Browse FMR1 in pancancer
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF05641 Agenet domain PF16098 Fragile X-related mental retardation protein C-terminal region 2 PF12235 Fragile X-related 1 protein core C terminal PF00013 KH domain |
||||||||||
| Function |
Multifunctional polyribosome-associated RNA-binding protein that plays a central role in neuronal development and synaptic plasticity through the regulation of alternative mRNA splicing, mRNA stability, mRNA dendritic transport and postsynaptic local protein synthesis of a subset of mRNAs (PubMed:16631377, PubMed:18653529, PubMed:19166269, PubMed:23235829, PubMed:25464849). Plays a role in the alternative splicing of its own mRNA (PubMed:18653529). Plays a role in mRNA nuclear export (By similarity). Together with export factor NXF2, is involved in the regulation of the NXF1 mRNA stability in neurons (By similarity). Stabilizes the scaffolding postsynaptic density protein DLG4/PSD-95 and the myelin basic protein (MBP) mRNAs in hippocampal neurons and glial cells, respectively; this stabilization is further increased in response to metabotropic glutamate receptor (mGluR) stimulation (By similarity). Plays a role in selective delivery of a subset of dendritic mRNAs to synaptic sites in response to mGluR activation in a kinesin-dependent manner (By similarity). Plays a role as a repressor of mRNA translation during the transport of dendritic mRNAs to postnyaptic dendritic spines (PubMed:11532944, PubMed:11157796, PubMed:12594214, PubMed:23235829). Component of the CYFIP1-EIF4E-FMR1 complex which blocks cap-dependent mRNA translation initiation (By similarity). Represses mRNA translation by stalling ribosomal translocation during elongation (By similarity). Reports are contradictory with regards to its ability to mediate translation inhibition of MBP mRNA in oligodendrocytes (PubMed:23891804). Also involved in the recruitment of the RNA helicase MOV10 to a subset of mRNAs and hence regulates microRNA (miRNA)-mediated translational repression by AGO2 (PubMed:14703574, PubMed:17057366, PubMed:25464849). Facilitates the assembly of miRNAs on specific target mRNAs (PubMed:17057366). Plays also a role as an activator of mRNA translation of a subset of dendritic mRNAs at synapses (PubMed:19097999, PubMed:19166269). In response to mGluR stimulation, FMR1-target mRNAs are rapidly derepressed, allowing for local translation at synapses (By similarity). Binds to a large subset of dendritic mRNAs that encode a myriad of proteins involved in pre- and postsynaptic functions (PubMed:7692601, PubMed:11719189, PubMed:11157796, PubMed:12594214, PubMed:17417632, PubMed:23235829, PubMed:24448548). Binds to 5'-ACU[GU]-3' and/or 5'-[AU]GGA-3' RNA consensus sequences within mRNA targets, mainly at coding sequence (CDS) and 3'-untranslated region (UTR) and less frequently at 5'-UTR (PubMed:23235829). Binds to intramolecular G-quadruplex structures in the 5'- or 3'-UTRs of mRNA targets (PubMed:11719189, PubMed:18579868, PubMed:25464849, PubMed:25692235). Binds to G-quadruplex structures in the 3'-UTR of its own mRNA (PubMed:7692601, PubMed:11532944, PubMed:12594214, PubMed:15282548, PubMed:18653529). Binds also to RNA ligands harboring a kissing complex (kc) structure; this binding may mediate the association of FMR1 with polyribosomes (PubMed:15805463). Binds mRNAs containing U-rich target sequences (PubMed:12927206). Binds to a triple stem-loop RNA structure, called Sod1 stem loop interacting with FMRP (SoSLIP), in the 5'-UTR region of superoxide dismutase SOD1 mRNA (PubMed:19166269). Binds to the dendritic, small non-coding brain cytoplasmic RNA 1 (BC1); which may increase the association of the CYFIP1-EIF4E-FMR1 complex to FMR1 target mRNAs at synapses (By similarity). Associates with export factor NXF1 mRNA-containing ribonucleoprotein particles (mRNPs) in a NXF2-dependent manner (By similarity). Binds to a subset of miRNAs in the brain (PubMed:14703574, PubMed:17057366). May associate with nascent transcripts in a nuclear protein NXF1-dependent manner (PubMed:18936162). In vitro, binds to RNA homopolymer; preferentially on poly(G) and to a lesser extent on poly(U), but not on poly(A) or poly(C) (PubMed:7688265, PubMed:7781595, PubMed:12950170, PubMed:15381419, PubMed:8156595). Moreover, plays a role in the modulation of the sodium-activated potassium channel KCNT1 gating activity (PubMed:20512134). Negatively regulates the voltage-dependent calcium channel current density in soma and presynaptic terminals of dorsal root ganglion (DRG) neurons, and hence regulates synaptic vesicle exocytosis (By similarity). Modulates the voltage-dependent calcium channel CACNA1B expression at the plasma membrane by targeting the channels for proteosomal degradation (By similarity). Plays a role in regulation of MAP1B-dependent microtubule dynamics during neuronal development (By similarity). Recently, has been shown to play a translation-independent role in the modulation of presynaptic action potential (AP) duration and neurotransmitter release via large-conductance calcium-activated potassium (BK) channels in hippocampal and cortical excitatory neurons (PubMed:25561520). Finally, FMR1 may be involved in the control of DNA damage response (DDR) mechanisms through the regulation of ATR-dependent signaling pathways such as histone H2AFX/H2A.x and BRCA1 phosphorylations (PubMed:24813610). ; FUNCTION: Isoform 10: binds to RNA homopolymer; preferentially on poly(G) and to a lesser extent on poly(U), but not on poly(A) or poly(C) (PubMed:24204304). May bind to RNA in Cajal bodies (PubMed:24204304). ; FUNCTION: Isoform 6: binds to RNA homopolymer; preferentially on poly(G) and to a lesser extent on poly(U), but not on poly(A) or poly(C) (PubMed:24204304). May bind to RNA in Cajal bodies (PubMed:24204304). ; FUNCTION: (Microbial infection) Acts as a positive regulator of influenza A virus (IAV) replication. Required for the assembly and nuclear export of the viral ribonucleoprotein (vRNP) components. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000375 RNA splicing, via transesterification reactions GO:0000377 RNA splicing, via transesterification reactions with bulged adenosine as nucleophile GO:0000380 alternative mRNA splicing, via spliceosome GO:0000381 regulation of alternative mRNA splicing, via spliceosome GO:0000398 mRNA splicing, via spliceosome GO:0001505 regulation of neurotransmitter levels GO:0001508 action potential GO:0002090 regulation of receptor internalization GO:0002092 positive regulation of receptor internalization GO:0002181 cytoplasmic translation GO:0006397 mRNA processing GO:0006403 RNA localization GO:0006413 translational initiation GO:0006417 regulation of translation GO:0006446 regulation of translational initiation GO:0006816 calcium ion transport GO:0006836 neurotransmitter transport GO:0006887 exocytosis GO:0006898 receptor-mediated endocytosis GO:0007215 glutamate receptor signaling pathway GO:0007269 neurotransmitter secretion GO:0008380 RNA splicing GO:0009314 response to radiation GO:0009411 response to UV GO:0009416 response to light stimulus GO:0009615 response to virus GO:0009894 regulation of catabolic process GO:0009896 positive regulation of catabolic process GO:0010035 response to inorganic substance GO:0010498 proteasomal protein catabolic process GO:0010608 posttranscriptional regulation of gene expression GO:0010720 positive regulation of cell development GO:0010959 regulation of metal ion transport GO:0010975 regulation of neuron projection development GO:0010976 positive regulation of neuron projection development GO:0015931 nucleobase-containing compound transport GO:0016079 synaptic vesicle exocytosis GO:0016358 dendrite development GO:0016441 posttranscriptional gene silencing GO:0016458 gene silencing GO:0016570 histone modification GO:0016572 histone phosphorylation GO:0017148 negative regulation of translation GO:0017156 calcium ion regulated exocytosis GO:0017157 regulation of exocytosis GO:0017158 regulation of calcium ion-dependent exocytosis GO:0019058 viral life cycle GO:0019079 viral genome replication GO:0019226 transmission of nerve impulse GO:0019228 neuronal action potential GO:0022898 regulation of transmembrane transporter activity GO:0023061 signal release GO:0030031 cell projection assembly GO:0030100 regulation of endocytosis GO:0031047 gene silencing by RNA GO:0031056 regulation of histone modification GO:0031058 positive regulation of histone modification GO:0031329 regulation of cellular catabolic process GO:0031331 positive regulation of cellular catabolic process GO:0031346 positive regulation of cell projection organization GO:0031623 receptor internalization GO:0031644 regulation of neurological system process GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0032409 regulation of transporter activity GO:0032410 negative regulation of transporter activity GO:0032412 regulation of ion transmembrane transporter activity GO:0032413 negative regulation of ion transmembrane transporter activity GO:0033127 regulation of histone phosphorylation GO:0033129 positive regulation of histone phosphorylation GO:0034248 regulation of cellular amide metabolic process GO:0034249 negative regulation of cellular amide metabolic process GO:0034250 positive regulation of cellular amide metabolic process GO:0034644 cellular response to UV GO:0034762 regulation of transmembrane transport GO:0034763 negative regulation of transmembrane transport GO:0034765 regulation of ion transmembrane transport GO:0034766 negative regulation of ion transmembrane transport GO:0035194 posttranscriptional gene silencing by RNA GO:0035195 gene silencing by miRNA GO:0035637 multicellular organismal signaling GO:0035821 modification of morphology or physiology of other organism GO:0039694 viral RNA genome replication GO:0039703 RNA replication GO:0040029 regulation of gene expression, epigenetic GO:0042176 regulation of protein catabolic process GO:0042391 regulation of membrane potential GO:0043112 receptor metabolic process GO:0043271 negative regulation of ion transport GO:0043484 regulation of RNA splicing GO:0043487 regulation of RNA stability GO:0043488 regulation of mRNA stability GO:0043900 regulation of multi-organism process GO:0043902 positive regulation of multi-organism process GO:0043903 regulation of symbiosis, encompassing mutualism through parasitism GO:0044033 multi-organism metabolic process GO:0044057 regulation of system process GO:0044089 positive regulation of cellular component biogenesis GO:0044766 multi-organism transport GO:0044788 modulation by host of viral process GO:0044827 modulation by host of viral genome replication GO:0044830 modulation by host of viral RNA genome replication GO:0045055 regulated exocytosis GO:0045069 regulation of viral genome replication GO:0045666 positive regulation of neuron differentiation GO:0045727 positive regulation of translation GO:0045732 positive regulation of protein catabolic process GO:0045807 positive regulation of endocytosis GO:0045862 positive regulation of proteolysis GO:0045920 negative regulation of exocytosis GO:0045947 negative regulation of translational initiation GO:0045955 negative regulation of calcium ion-dependent exocytosis GO:0046794 transport of virus GO:0046847 filopodium assembly GO:0046928 regulation of neurotransmitter secretion GO:0046929 negative regulation of neurotransmitter secretion GO:0048024 regulation of mRNA splicing, via spliceosome GO:0048167 regulation of synaptic plasticity GO:0048259 regulation of receptor-mediated endocytosis GO:0048260 positive regulation of receptor-mediated endocytosis GO:0048489 synaptic vesicle transport GO:0048524 positive regulation of viral process GO:0050657 nucleic acid transport GO:0050658 RNA transport GO:0050684 regulation of mRNA processing GO:0050769 positive regulation of neurogenesis GO:0050773 regulation of dendrite development GO:0050792 regulation of viral process GO:0050804 modulation of synaptic transmission GO:0050805 negative regulation of synaptic transmission GO:0050806 positive regulation of synaptic transmission GO:0051028 mRNA transport GO:0051048 negative regulation of secretion GO:0051051 negative regulation of transport GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051236 establishment of RNA localization GO:0051489 regulation of filopodium assembly GO:0051491 positive regulation of filopodium assembly GO:0051588 regulation of neurotransmitter transport GO:0051589 negative regulation of neurotransmitter transport GO:0051640 organelle localization GO:0051648 vesicle localization GO:0051650 establishment of vesicle localization GO:0051656 establishment of organelle localization GO:0051702 interaction with symbiont GO:0051817 modification of morphology or physiology of other organism involved in symbiotic interaction GO:0051851 modification by host of symbiont morphology or physiology GO:0051924 regulation of calcium ion transport GO:0051926 negative regulation of calcium ion transport GO:0051962 positive regulation of nervous system development GO:0051969 regulation of transmission of nerve impulse GO:0060147 regulation of posttranscriptional gene silencing GO:0060148 positive regulation of posttranscriptional gene silencing GO:0060292 long term synaptic depression GO:0060491 regulation of cell projection assembly GO:0060627 regulation of vesicle-mediated transport GO:0060964 regulation of gene silencing by miRNA GO:0060966 regulation of gene silencing by RNA GO:0060968 regulation of gene silencing GO:0060996 dendritic spine development GO:0060998 regulation of dendritic spine development GO:0060999 positive regulation of dendritic spine development GO:0061136 regulation of proteasomal protein catabolic process GO:0070588 calcium ion transmembrane transport GO:0070838 divalent metal ion transport GO:0071214 cellular response to abiotic stimulus GO:0071241 cellular response to inorganic substance GO:0071478 cellular response to radiation GO:0071482 cellular response to light stimulus GO:0072511 divalent inorganic cation transport GO:0072710 response to hydroxyurea GO:0072711 cellular response to hydroxyurea GO:0075733 intracellular transport of virus GO:0097479 synaptic vesicle localization GO:0097480 establishment of synaptic vesicle localization GO:0098586 cellular response to virus GO:0098900 regulation of action potential GO:0098908 regulation of neuronal action potential GO:0099003 vesicle-mediated transport in synapse GO:0099504 synaptic vesicle cycle GO:0099531 presynaptic process involved in chemical synaptic transmission GO:0099643 signal release from synapse GO:1900006 positive regulation of dendrite development GO:1900452 regulation of long term synaptic depression GO:1900453 negative regulation of long term synaptic depression GO:1901019 regulation of calcium ion transmembrane transporter activity GO:1901020 negative regulation of calcium ion transmembrane transporter activity GO:1901252 regulation of intracellular transport of viral material GO:1901254 positive regulation of intracellular transport of viral material GO:1901385 regulation of voltage-gated calcium channel activity GO:1901386 negative regulation of voltage-gated calcium channel activity GO:1901800 positive regulation of proteasomal protein catabolic process GO:1902275 regulation of chromatin organization GO:1902415 regulation of mRNA binding GO:1902416 positive regulation of mRNA binding GO:1902579 multi-organism localization GO:1902581 multi-organism cellular localization GO:1902583 multi-organism intracellular transport GO:1902803 regulation of synaptic vesicle transport GO:1902804 negative regulation of synaptic vesicle transport GO:1903050 regulation of proteolysis involved in cellular protein catabolic process GO:1903052 positive regulation of proteolysis involved in cellular protein catabolic process GO:1903169 regulation of calcium ion transmembrane transport GO:1903170 negative regulation of calcium ion transmembrane transport GO:1903305 regulation of regulated secretory pathway GO:1903306 negative regulation of regulated secretory pathway GO:1903311 regulation of mRNA metabolic process GO:1903362 regulation of cellular protein catabolic process GO:1903364 positive regulation of cellular protein catabolic process GO:1903531 negative regulation of secretion by cell GO:1903900 regulation of viral life cycle GO:1903902 positive regulation of viral life cycle GO:1904062 regulation of cation transmembrane transport GO:1904063 negative regulation of cation transmembrane transport GO:1905214 regulation of RNA binding GO:1905216 positive regulation of RNA binding GO:1905269 positive regulation of chromatin organization GO:2000300 regulation of synaptic vesicle exocytosis GO:2000301 negative regulation of synaptic vesicle exocytosis GO:2000637 positive regulation of gene silencing by miRNA GO:2000765 regulation of cytoplasmic translation GO:2000766 negative regulation of cytoplasmic translation GO:2001020 regulation of response to DNA damage stimulus GO:2001022 positive regulation of response to DNA damage stimulus GO:2001257 regulation of cation channel activity GO:2001258 negative regulation of cation channel activity |
| Molecular Function |
GO:0002151 G-quadruplex RNA binding GO:0003682 chromatin binding GO:0003725 double-stranded RNA binding GO:0003727 single-stranded RNA binding GO:0003729 mRNA binding GO:0003730 mRNA 3'-UTR binding GO:0008017 microtubule binding GO:0008187 poly-pyrimidine tract binding GO:0008266 poly(U) RNA binding GO:0015631 tubulin binding GO:0030371 translation repressor activity GO:0031369 translation initiation factor binding GO:0033592 RNA strand annealing activity GO:0034046 poly(G) binding GO:0035064 methylated histone binding GO:0035197 siRNA binding GO:0035198 miRNA binding GO:0035613 RNA stem-loop binding GO:0042393 histone binding GO:0043021 ribonucleoprotein complex binding GO:0043022 ribosome binding GO:0044325 ion channel binding GO:0045182 translation regulator activity GO:0045502 dynein binding GO:0046982 protein heterodimerization activity GO:0048027 mRNA 5'-UTR binding GO:0070717 poly-purine tract binding GO:0097617 annealing activity GO:1990825 sequence-specific mRNA binding |
| Cellular Component |
GO:0000775 chromosome, centromeric region GO:0005840 ribosome GO:0005844 polysome GO:0005845 mRNA cap binding complex GO:0010369 chromocenter GO:0014069 postsynaptic density GO:0015030 Cajal body GO:0016604 nuclear body GO:0018995 host GO:0019034 viral replication complex GO:0019897 extrinsic component of plasma membrane GO:0019898 extrinsic component of membrane GO:0030175 filopodium GO:0030424 axon GO:0030425 dendrite GO:0030426 growth cone GO:0030427 site of polarized growth GO:0032433 filopodium tip GO:0032797 SMN complex GO:0032838 cell projection cytoplasm GO:0033267 axon part GO:0033643 host cell part GO:0033646 host intracellular part GO:0033647 host intracellular organelle GO:0033648 host intracellular membrane-bounded organelle GO:0034518 RNA cap binding complex GO:0035770 ribonucleoprotein granule GO:0036464 cytoplasmic ribonucleoprotein granule GO:0042025 host cell nucleus GO:0042734 presynaptic membrane GO:0042788 polysomal ribosome GO:0043025 neuronal cell body GO:0043197 dendritic spine GO:0043204 perikaryon GO:0043656 intracellular region of host GO:0043657 host cell GO:0043679 axon terminus GO:0044094 host cell nuclear part GO:0044215 other organism GO:0044216 other organism cell GO:0044217 other organism part GO:0044297 cell body GO:0044306 neuron projection terminus GO:0044309 neuron spine GO:0045211 postsynaptic membrane GO:0060076 excitatory synapse GO:0071598 neuronal ribonucleoprotein granule GO:0097060 synaptic membrane GO:0097386 glial cell projection GO:0098687 chromosomal region GO:0098793 presynapse GO:0098794 postsynapse GO:0098858 actin-based cell projection GO:0099568 cytoplasmic region GO:0099572 postsynaptic specialization GO:1902737 dendritic filopodium GO:1990812 growth cone filopodium |
| KEGG |
hsa03013 RNA transport |
| Reactome | - |
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for FMR1. |
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
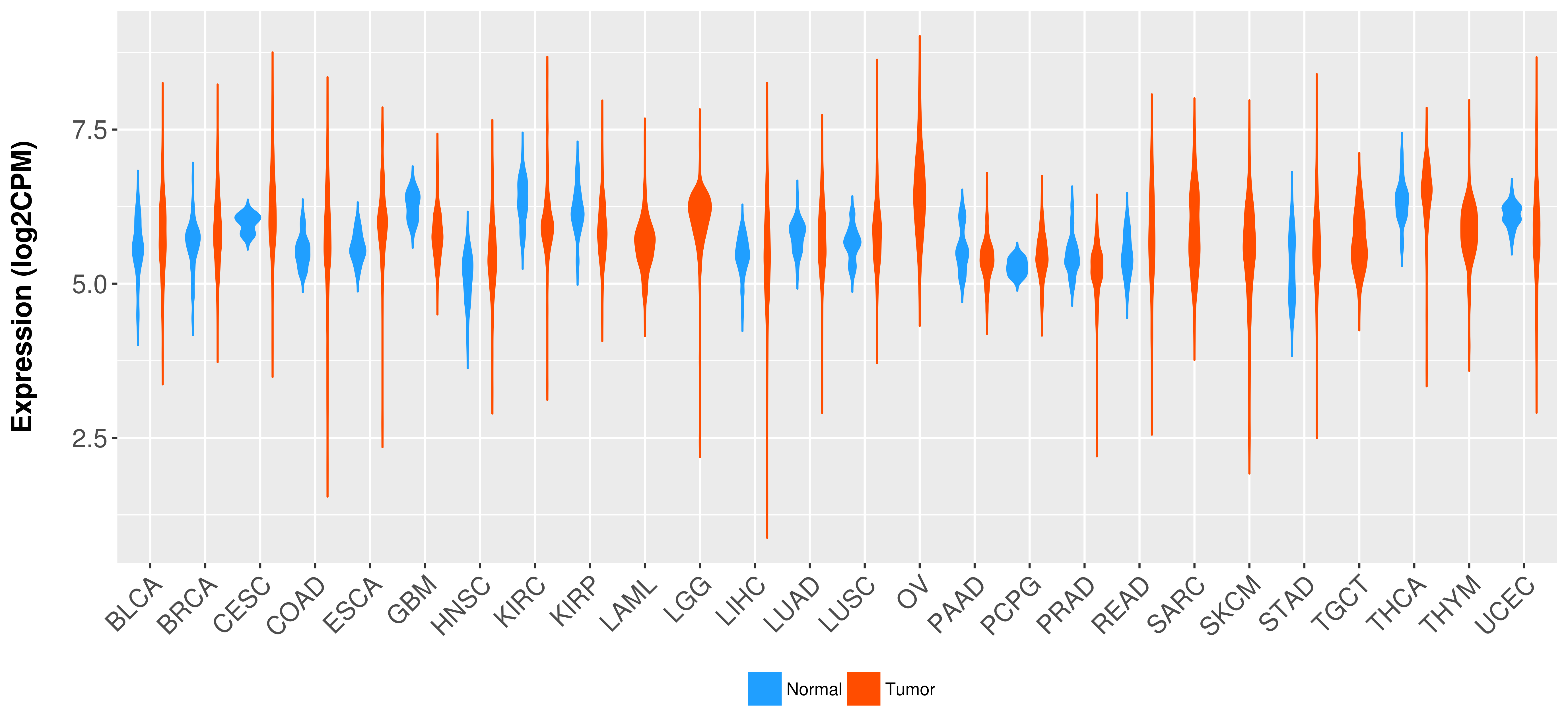
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
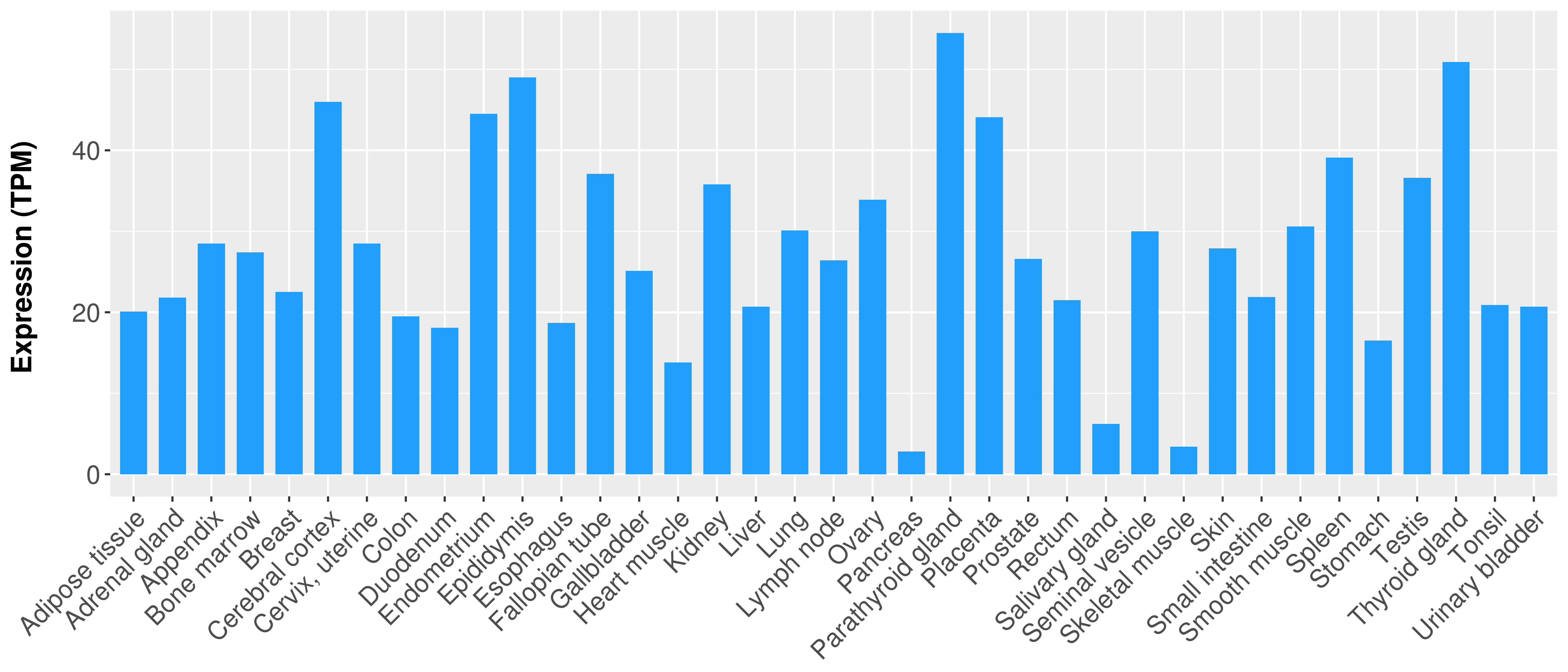
|
| There is no record. |
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
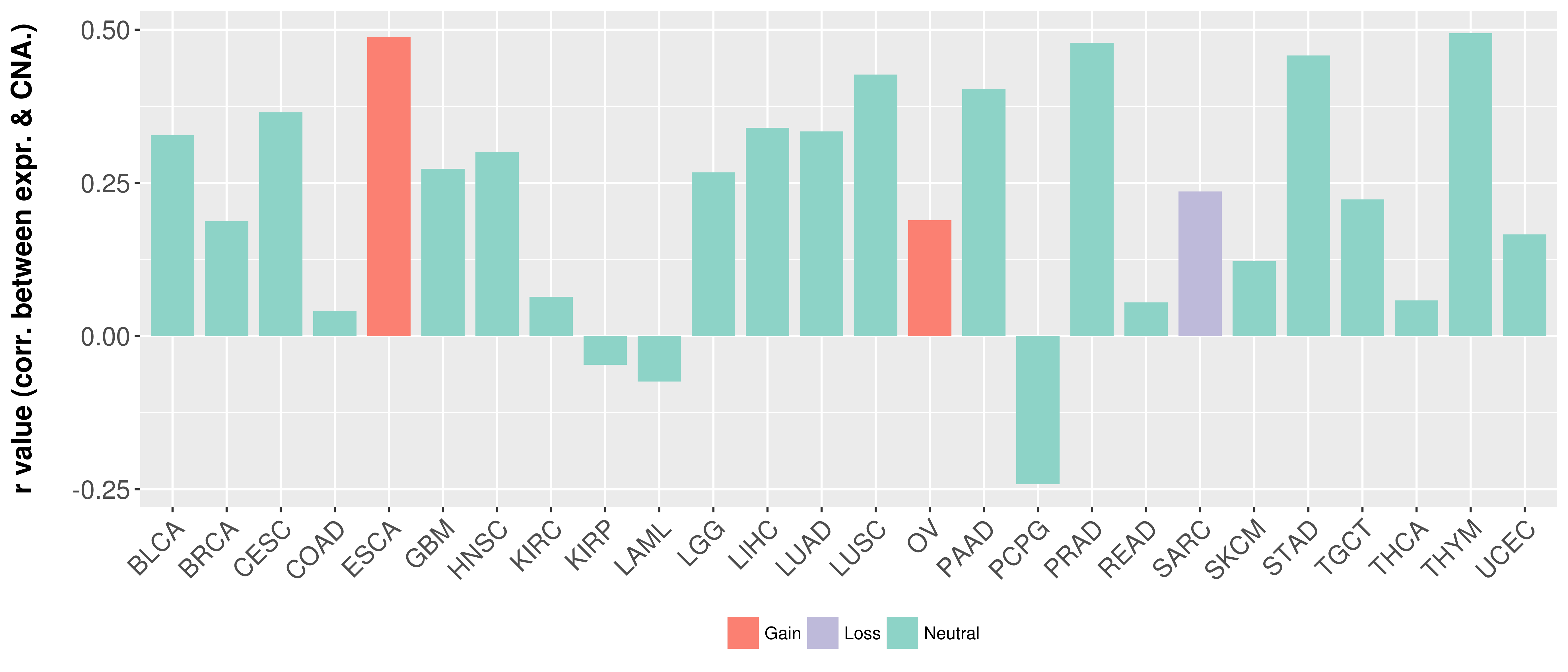
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
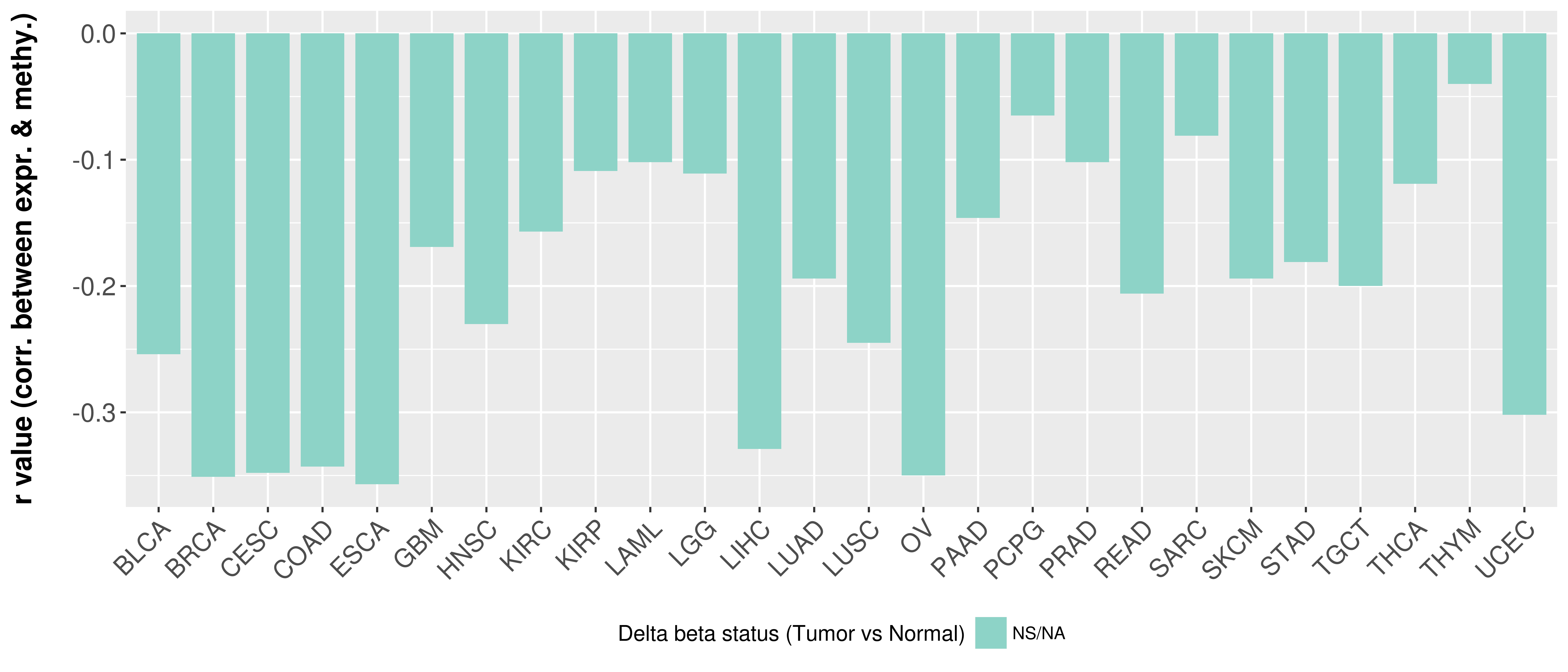
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
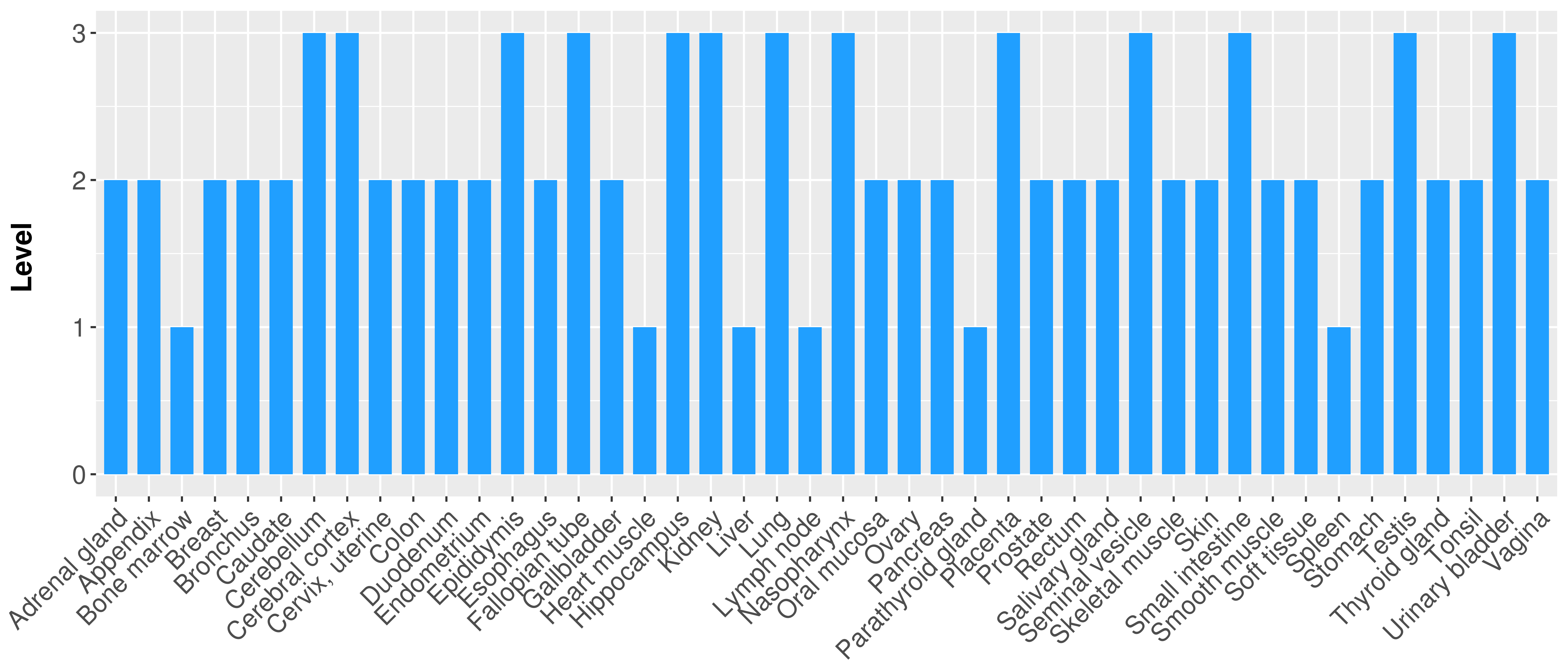
|
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
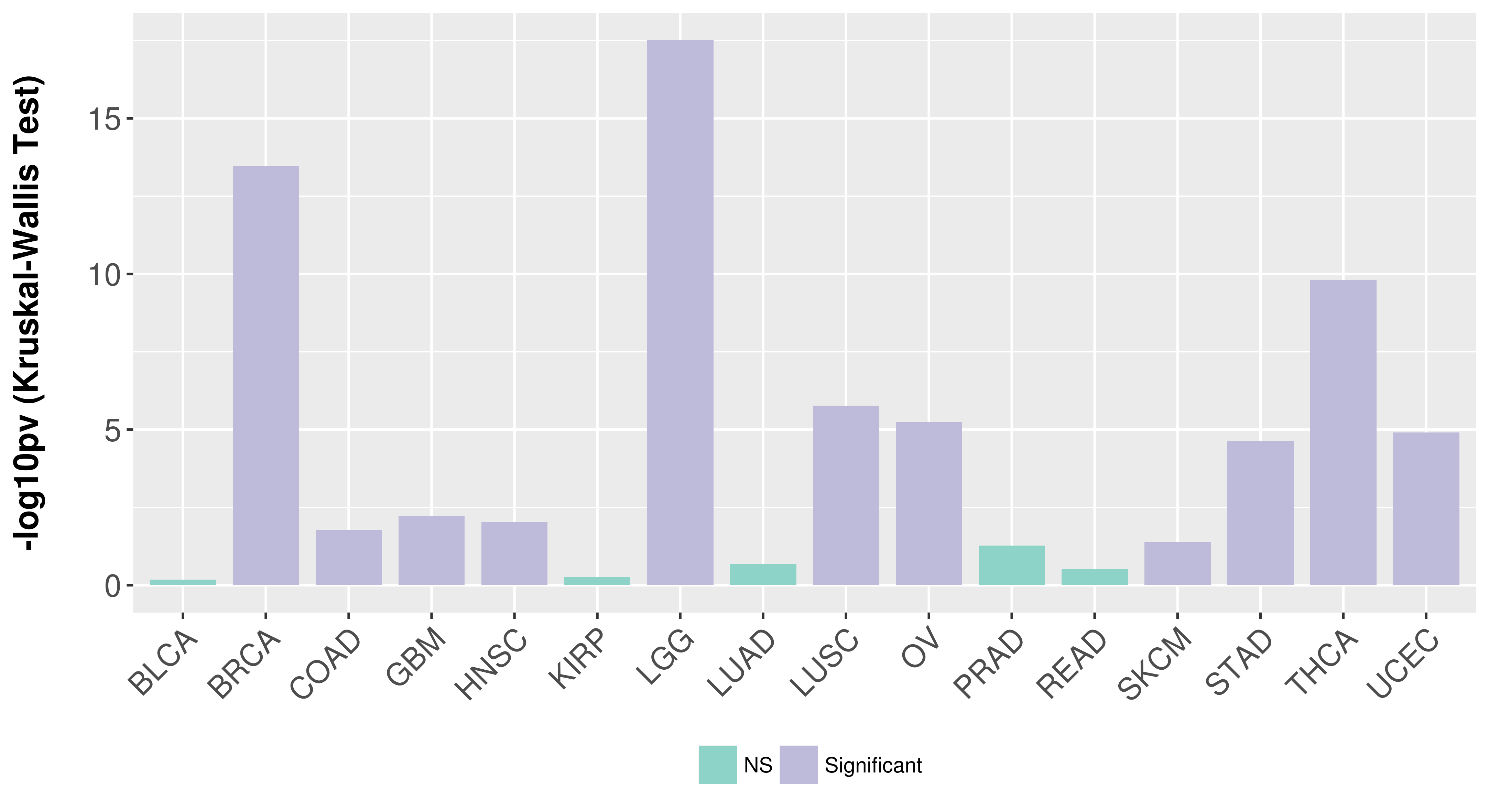
Association between expresson and subtype.
|
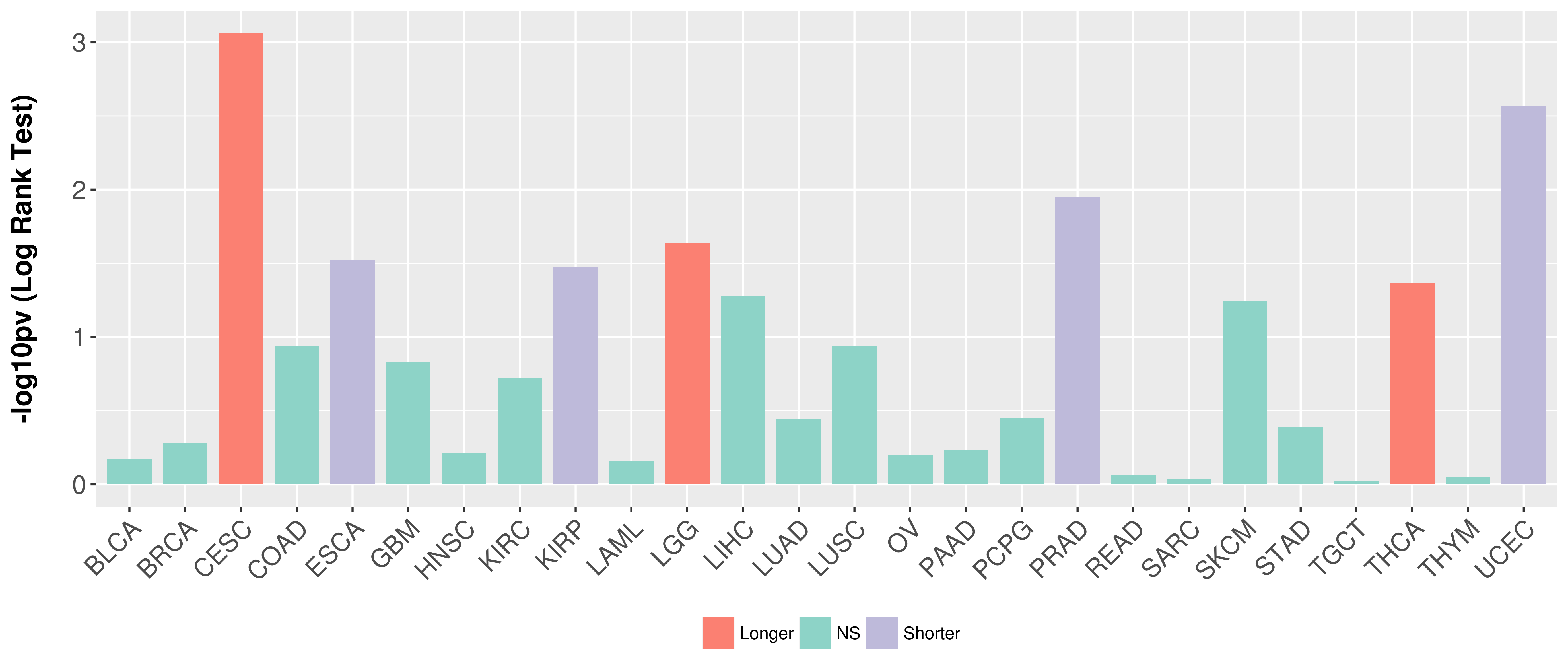
Overall survival analysis based on expression.
|
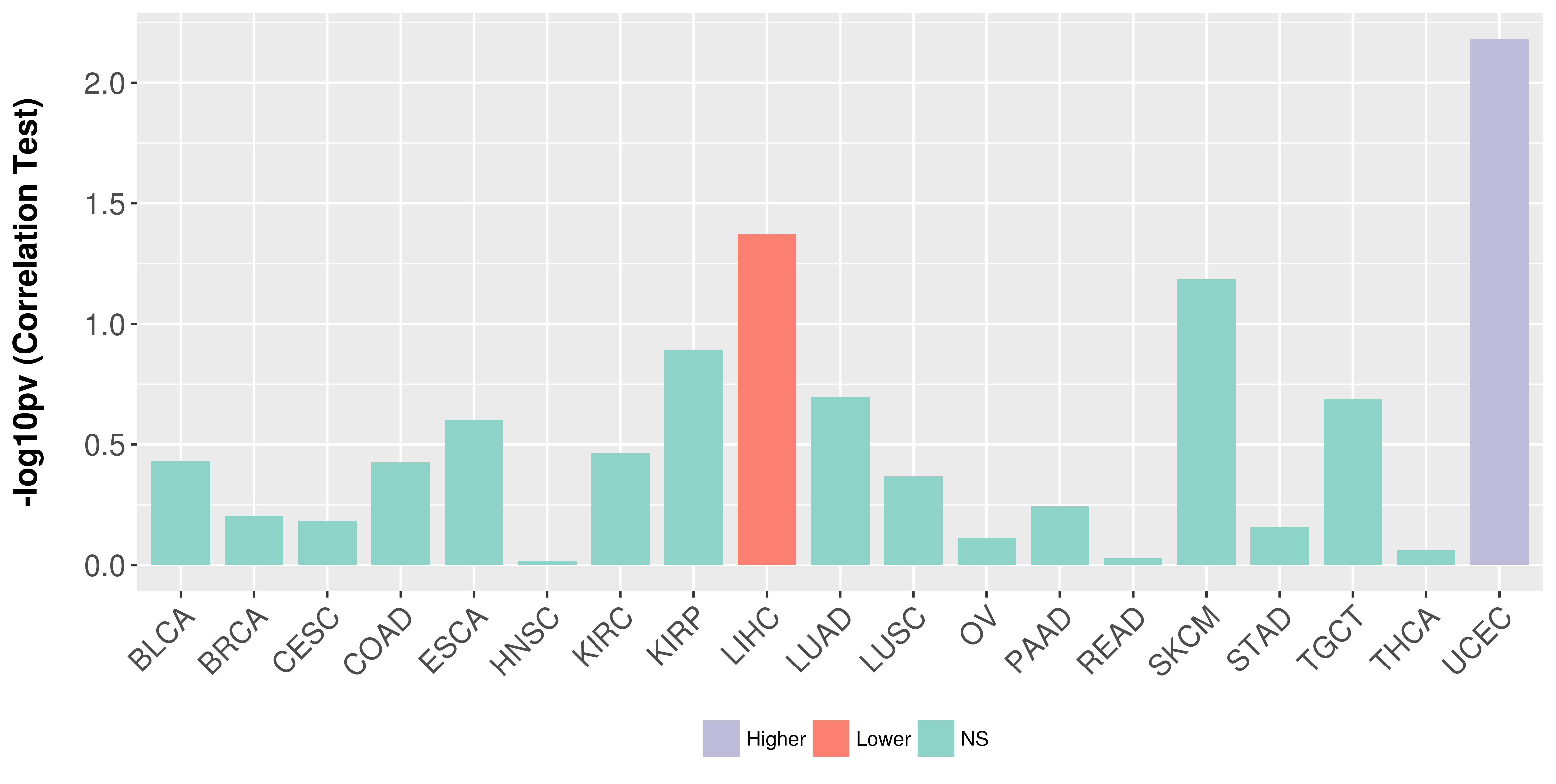
Association between expresson and stage.
|
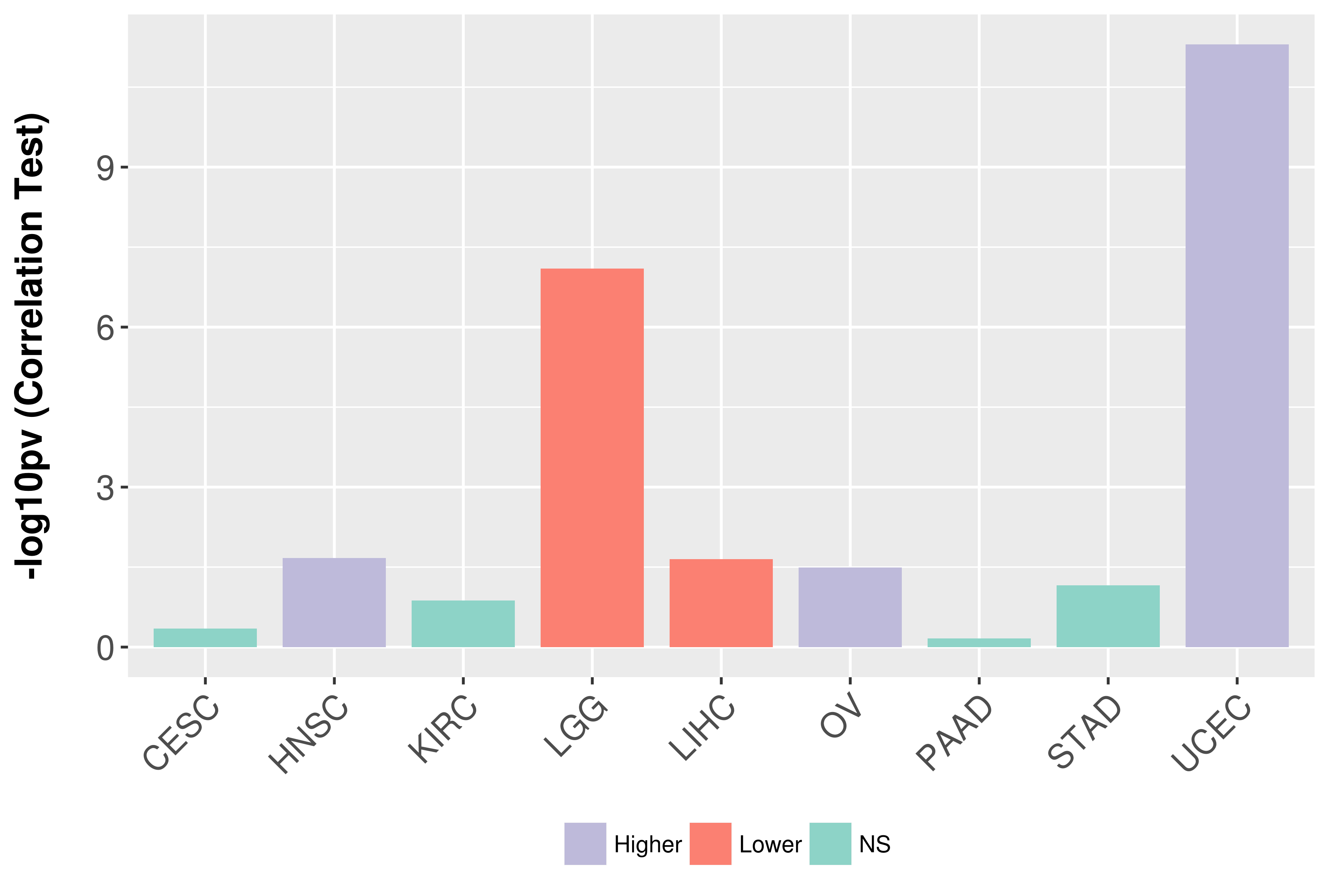
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Drugs from DrugBank database |
| There is no record for FMR1. |
| Summary | |
|---|---|
| Symbol | FMR1 |
| Name | fragile X mental retardation 1 |
| Aliases | FMRP; MGC87458; POF1; premature ovarian failure 1; Protein FMR-1; Fragile X mental retardation protein 1 |
| Location | Xq27.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for FMR1. |