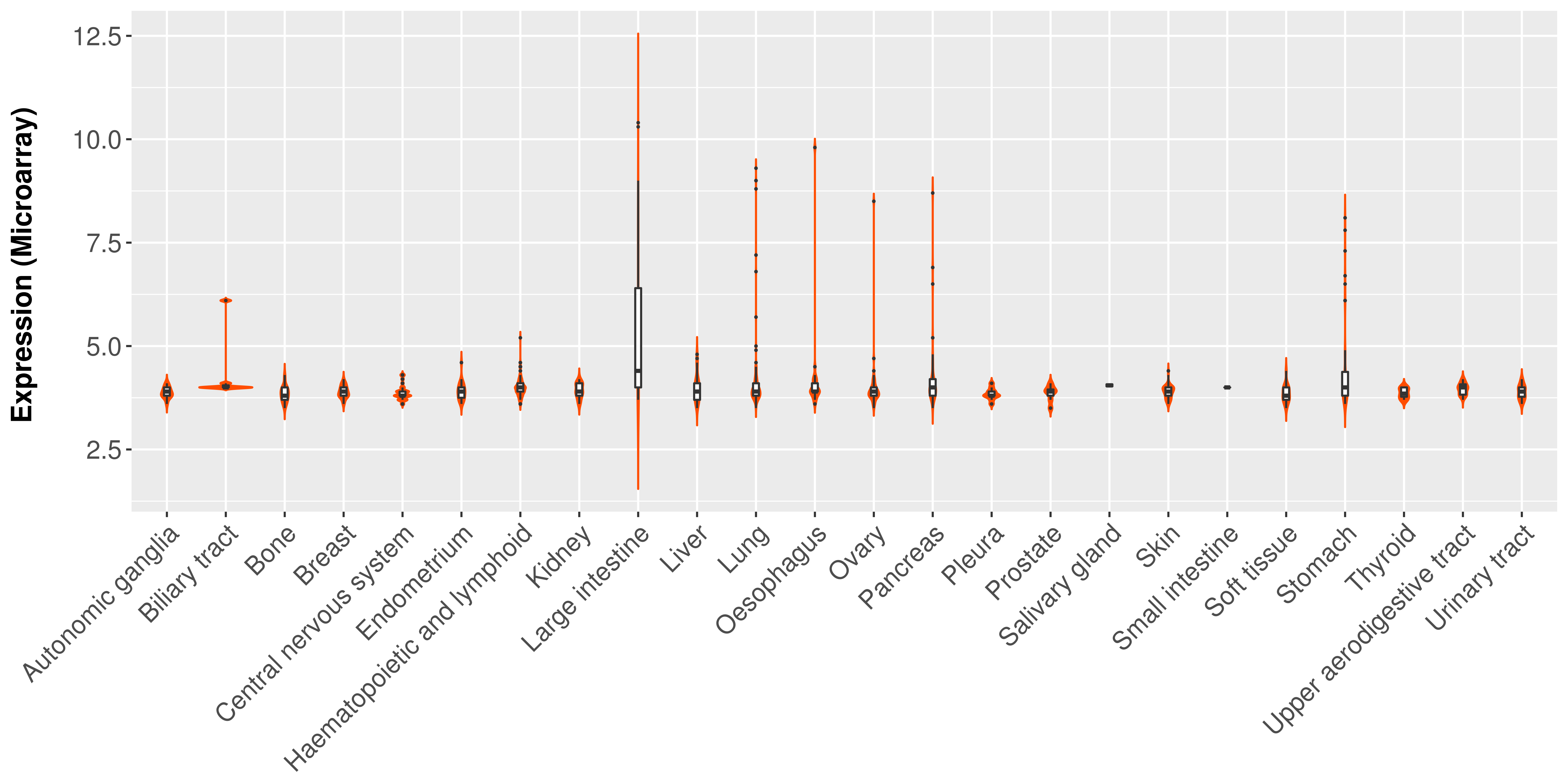
|
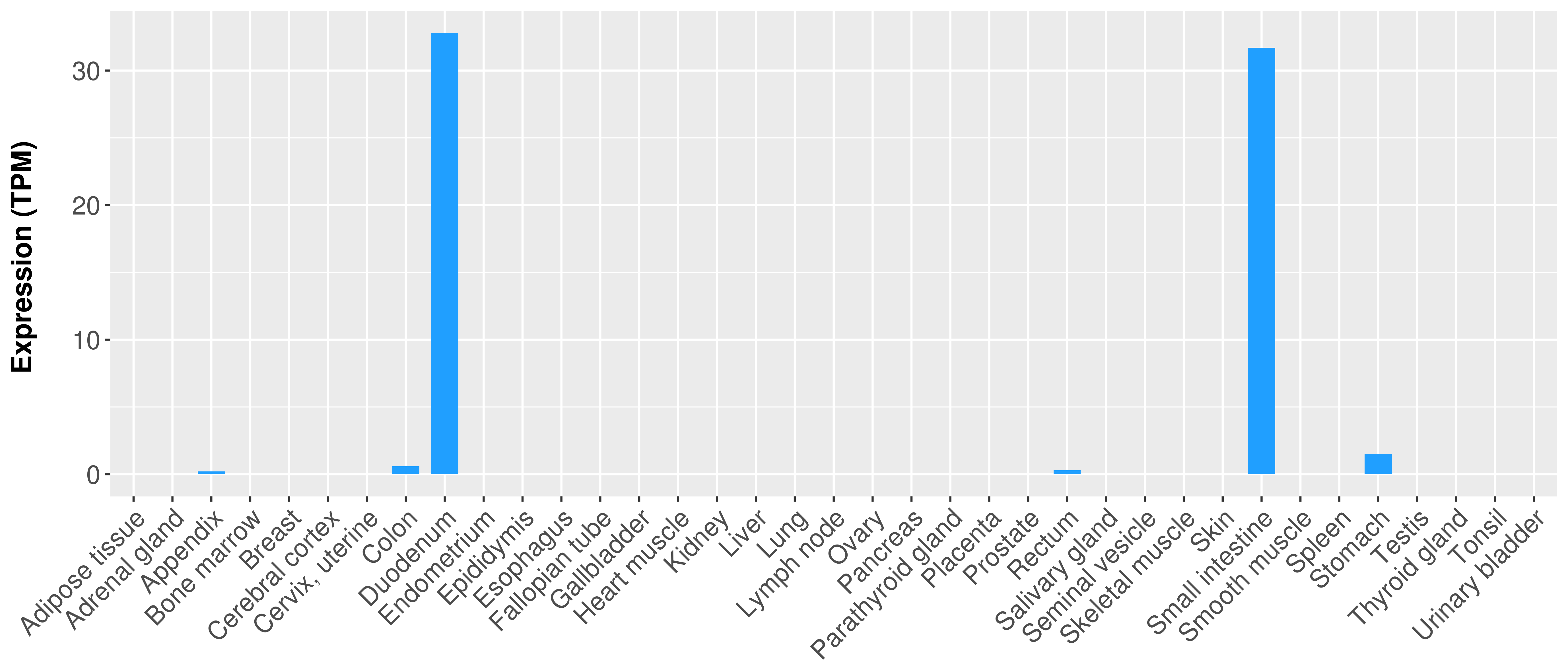
|
|
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
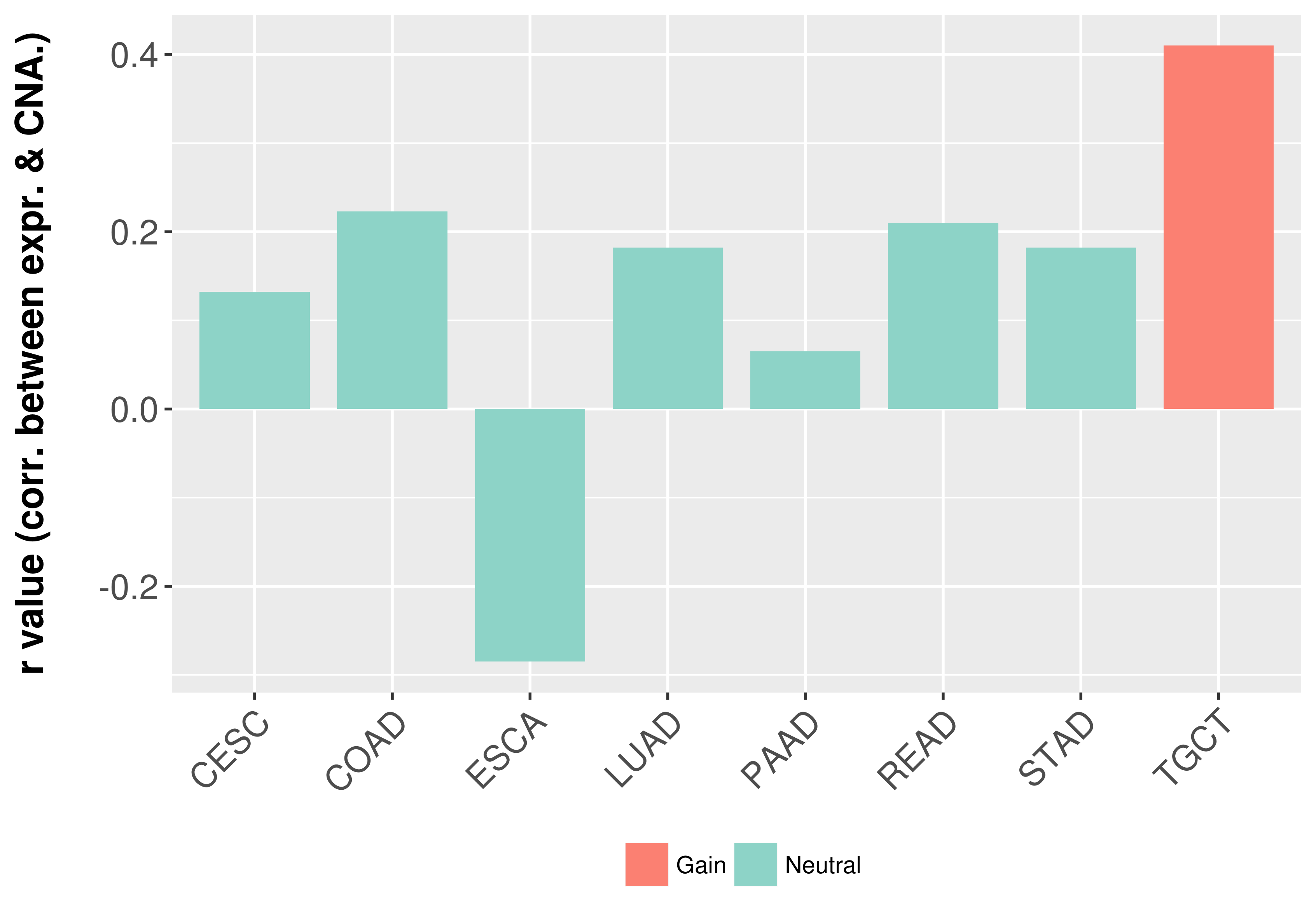
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
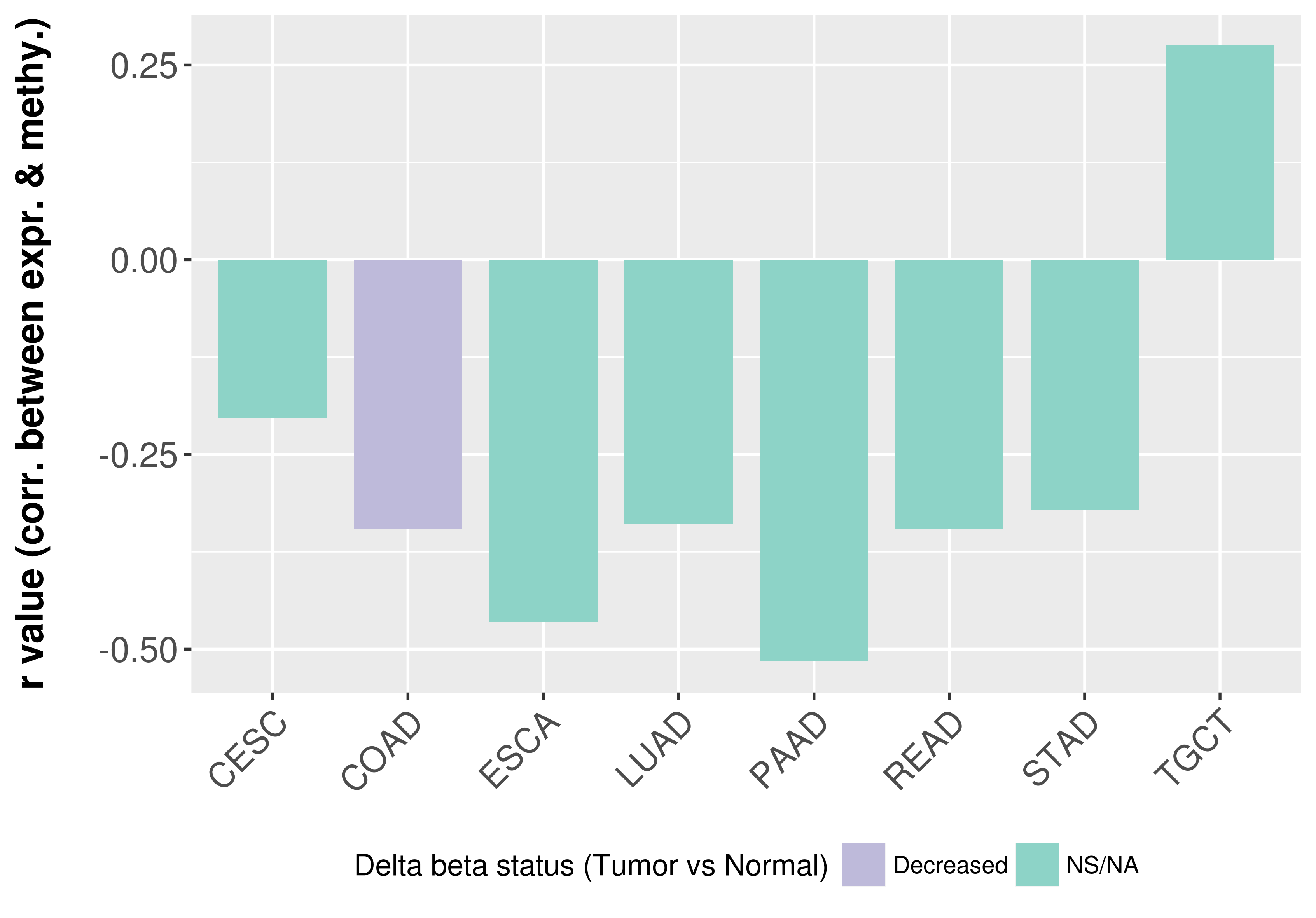
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
| There is no antibody staining data. |
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
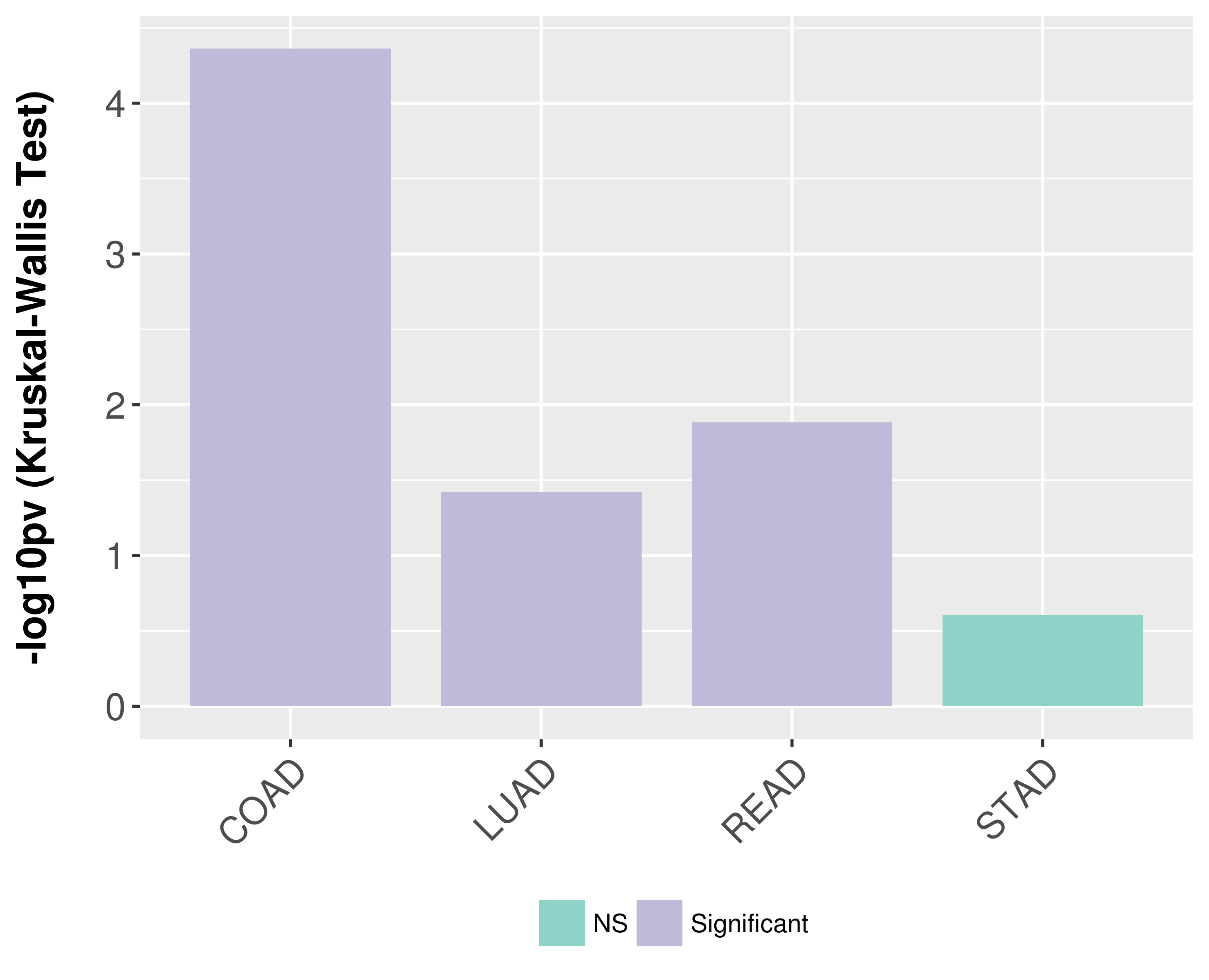
Association between expresson and subtype.
|
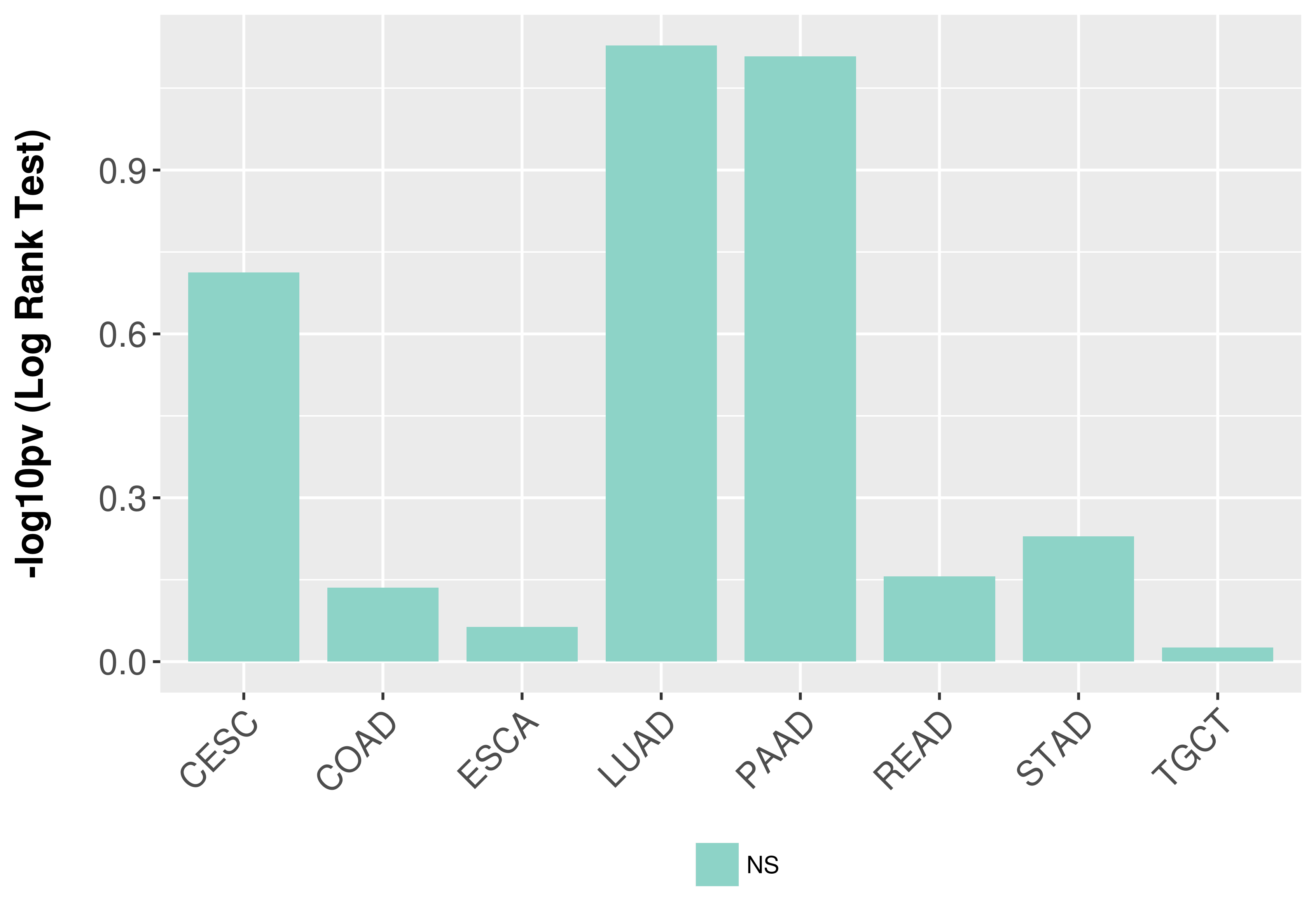
Overall survival analysis based on expression.
|
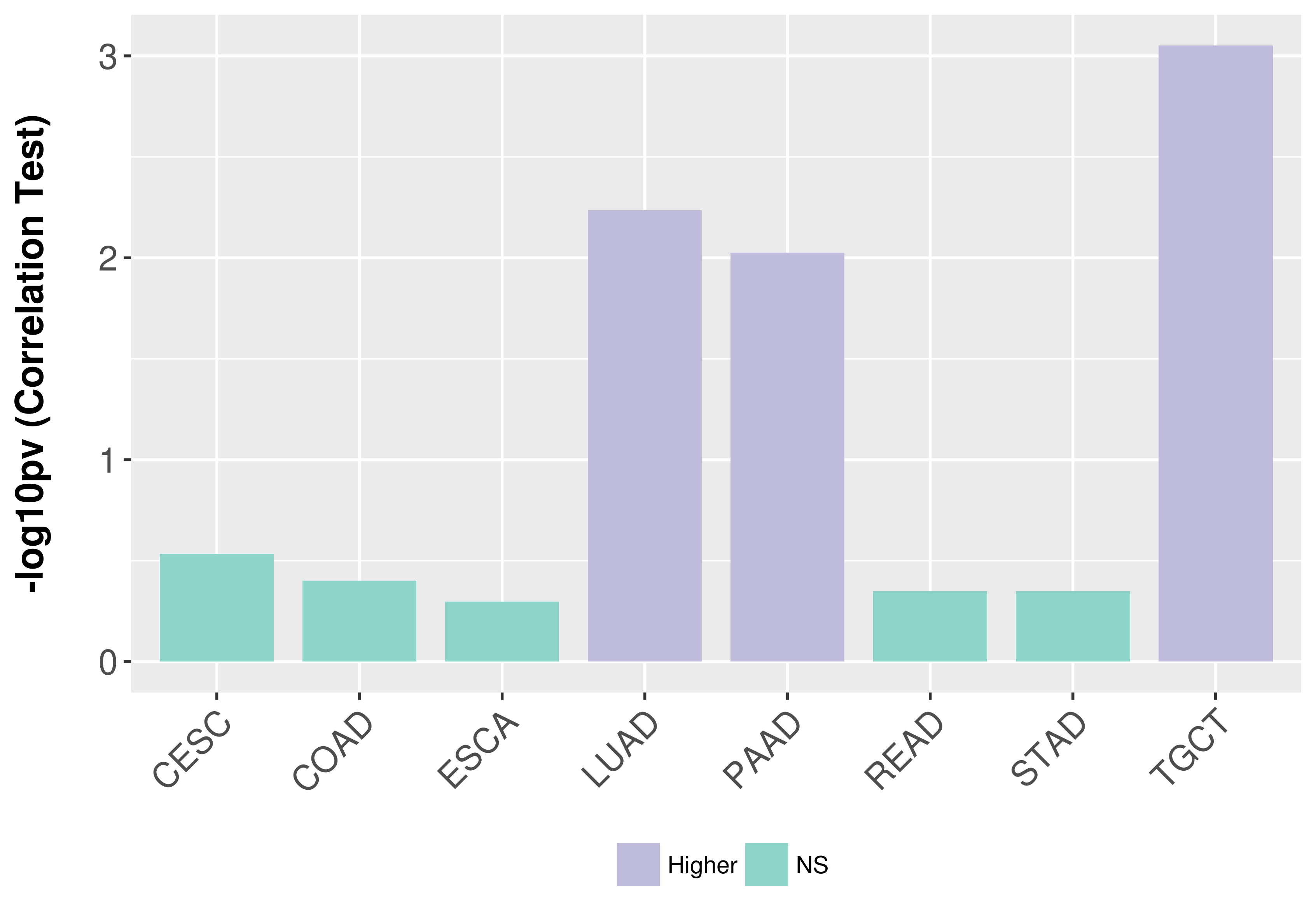
Association between expresson and stage.
|
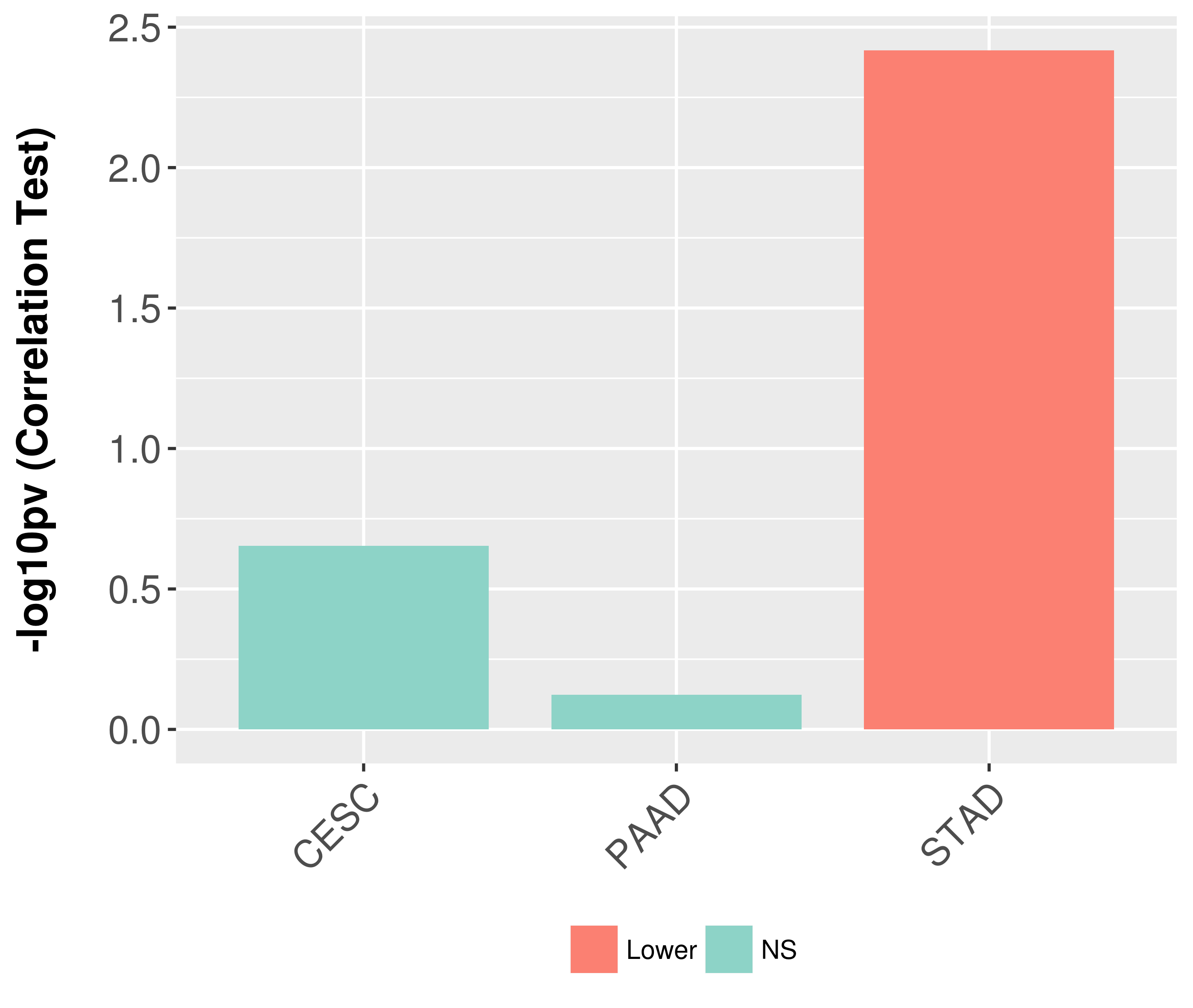
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for APOBEC1. |
| Summary | |
|---|---|
| Symbol | APOBEC1 |
| Name | apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| Aliases | CDAR1; APOBEC-1; HEPR; apolipoprotein B mRNA editing enzyme complex-1; apolipoprotein B mRNA-editing enzyme ...... |
| Location | 12p13.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for APOBEC1. |