Browse CLOCK in pancancer
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00010 Helix-loop-helix DNA-binding domain PF00989 PAS fold |
||||||||||
| Function |
Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. CLOCK has an intrinsic acetyltransferase activity, which enables circadian chromatin remodeling by acetylating histones and nonhistone proteins, including its own partner ARNTL/BMAL1. Regulates the circadian expression of ICAM1, VCAM1, CCL2, THPO and MPL and also acts as an enhancer of the transactivation potential of NF-kappaB. Plays an important role in the homeostatic regulation of sleep. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Represses glucocorticoid receptor NR3C1/GR-induced transcriptional activity by reducing the association of NR3C1/GR to glucocorticoid response elements (GREs) via the acetylation of multiple lysine residues located in its hinge region. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The CLOCK-ARNTL2/BMAL2 heterodimer activates the transcription of SERPINE1/PAI1 and BHLHE40/DEC1. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence (PubMed:23229515). CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3' (PubMed:23229515). The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3' (PubMed:23229515). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000075 cell cycle checkpoint GO:0000077 DNA damage checkpoint GO:0002064 epithelial cell development GO:0002065 columnar/cuboidal epithelial cell differentiation GO:0002066 columnar/cuboidal epithelial cell development GO:0002067 glandular epithelial cell differentiation GO:0002068 glandular epithelial cell development GO:0002790 peptide secretion GO:0002791 regulation of peptide secretion GO:0003309 type B pancreatic cell differentiation GO:0003323 type B pancreatic cell development GO:0006473 protein acetylation GO:0006475 internal protein amino acid acetylation GO:0007283 spermatogenesis GO:0007623 circadian rhythm GO:0009306 protein secretion GO:0009314 response to radiation GO:0009416 response to light stimulus GO:0009648 photoperiodism GO:0009755 hormone-mediated signaling pathway GO:0009914 hormone transport GO:0010212 response to ionizing radiation GO:0010498 proteasomal protein catabolic process GO:0010817 regulation of hormone levels GO:0015833 peptide transport GO:0016570 histone modification GO:0016573 histone acetylation GO:0018205 peptidyl-lysine modification GO:0018393 internal peptidyl-lysine acetylation GO:0018394 peptidyl-lysine acetylation GO:0023061 signal release GO:0030072 peptide hormone secretion GO:0030073 insulin secretion GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0030856 regulation of epithelial cell differentiation GO:0031016 pancreas development GO:0031018 endocrine pancreas development GO:0031349 positive regulation of defense response GO:0031570 DNA integrity checkpoint GO:0031958 corticosteroid receptor signaling pathway GO:0032103 positive regulation of response to external stimulus GO:0032922 circadian regulation of gene expression GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0033144 negative regulation of intracellular steroid hormone receptor signaling pathway GO:0035270 endocrine system development GO:0035883 enteroendocrine cell differentiation GO:0042303 molting cycle GO:0042633 hair cycle GO:0042634 regulation of hair cycle GO:0042886 amide transport GO:0042921 glucocorticoid receptor signaling pathway GO:0043161 proteasome-mediated ubiquitin-dependent protein catabolic process GO:0043401 steroid hormone mediated signaling pathway GO:0043543 protein acylation GO:0046879 hormone secretion GO:0046883 regulation of hormone secretion GO:0048232 male gamete generation GO:0048511 rhythmic process GO:0048545 response to steroid hormone GO:0050708 regulation of protein secretion GO:0050727 regulation of inflammatory response GO:0050729 positive regulation of inflammatory response GO:0050796 regulation of insulin secretion GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051092 positive regulation of NF-kappaB transcription factor activity GO:0051775 response to redox state GO:0071214 cellular response to abiotic stimulus GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071478 cellular response to radiation GO:0071479 cellular response to ionizing radiation GO:0090087 regulation of peptide transport GO:0090276 regulation of peptide hormone secretion GO:2000074 regulation of type B pancreatic cell development GO:2000322 regulation of glucocorticoid receptor signaling pathway GO:2000323 negative regulation of glucocorticoid receptor signaling pathway |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000982 transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001046 core promoter sequence-specific DNA binding GO:0001047 core promoter binding GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001159 core promoter proximal region DNA binding GO:0001190 transcriptional activator activity, RNA polymerase II transcription factor binding GO:0001191 transcriptional repressor activity, RNA polymerase II transcription factor binding GO:0003682 chromatin binding GO:0004402 histone acetyltransferase activity GO:0008080 N-acetyltransferase activity GO:0016407 acetyltransferase activity GO:0016410 N-acyltransferase activity GO:0016746 transferase activity, transferring acyl groups GO:0016747 transferase activity, transferring acyl groups other than amino-acyl groups GO:0031490 chromatin DNA binding GO:0034212 peptide N-acetyltransferase activity GO:0043566 structure-specific DNA binding GO:0061733 peptide-lysine-N-acetyltransferase activity GO:0070888 E-box binding GO:0098811 transcriptional repressor activity, RNA polymerase II activating transcription factor binding |
| Cellular Component |
GO:0005667 transcription factor complex GO:0033391 chromatoid body GO:0035770 ribonucleoprotein granule GO:0036464 cytoplasmic ribonucleoprotein granule |
| KEGG |
hsa04710 Circadian rhythm hsa04728 Dopaminergic synapse |
| Reactome |
R-HSA-1368108: BMAL1 R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-3214847: HATs acetylate histones R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-1368071: NR1D1 (REV-ERBA) represses gene expression R-HSA-1989781: PPARA activates gene expression R-HSA-1368082: RORA activates gene expression R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) |
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
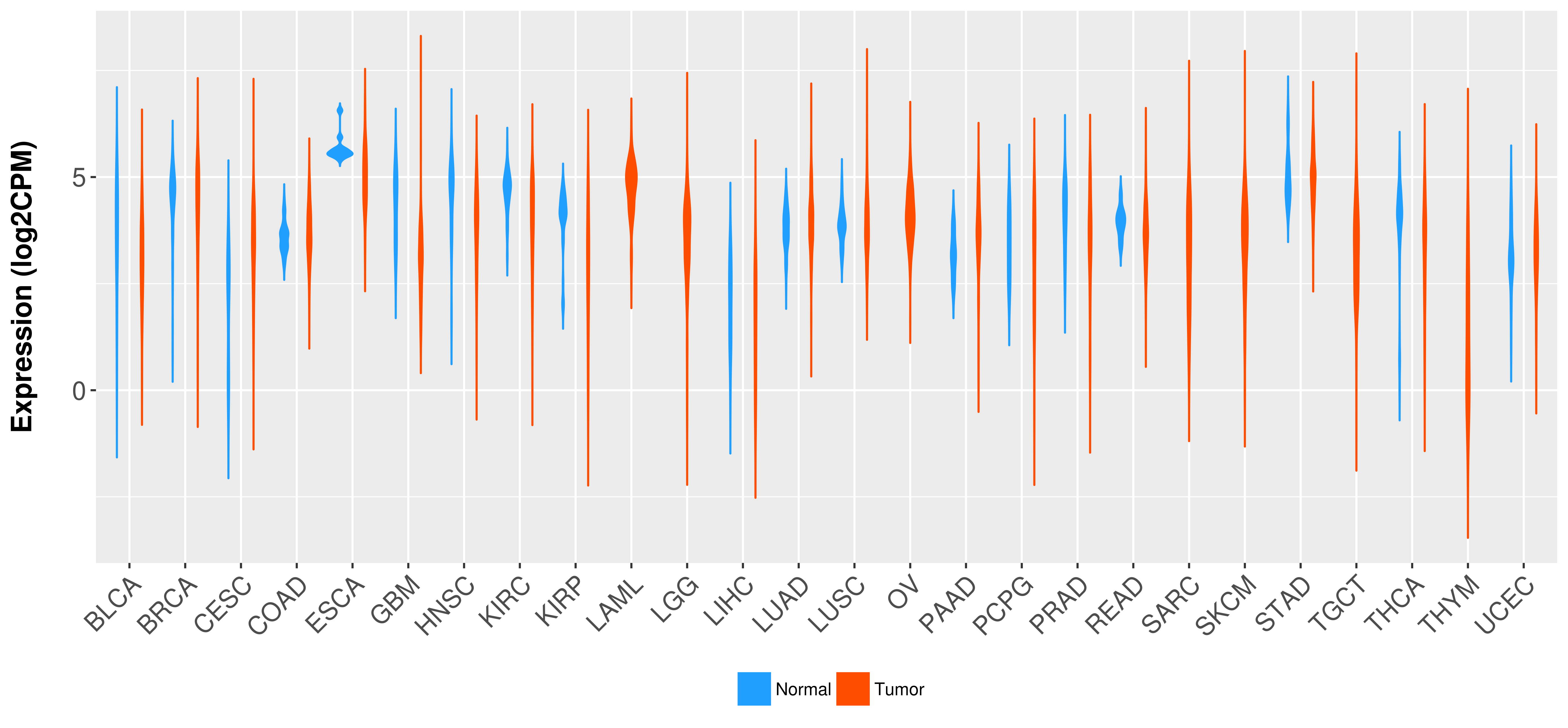
Differential expression analysis for cancers with more than 10 normal samples
|
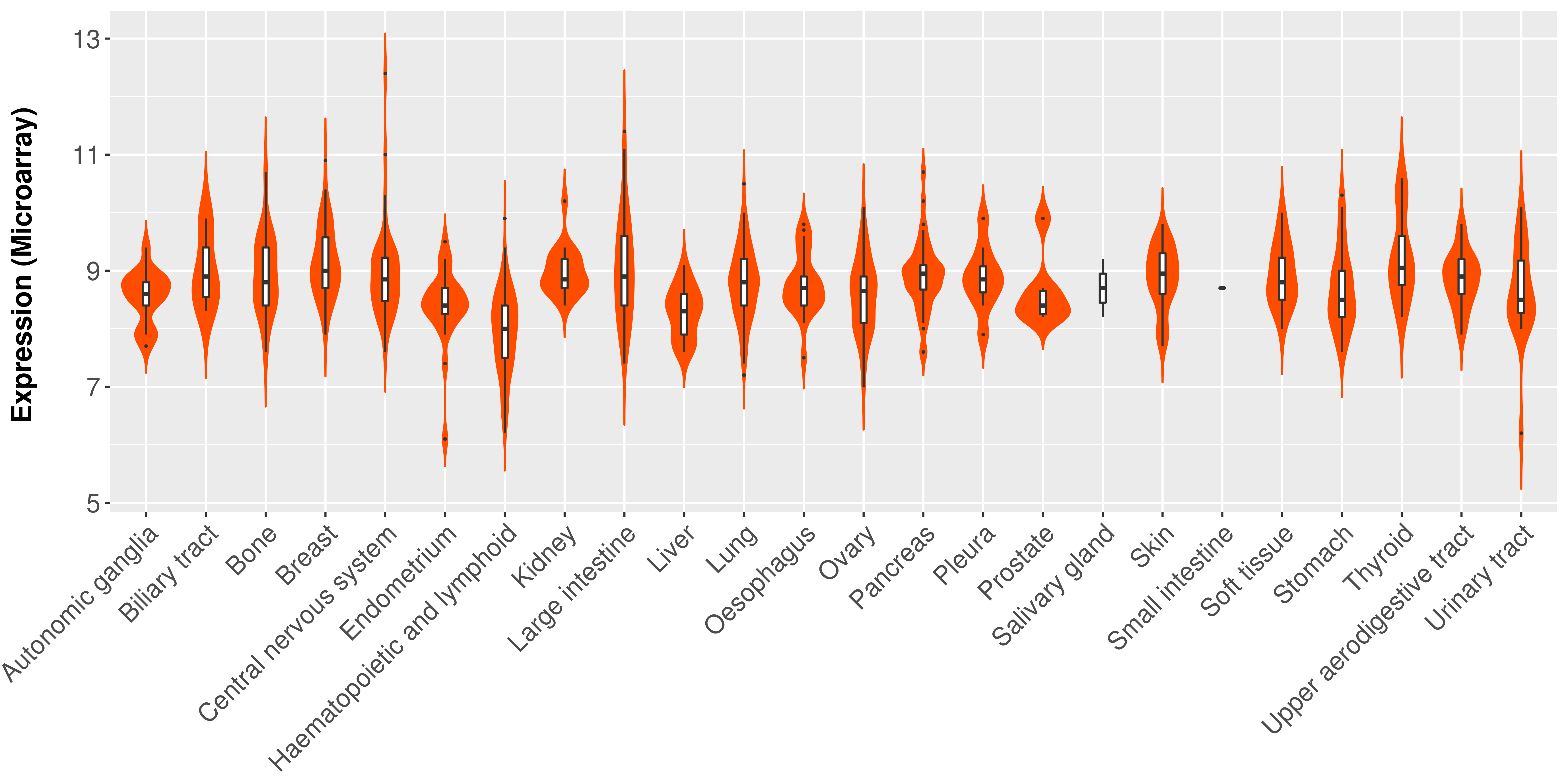
|
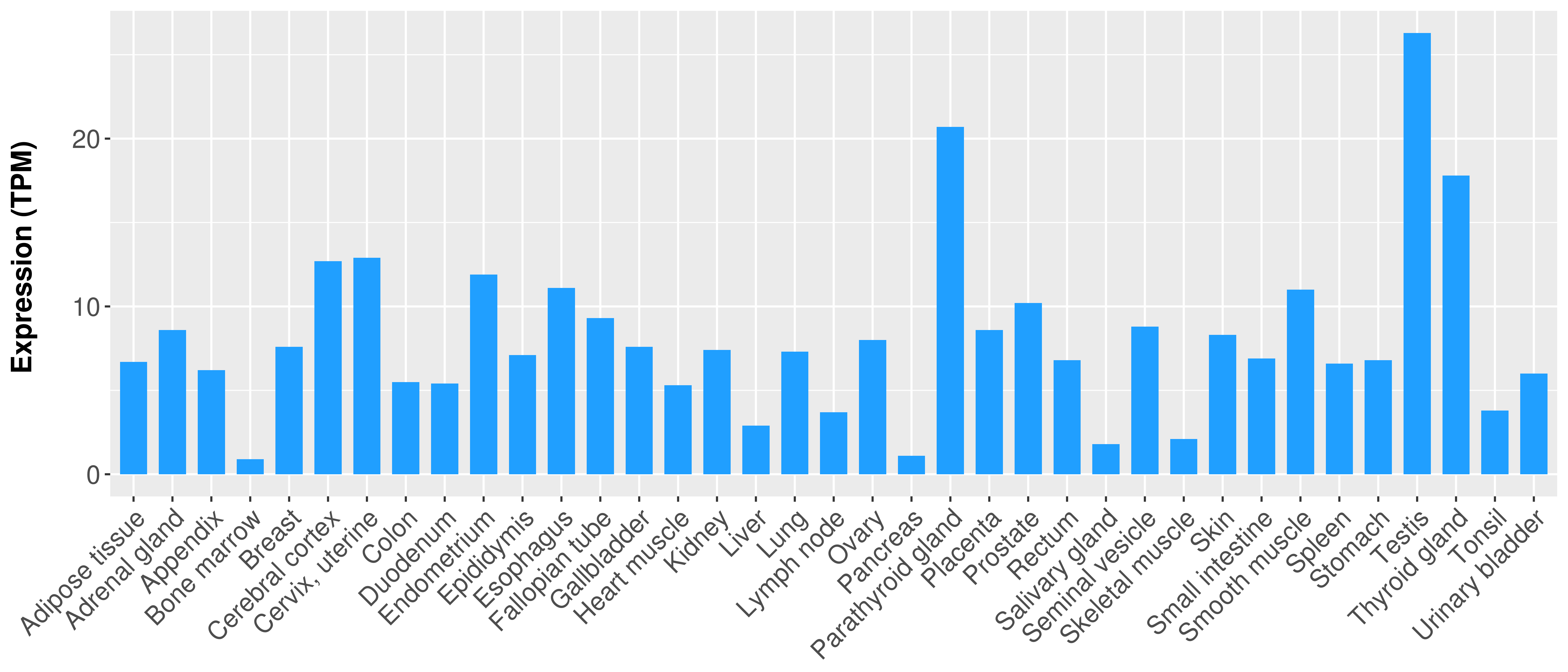
|
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
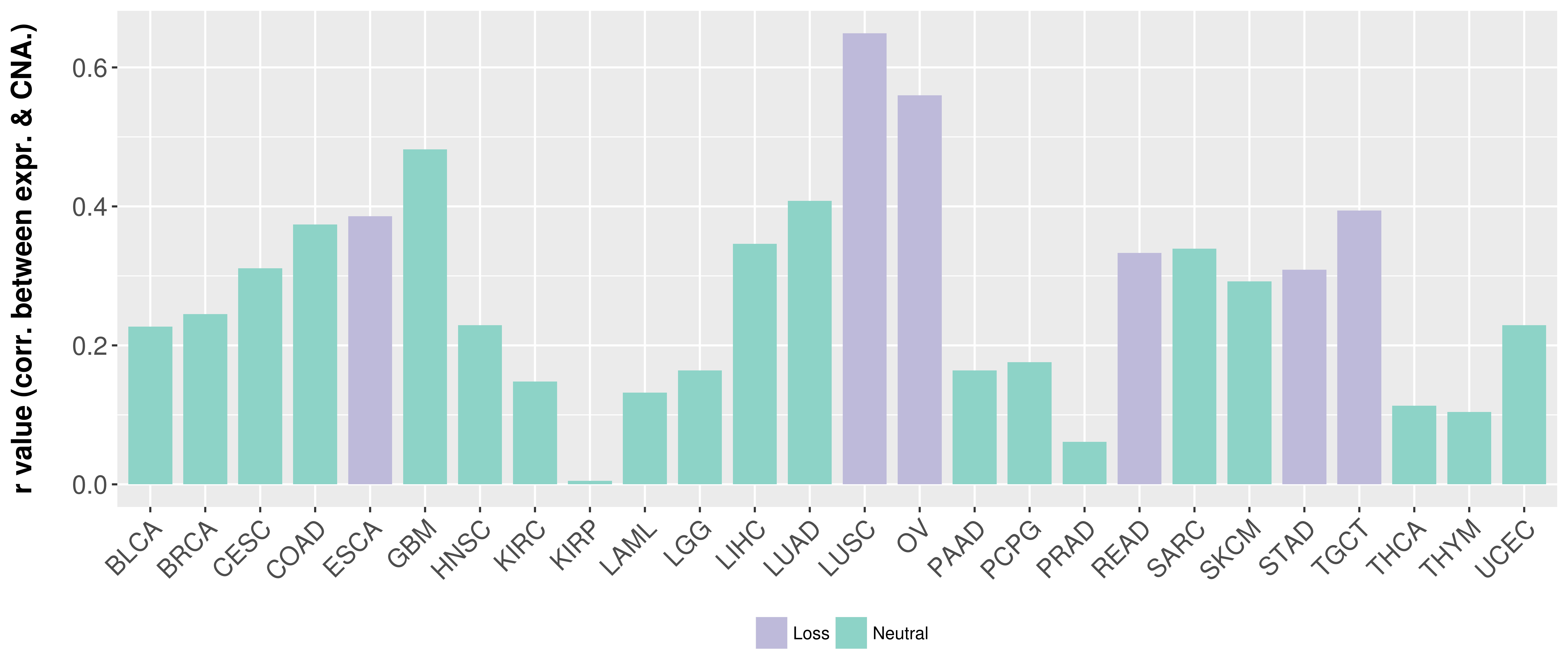
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
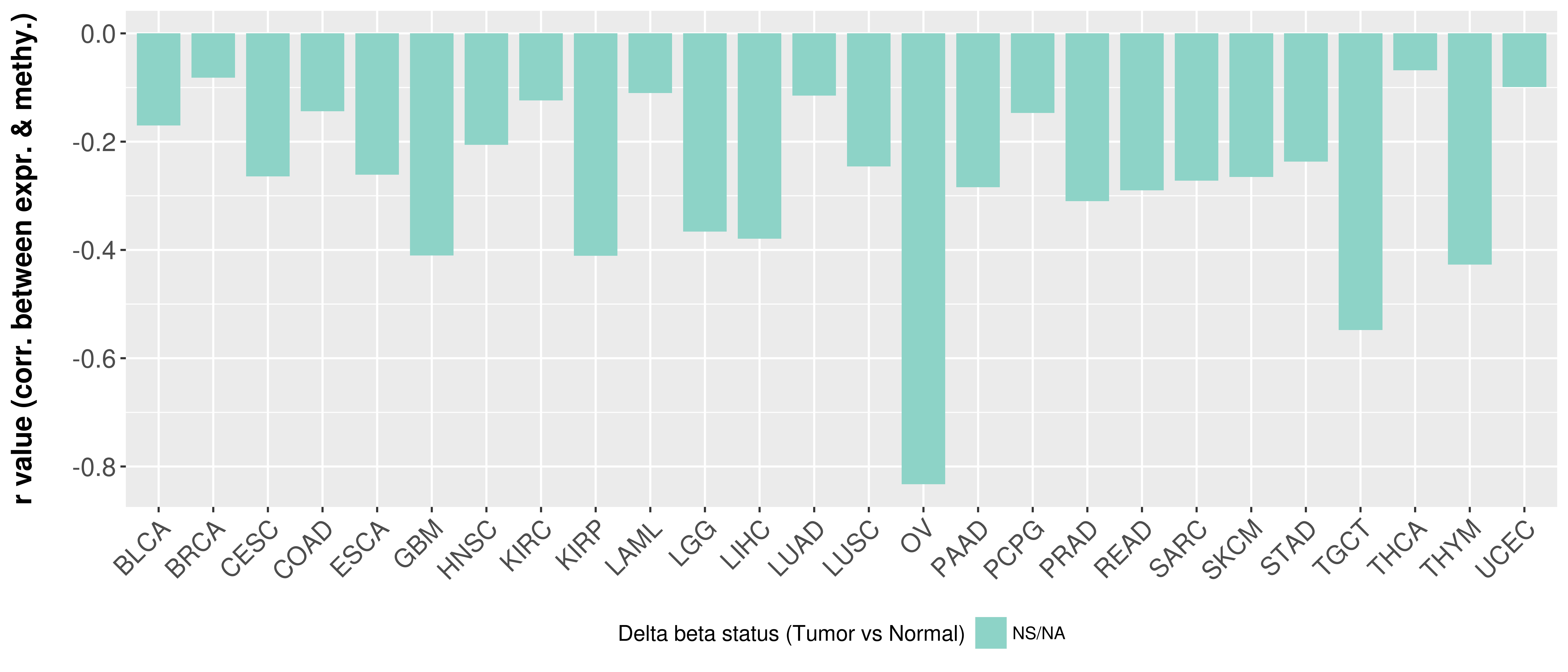
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
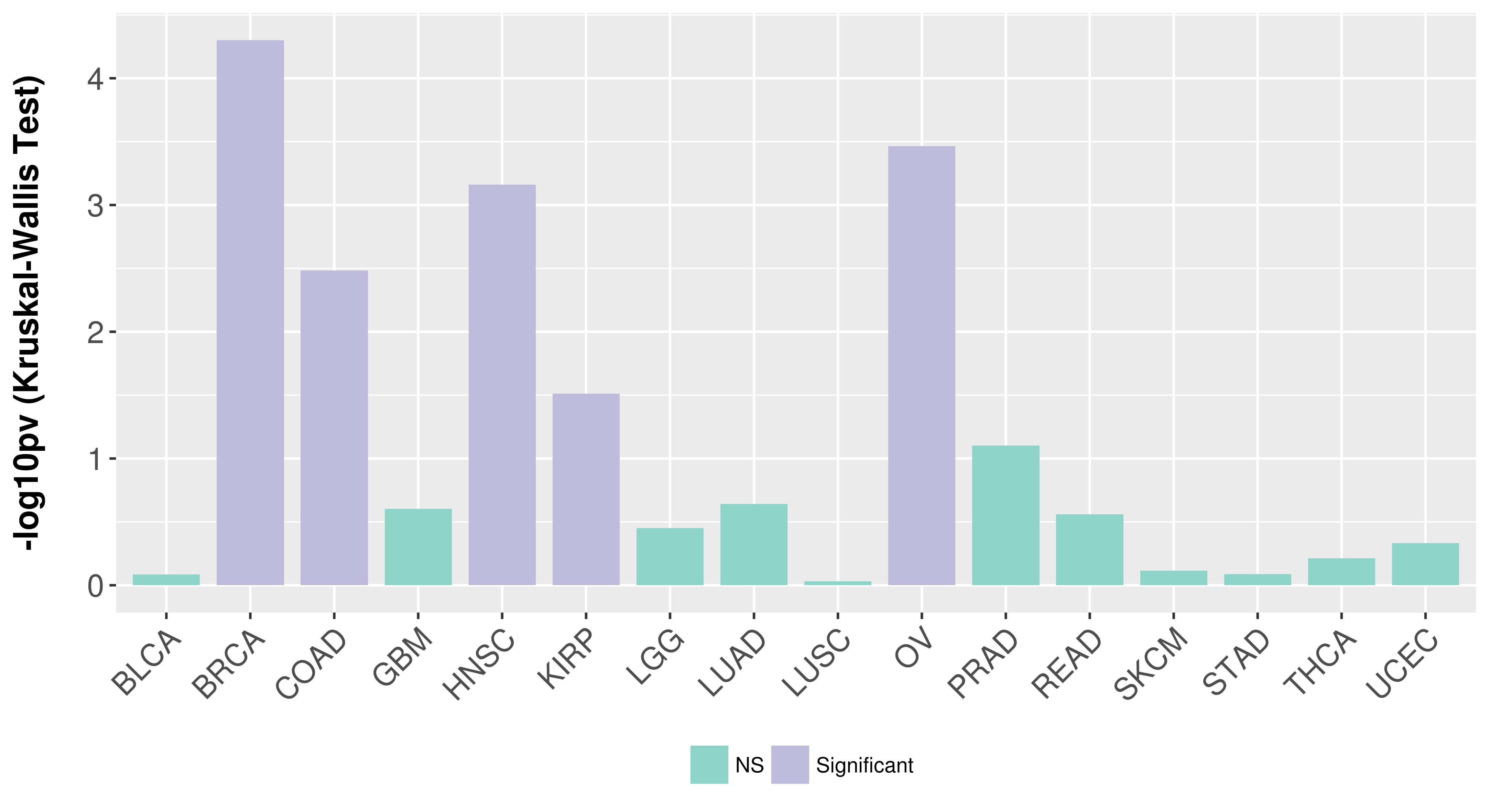
Association between expresson and subtype.
|
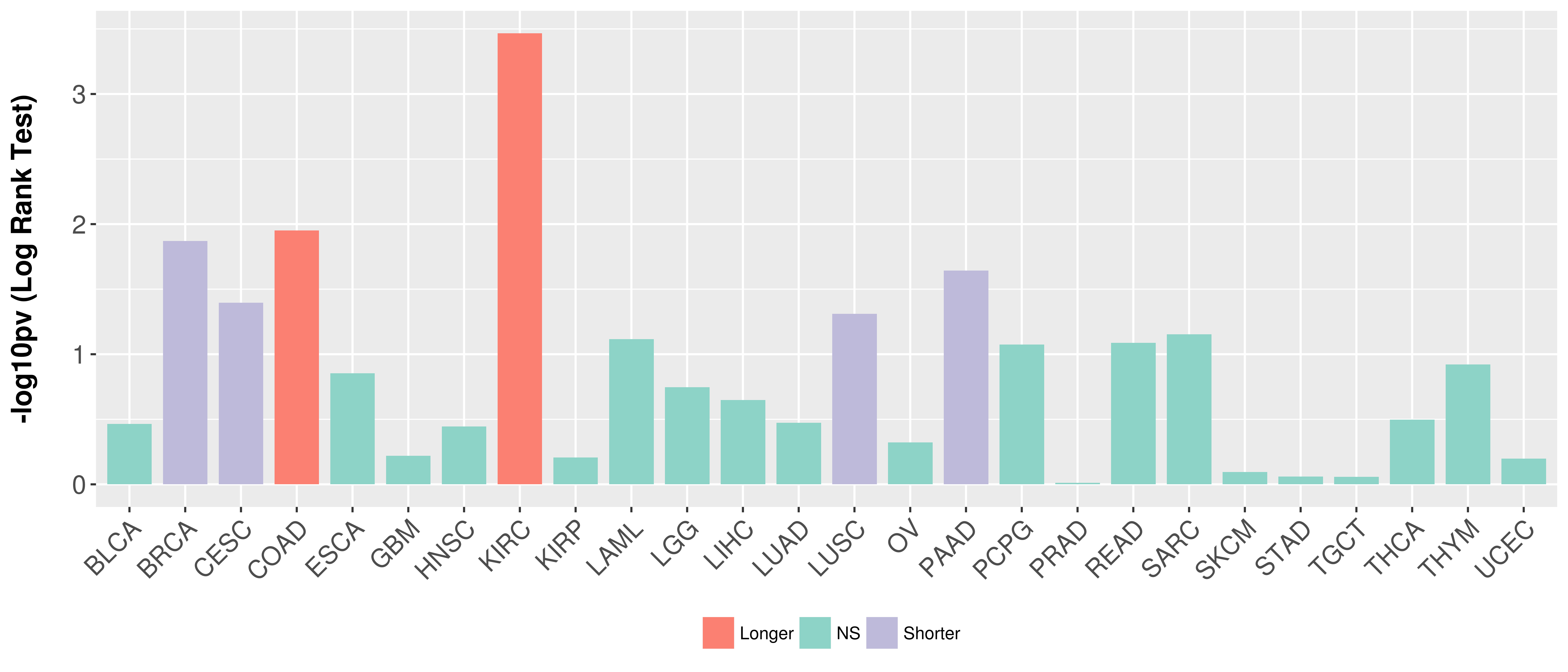
Overall survival analysis based on expression.
|
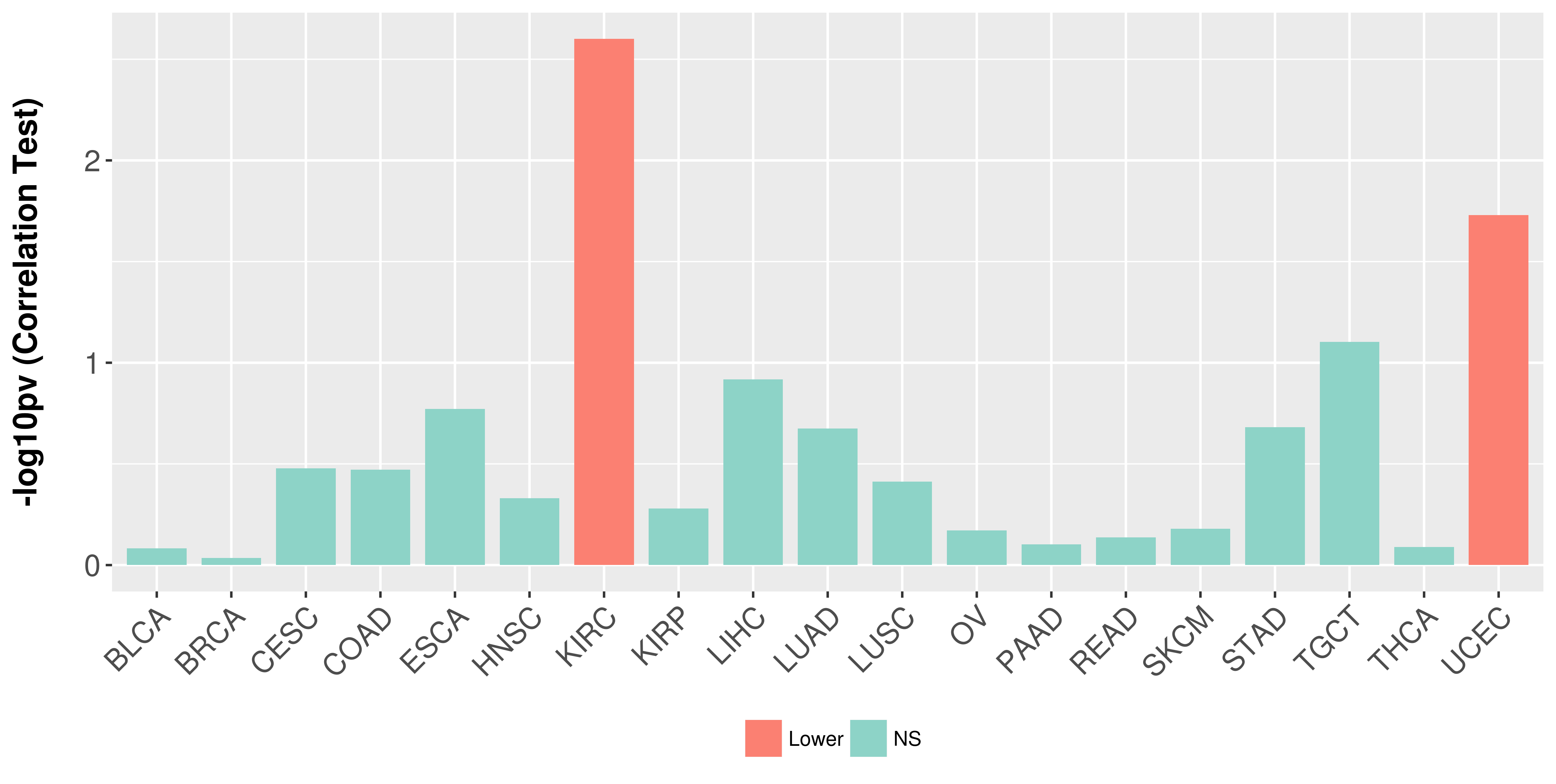
Association between expresson and stage.
|
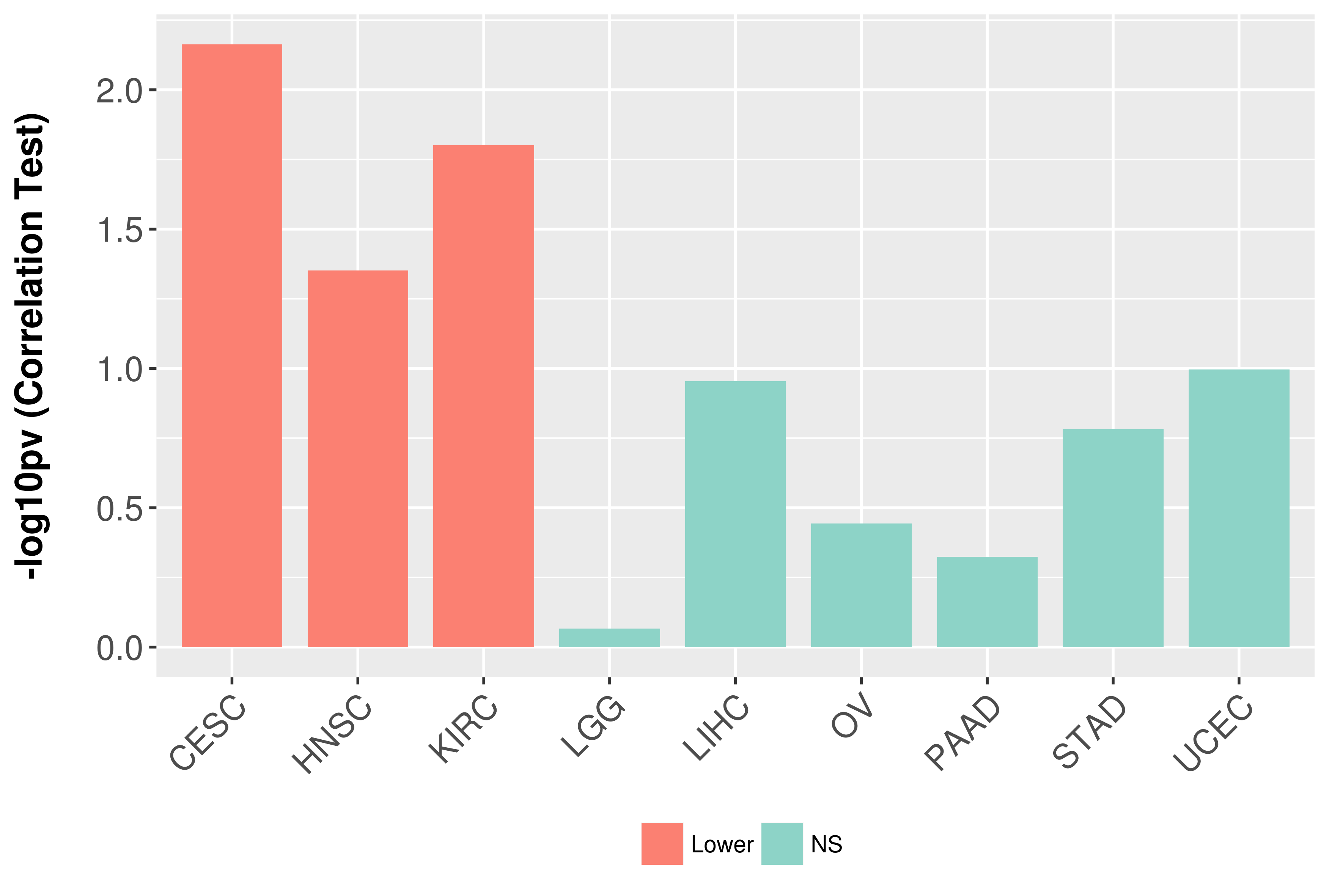
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for CLOCK. |
| Summary | |
|---|---|
| Symbol | CLOCK |
| Name | clock circadian regulator |
| Aliases | KIAA0334; KAT13D; bHLHe8; clock (mouse) homolog; clock homolog (mouse); circadian locomoter output cycles ka ...... |
| Location | 4q12 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|