Browse DUSP1 in pancancer
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00782 Dual specificity phosphatase PF00581 Rhodanese-like domain |
||||||||||
| Function |
Dual specificity phosphatase that dephosphorylates MAP kinase MAPK1/ERK2 on both 'Thr-183' and 'Tyr-185', regulating its activity during the meiotic cell cycle. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000070 mitotic sister chromatid segregation GO:0000075 cell cycle checkpoint GO:0000188 inactivation of MAPK activity GO:0000302 response to reactive oxygen species GO:0000819 sister chromatid segregation GO:0001101 response to acid chemical GO:0001704 formation of primary germ layer GO:0001706 endoderm formation GO:0001933 negative regulation of protein phosphorylation GO:0006469 negative regulation of protein kinase activity GO:0006470 protein dephosphorylation GO:0006979 response to oxidative stress GO:0007050 cell cycle arrest GO:0007059 chromosome segregation GO:0007067 mitotic nuclear division GO:0007088 regulation of mitotic nuclear division GO:0007091 metaphase/anaphase transition of mitotic cell cycle GO:0007093 mitotic cell cycle checkpoint GO:0007094 mitotic spindle assembly checkpoint GO:0007346 regulation of mitotic cell cycle GO:0007369 gastrulation GO:0007492 endoderm development GO:0009314 response to radiation GO:0009416 response to light stimulus GO:0010035 response to inorganic substance GO:0010038 response to metal ion GO:0010639 negative regulation of organelle organization GO:0010948 negative regulation of cell cycle process GO:0010965 regulation of mitotic sister chromatid separation GO:0014074 response to purine-containing compound GO:0016311 dephosphorylation GO:0030071 regulation of mitotic metaphase/anaphase transition GO:0031577 spindle checkpoint GO:0031960 response to corticosteroid GO:0032355 response to estradiol GO:0032526 response to retinoic acid GO:0033044 regulation of chromosome organization GO:0033045 regulation of sister chromatid segregation GO:0033046 negative regulation of sister chromatid segregation GO:0033047 regulation of mitotic sister chromatid segregation GO:0033048 negative regulation of mitotic sister chromatid segregation GO:0033574 response to testosterone GO:0033673 negative regulation of kinase activity GO:0035335 peptidyl-tyrosine dephosphorylation GO:0035970 peptidyl-threonine dephosphorylation GO:0042326 negative regulation of phosphorylation GO:0042542 response to hydrogen peroxide GO:0043405 regulation of MAP kinase activity GO:0043407 negative regulation of MAP kinase activity GO:0043409 negative regulation of MAPK cascade GO:0044770 cell cycle phase transition GO:0044772 mitotic cell cycle phase transition GO:0044784 metaphase/anaphase transition of cell cycle GO:0045786 negative regulation of cell cycle GO:0045839 negative regulation of mitotic nuclear division GO:0045841 negative regulation of mitotic metaphase/anaphase transition GO:0045930 negative regulation of mitotic cell cycle GO:0046683 response to organophosphorus GO:0048545 response to steroid hormone GO:0051052 regulation of DNA metabolic process GO:0051053 negative regulation of DNA metabolic process GO:0051304 chromosome separation GO:0051306 mitotic sister chromatid separation GO:0051321 meiotic cell cycle GO:0051348 negative regulation of transferase activity GO:0051384 response to glucocorticoid GO:0051445 regulation of meiotic cell cycle GO:0051447 negative regulation of meiotic cell cycle GO:0051591 response to cAMP GO:0051592 response to calcium ion GO:0051783 regulation of nuclear division GO:0051784 negative regulation of nuclear division GO:0051983 regulation of chromosome segregation GO:0051985 negative regulation of chromosome segregation GO:0070371 ERK1 and ERK2 cascade GO:0070372 regulation of ERK1 and ERK2 cascade GO:0070373 negative regulation of ERK1 and ERK2 cascade GO:0071173 spindle assembly checkpoint GO:0071174 mitotic spindle checkpoint GO:0071850 mitotic cell cycle arrest GO:0071897 DNA biosynthetic process GO:0071900 regulation of protein serine/threonine kinase activity GO:0071901 negative regulation of protein serine/threonine kinase activity GO:0090231 regulation of spindle checkpoint GO:0090266 regulation of mitotic cell cycle spindle assembly checkpoint GO:0098813 nuclear chromosome segregation GO:1901654 response to ketone GO:1901976 regulation of cell cycle checkpoint GO:1901987 regulation of cell cycle phase transition GO:1901988 negative regulation of cell cycle phase transition GO:1901990 regulation of mitotic cell cycle phase transition GO:1901991 negative regulation of mitotic cell cycle phase transition GO:1902099 regulation of metaphase/anaphase transition of cell cycle GO:1902100 negative regulation of metaphase/anaphase transition of cell cycle GO:1902532 negative regulation of intracellular signal transduction GO:1903504 regulation of mitotic spindle checkpoint GO:2000241 regulation of reproductive process GO:2000242 negative regulation of reproductive process GO:2000278 regulation of DNA biosynthetic process GO:2000279 negative regulation of DNA biosynthetic process GO:2000816 negative regulation of mitotic sister chromatid separation GO:2001251 negative regulation of chromosome organization |
| Molecular Function |
GO:0004721 phosphoprotein phosphatase activity GO:0004725 protein tyrosine phosphatase activity GO:0004726 non-membrane spanning protein tyrosine phosphatase activity GO:0008138 protein tyrosine/serine/threonine phosphatase activity GO:0008330 protein tyrosine/threonine phosphatase activity GO:0016791 phosphatase activity GO:0017017 MAP kinase tyrosine/serine/threonine phosphatase activity GO:0019838 growth factor binding GO:0033549 MAP kinase phosphatase activity GO:0042578 phosphoric ester hydrolase activity |
| Cellular Component | - |
| KEGG |
hsa04010 MAPK signaling pathway hsa04726 Serotonergic synapse |
| Reactome |
R-HSA-170984: ARMS-mediated activation R-HSA-422475: Axon guidance R-HSA-1280215: Cytokine Signaling in Immune system R-HSA-2172127: DAP12 interactions R-HSA-2424491: DAP12 signaling R-HSA-1266738: Developmental Biology R-HSA-186763: Downstream signal transduction R-HSA-2871796: FCERI mediated MAPK activation R-HSA-2454202: Fc epsilon receptor (FCERI) signaling R-HSA-170968: Frs2-mediated activation R-HSA-179812: GRB2 events in EGFR signaling R-HSA-881907: Gastrin-CREB signalling pathway via PKC and MAPK R-HSA-2428924: IGF1R signaling cascade R-HSA-112399: IRS-mediated signalling R-HSA-2428928: IRS-related events triggered by IGF1R R-HSA-168256: Immune System R-HSA-168249: Innate Immune System R-HSA-74751: Insulin receptor signalling cascade R-HSA-912526: Interleukin receptor SHC signaling R-HSA-451927: Interleukin-2 signaling R-HSA-512988: Interleukin-3, 5 and GM-CSF signaling R-HSA-5683057: MAPK family signaling cascades R-HSA-5684996: MAPK1/MAPK3 signaling R-HSA-375165: NCAM signaling for neurite out-growth R-HSA-187037: NGF signalling via TRKA from the plasma membrane R-HSA-5675221: Negative regulation of MAPK pathway R-HSA-169893: Prolonged ERK activation events R-HSA-112409: RAF-independent MAPK1/3 activation R-HSA-5673001: RAF/MAP kinase cascade R-HSA-8853659: RET signaling R-HSA-180336: SHC1 events in EGFR signaling R-HSA-112412: SOS-mediated signalling R-HSA-162582: Signal Transduction R-HSA-177929: Signaling by EGFR R-HSA-372790: Signaling by GPCR R-HSA-74752: Signaling by Insulin receptor R-HSA-449147: Signaling by Interleukins R-HSA-2586552: Signaling by Leptin R-HSA-186797: Signaling by PDGF R-HSA-1433557: Signaling by SCF-KIT R-HSA-2404192: Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) R-HSA-194138: Signaling by VEGF R-HSA-166520: Signalling by NGF R-HSA-187687: Signalling to ERKs R-HSA-167044: Signalling to RAS R-HSA-187706: Signalling to p38 via RIT and RIN R-HSA-4420097: VEGFA-VEGFR2 Pathway R-HSA-5218921: VEGFR2 mediated cell proliferation |
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for DUSP1. |
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |

Differential expression analysis for cancers with more than 10 normal samples
|
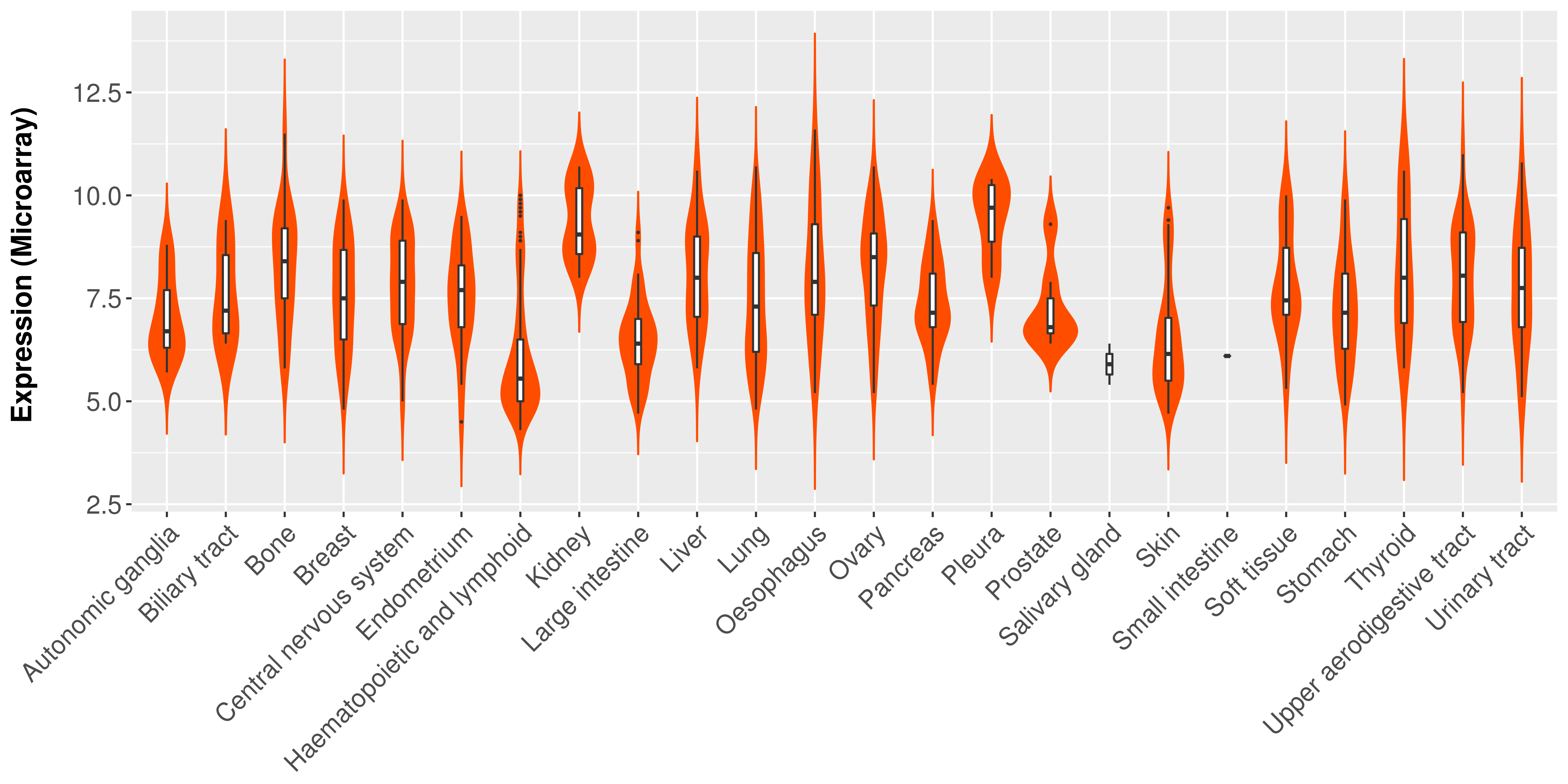
|
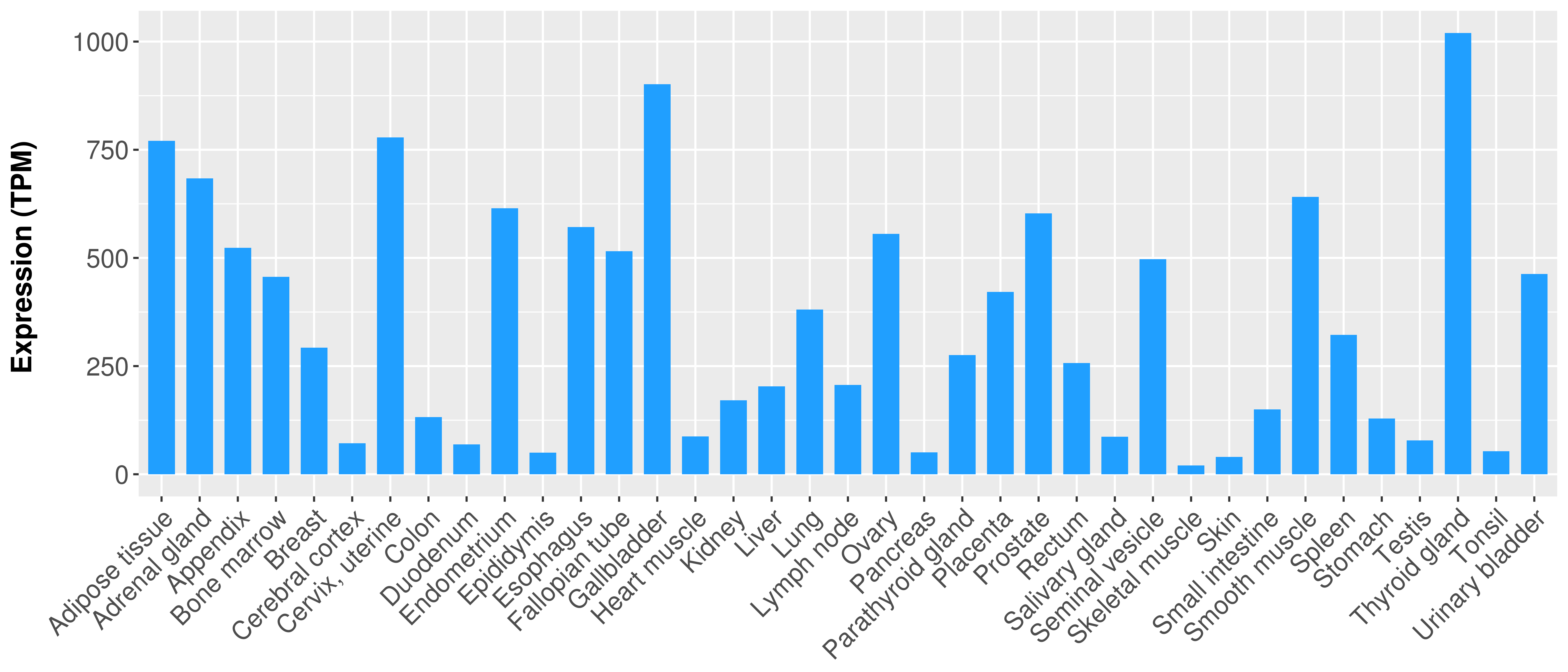
|
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
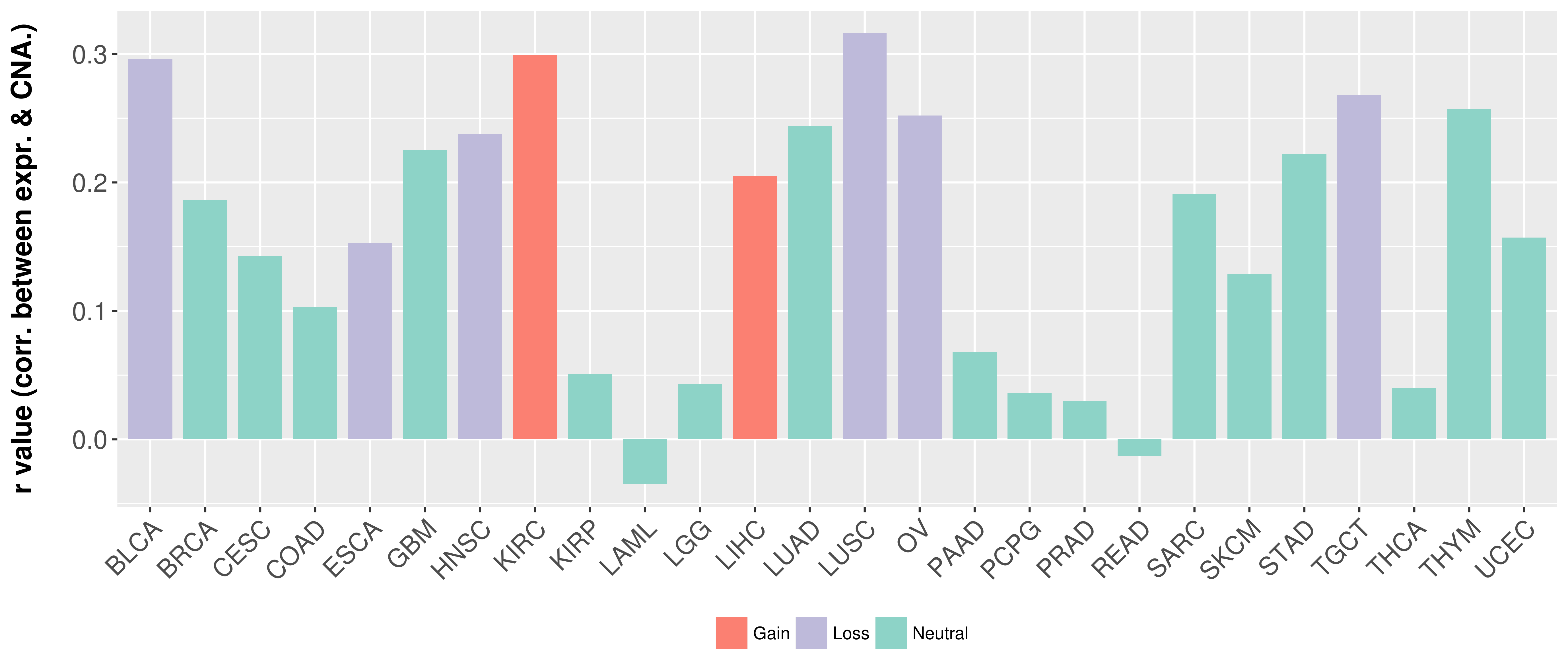
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |

Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
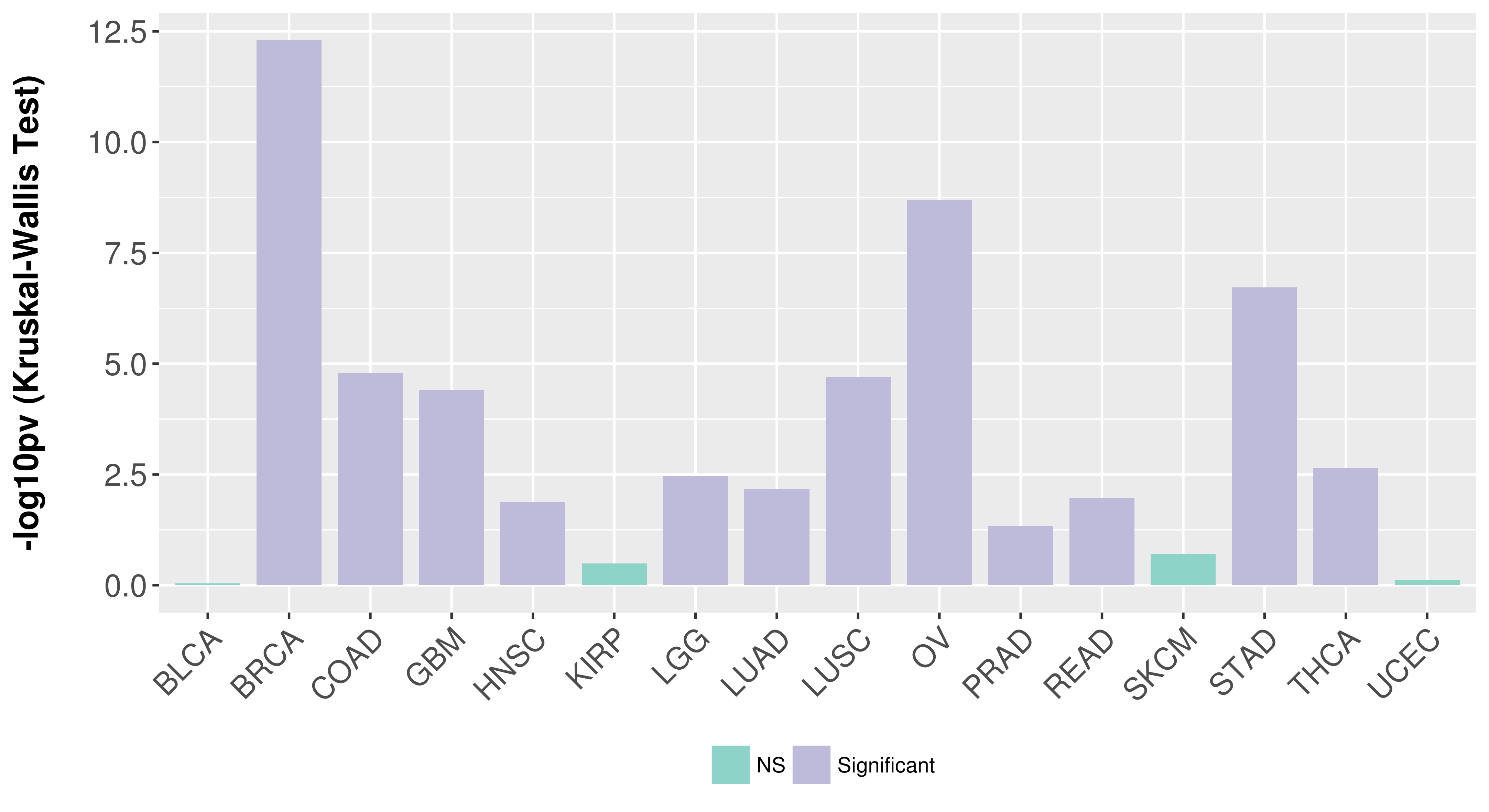
Association between expresson and subtype.
|
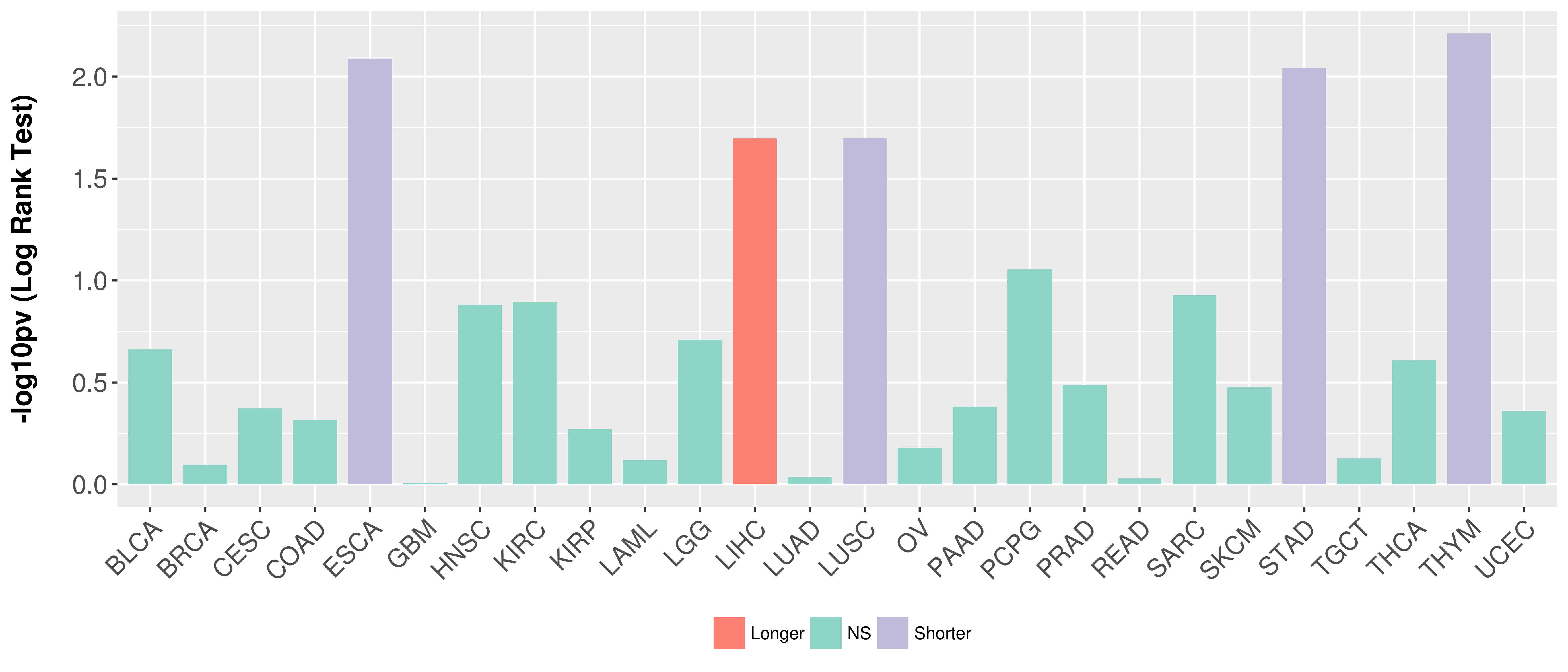
Overall survival analysis based on expression.
|
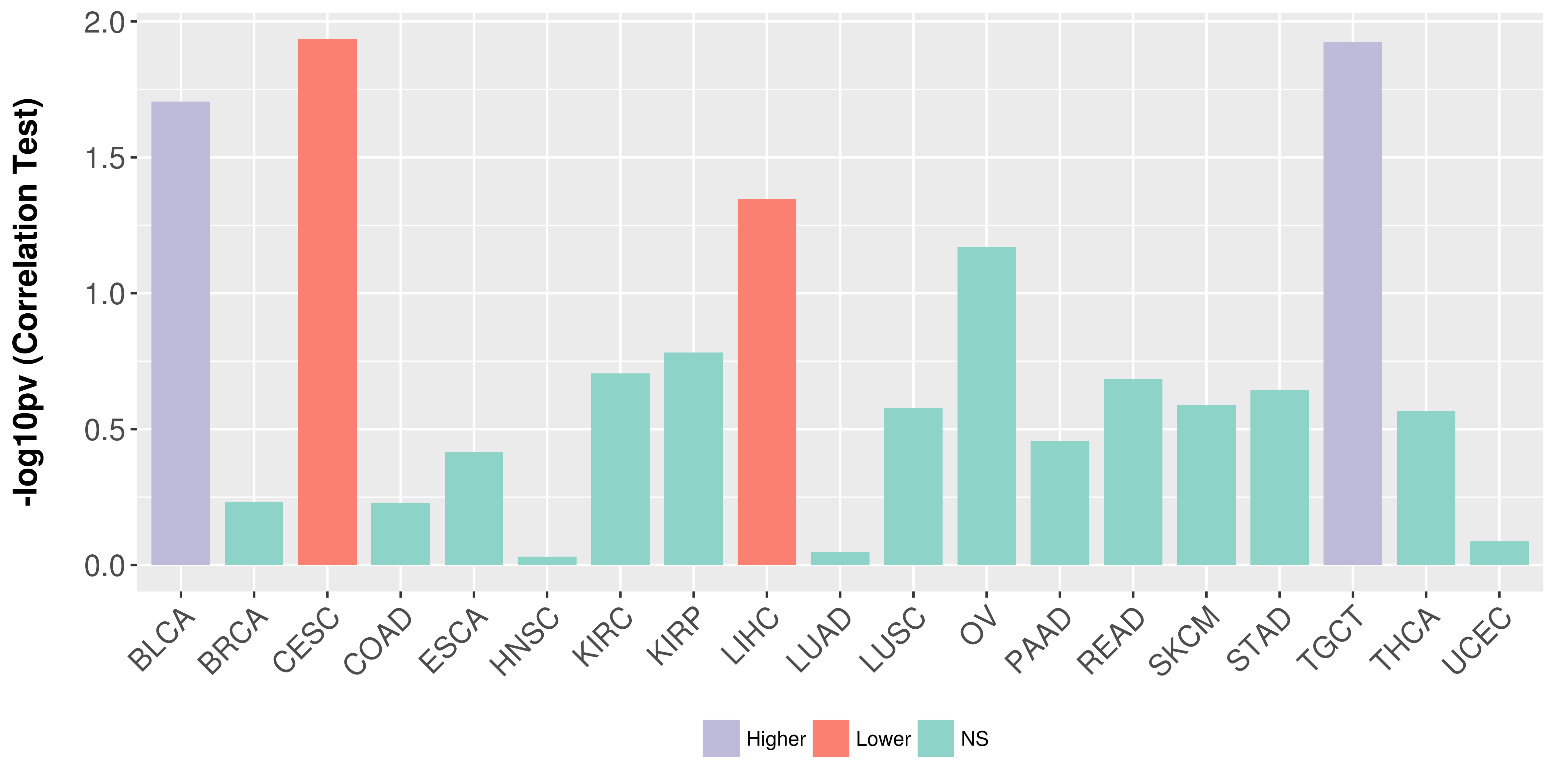
Association between expresson and stage.
|
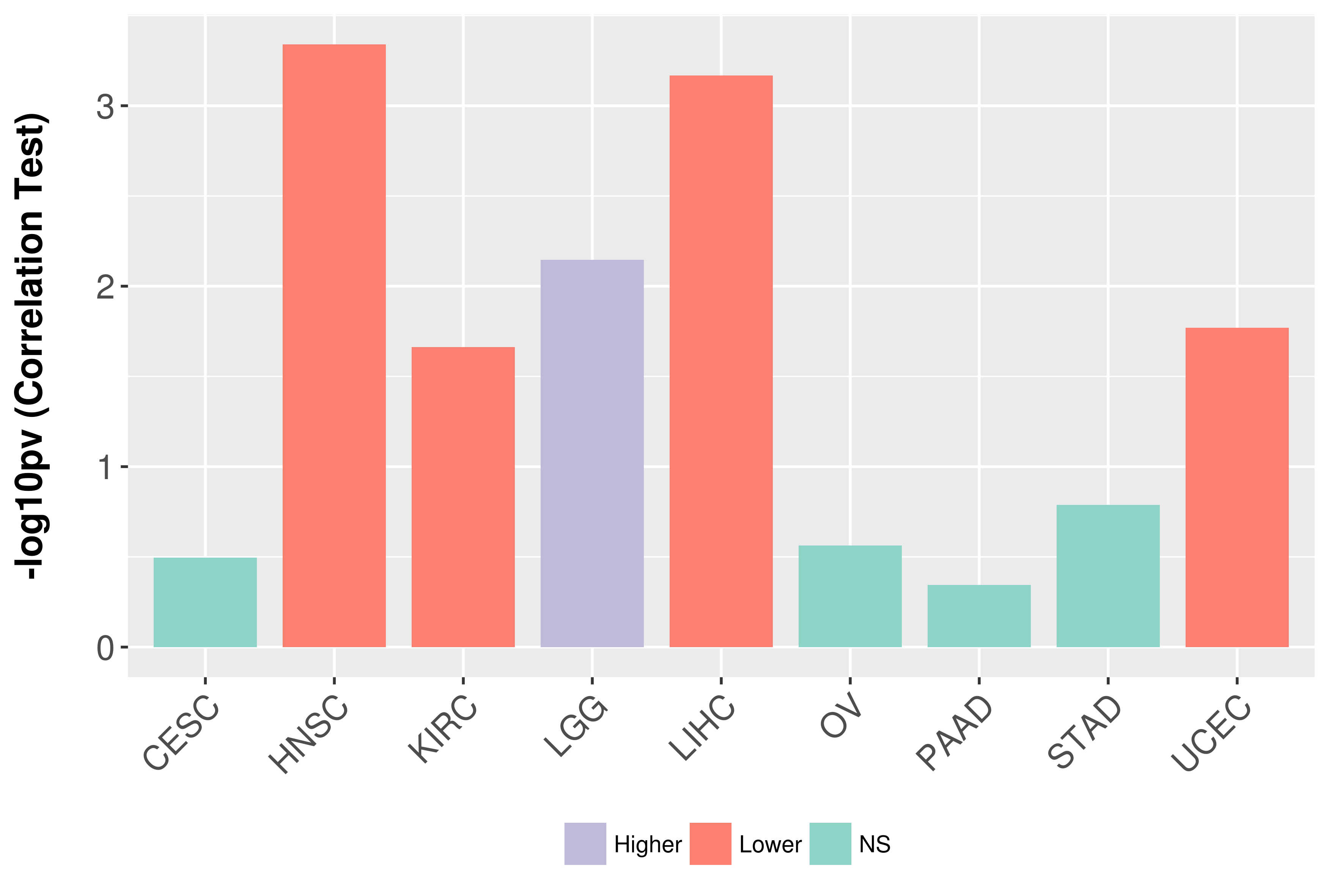
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for DUSP1. |
| Summary | |
|---|---|
| Symbol | DUSP1 |
| Name | dual specificity phosphatase 1 |
| Aliases | HVH1; CL100; MKP-1; PTPN10; MKP1; CL 100; MAP kinase phosphatase 1; dual specificity protein phosphatase hVH ...... |
| Location | 5q35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|