Browse HDAC1 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00850 Histone deacetylase domain |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Deacetylates SP proteins, SP1 and SP3, and regulates their function. Component of the BRG1-RB1-HDAC1 complex, which negatively regulates the CREST-mediated transcription in resting neurons. Upon calcium stimulation, HDAC1 is released from the complex and CREBBP is recruited, which facilitates transcriptional activation. Deacetylates TSHZ3 and regulates its transcriptional repressor activity. Deacetylates 'Lys-310' in RELA and thereby inhibits the transcriptional activity of NF-kappa-B. Deacetylates NR1D2 and abrogates the effect of KAT5-mediated relieving of NR1D2 transcription repression activity. Component of a RCOR/GFI/KDM1A/HDAC complex that suppresses, via histone deacetylase (HDAC) recruitment, a number of genes implicated in multilineage blood cell development. Involved in CIART-mediated transcriptional repression of the circadian transcriptional activator: CLOCK-ARNTL/BMAL1 heterodimer. Required for the transcriptional repression of circadian target genes, such as PER1, mediated by the large PER complex or CRY1 through histone deacetylation. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001654 eye development GO:0001819 positive regulation of cytokine production GO:0001942 hair follicle development GO:0001975 response to amphetamine GO:0002237 response to molecule of bacterial origin GO:0002790 peptide secretion GO:0002791 regulation of peptide secretion GO:0002792 negative regulation of peptide secretion GO:0006338 chromatin remodeling GO:0006342 chromatin silencing GO:0006346 methylation-dependent chromatin silencing GO:0006473 protein acetylation GO:0006476 protein deacetylation GO:0006979 response to oxidative stress GO:0007259 JAK-STAT cascade GO:0007260 tyrosine phosphorylation of STAT protein GO:0007423 sensory organ development GO:0007596 blood coagulation GO:0007599 hemostasis GO:0007623 circadian rhythm GO:0008544 epidermis development GO:0009306 protein secretion GO:0009755 hormone-mediated signaling pathway GO:0009913 epidermal cell differentiation GO:0009914 hormone transport GO:0010001 glial cell differentiation GO:0010720 positive regulation of cell development GO:0010817 regulation of hormone levels GO:0010869 regulation of receptor biosynthetic process GO:0010870 positive regulation of receptor biosynthetic process GO:0014013 regulation of gliogenesis GO:0014015 positive regulation of gliogenesis GO:0014074 response to purine-containing compound GO:0014075 response to amine GO:0015833 peptide transport GO:0016055 Wnt signaling pathway GO:0016458 gene silencing GO:0016570 histone modification GO:0016575 histone deacetylation GO:0018108 peptidyl-tyrosine phosphorylation GO:0018205 peptidyl-lysine modification GO:0018212 peptidyl-tyrosine modification GO:0018394 peptidyl-lysine acetylation GO:0019058 viral life cycle GO:0019080 viral gene expression GO:0019083 viral transcription GO:0022404 molting cycle process GO:0022405 hair cycle process GO:0023061 signal release GO:0030072 peptide hormone secretion GO:0030073 insulin secretion GO:0030111 regulation of Wnt signaling pathway GO:0030178 negative regulation of Wnt signaling pathway GO:0030326 embryonic limb morphogenesis GO:0030518 intracellular steroid hormone receptor signaling pathway GO:0030521 androgen receptor signaling pathway GO:0030522 intracellular receptor signaling pathway GO:0031000 response to caffeine GO:0032496 response to lipopolysaccharide GO:0032602 chemokine production GO:0032612 interleukin-1 production GO:0032640 tumor necrosis factor production GO:0032642 regulation of chemokine production GO:0032652 regulation of interleukin-1 production GO:0032680 regulation of tumor necrosis factor production GO:0032722 positive regulation of chemokine production GO:0032732 positive regulation of interleukin-1 production GO:0032760 positive regulation of tumor necrosis factor production GO:0032800 receptor biosynthetic process GO:0032897 negative regulation of viral transcription GO:0032922 circadian regulation of gene expression GO:0033143 regulation of intracellular steroid hormone receptor signaling pathway GO:0033144 negative regulation of intracellular steroid hormone receptor signaling pathway GO:0034599 cellular response to oxidative stress GO:0034612 response to tumor necrosis factor GO:0035107 appendage morphogenesis GO:0035108 limb morphogenesis GO:0035113 embryonic appendage morphogenesis GO:0035601 protein deacylation GO:0035821 modification of morphology or physiology of other organism GO:0036296 response to increased oxygen levels GO:0040029 regulation of gene expression, epigenetic GO:0042063 gliogenesis GO:0042303 molting cycle GO:0042475 odontogenesis of dentin-containing tooth GO:0042476 odontogenesis GO:0042493 response to drug GO:0042503 tyrosine phosphorylation of Stat3 protein GO:0042509 regulation of tyrosine phosphorylation of STAT protein GO:0042516 regulation of tyrosine phosphorylation of Stat3 protein GO:0042517 positive regulation of tyrosine phosphorylation of Stat3 protein GO:0042531 positive regulation of tyrosine phosphorylation of STAT protein GO:0042633 hair cycle GO:0042733 embryonic digit morphogenesis GO:0042886 amide transport GO:0043010 camera-type eye development GO:0043044 ATP-dependent chromatin remodeling GO:0043112 receptor metabolic process GO:0043279 response to alkaloid GO:0043401 steroid hormone mediated signaling pathway GO:0043523 regulation of neuron apoptotic process GO:0043524 negative regulation of neuron apoptotic process GO:0043543 protein acylation GO:0043586 tongue development GO:0043587 tongue morphogenesis GO:0043588 skin development GO:0043900 regulation of multi-organism process GO:0043901 negative regulation of multi-organism process GO:0043903 regulation of symbiosis, encompassing mutualism through parasitism GO:0043921 modulation by host of viral transcription GO:0043922 negative regulation by host of viral transcription GO:0044033 multi-organism metabolic process GO:0045685 regulation of glial cell differentiation GO:0045687 positive regulation of glial cell differentiation GO:0045814 negative regulation of gene expression, epigenetic GO:0046425 regulation of JAK-STAT cascade GO:0046427 positive regulation of JAK-STAT cascade GO:0046676 negative regulation of insulin secretion GO:0046782 regulation of viral transcription GO:0046879 hormone secretion GO:0046883 regulation of hormone secretion GO:0046888 negative regulation of hormone secretion GO:0048511 rhythmic process GO:0048525 negative regulation of viral process GO:0048545 response to steroid hormone GO:0048709 oligodendrocyte differentiation GO:0048713 regulation of oligodendrocyte differentiation GO:0048714 positive regulation of oligodendrocyte differentiation GO:0048736 appendage development GO:0050708 regulation of protein secretion GO:0050709 negative regulation of protein secretion GO:0050730 regulation of peptidyl-tyrosine phosphorylation GO:0050731 positive regulation of peptidyl-tyrosine phosphorylation GO:0050769 positive regulation of neurogenesis GO:0050792 regulation of viral process GO:0050796 regulation of insulin secretion GO:0050817 coagulation GO:0050878 regulation of body fluid levels GO:0051048 negative regulation of secretion GO:0051051 negative regulation of transport GO:0051224 negative regulation of protein transport GO:0051402 neuron apoptotic process GO:0051702 interaction with symbiont GO:0051817 modification of morphology or physiology of other organism involved in symbiotic interaction GO:0051851 modification by host of symbiont morphology or physiology GO:0051962 positive regulation of nervous system development GO:0052312 modulation of transcription in other organism involved in symbiotic interaction GO:0052472 modulation by host of symbiont transcription GO:0055093 response to hyperoxia GO:0060070 canonical Wnt signaling pathway GO:0060173 limb development GO:0060765 regulation of androgen receptor signaling pathway GO:0060766 negative regulation of androgen receptor signaling pathway GO:0060788 ectodermal placode formation GO:0060789 hair follicle placode formation GO:0060828 regulation of canonical Wnt signaling pathway GO:0061029 eyelid development in camera-type eye GO:0061196 fungiform papilla development GO:0061197 fungiform papilla morphogenesis GO:0061198 fungiform papilla formation GO:0070482 response to oxygen levels GO:0070932 histone H3 deacetylation GO:0070933 histone H4 deacetylation GO:0070997 neuron death GO:0071356 cellular response to tumor necrosis factor GO:0071383 cellular response to steroid hormone stimulus GO:0071396 cellular response to lipid GO:0071407 cellular response to organic cyclic compound GO:0071696 ectodermal placode development GO:0071697 ectodermal placode morphogenesis GO:0071706 tumor necrosis factor superfamily cytokine production GO:0072331 signal transduction by p53 class mediator GO:0072567 chemokine (C-X-C motif) ligand 2 production GO:0090087 regulation of peptide transport GO:0090090 negative regulation of canonical Wnt signaling pathway GO:0090276 regulation of peptide hormone secretion GO:0090278 negative regulation of peptide hormone secretion GO:0090596 sensory organ morphogenesis GO:0097050 type B pancreatic cell apoptotic process GO:0097696 STAT cascade GO:0098732 macromolecule deacylation GO:0098773 skin epidermis development GO:0198738 cell-cell signaling by wnt GO:1901214 regulation of neuron death GO:1901215 negative regulation of neuron death GO:1901796 regulation of signal transduction by p53 class mediator GO:1901983 regulation of protein acetylation GO:1901984 negative regulation of protein acetylation GO:1903531 negative regulation of secretion by cell GO:1903555 regulation of tumor necrosis factor superfamily cytokine production GO:1903557 positive regulation of tumor necrosis factor superfamily cytokine production GO:1903900 regulation of viral life cycle GO:1903901 negative regulation of viral life cycle GO:1904019 epithelial cell apoptotic process GO:1904035 regulation of epithelial cell apoptotic process GO:1904037 positive regulation of epithelial cell apoptotic process GO:1904837 beta-catenin-TCF complex assembly GO:1904892 regulation of STAT cascade GO:1904894 positive regulation of STAT cascade GO:1904950 negative regulation of establishment of protein localization GO:2000341 regulation of chemokine (C-X-C motif) ligand 2 production GO:2000343 positive regulation of chemokine (C-X-C motif) ligand 2 production GO:2000674 regulation of type B pancreatic cell apoptotic process GO:2000676 positive regulation of type B pancreatic cell apoptotic process GO:2000756 regulation of peptidyl-lysine acetylation GO:2000757 negative regulation of peptidyl-lysine acetylation |
| Molecular Function |
GO:0000978 RNA polymerase II core promoter proximal region sequence-specific DNA binding GO:0000980 RNA polymerase II distal enhancer sequence-specific DNA binding GO:0000987 core promoter proximal region sequence-specific DNA binding GO:0001047 core promoter binding GO:0001076 transcription factor activity, RNA polymerase II transcription factor binding GO:0001085 RNA polymerase II transcription factor binding GO:0001103 RNA polymerase II repressing transcription factor binding GO:0001104 RNA polymerase II transcription cofactor activity GO:0001106 RNA polymerase II transcription corepressor activity GO:0001158 enhancer sequence-specific DNA binding GO:0001159 core promoter proximal region DNA binding GO:0001191 transcriptional repressor activity, RNA polymerase II transcription factor binding GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0031490 chromatin DNA binding GO:0031491 nucleosome binding GO:0031492 nucleosomal DNA binding GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0033558 protein deacetylase activity GO:0033613 activating transcription factor binding GO:0034979 NAD-dependent protein deacetylase activity GO:0035326 enhancer binding GO:0042826 histone deacetylase binding GO:0043566 structure-specific DNA binding GO:0047485 protein N-terminus binding GO:0051059 NF-kappaB binding GO:0070491 repressing transcription factor binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0000785 chromatin GO:0000790 nuclear chromatin GO:0016580 Sin3 complex GO:0016581 NuRD complex GO:0017053 transcriptional repressor complex GO:0044454 nuclear chromosome part GO:0070603 SWI/SNF superfamily-type complex GO:0070822 Sin3-type complex GO:0090545 CHD-type complex GO:0090568 nuclear transcriptional repressor complex |
| KEGG |
hsa04110 Cell cycle hsa04330 Notch signaling pathway hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-3769402: Deactivation of the beta-catenin transactivating complex R-HSA-195253: Degradation of beta-catenin by the destruction complex R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-2173795: Downregulation of SMAD2/3 R-HSA-427389: ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression R-HSA-212165: Epigenetic regulation of gene expression R-HSA-983231: Factors involved in megakaryocyte development and platelet production R-HSA-201722: Formation of the beta-catenin R-HSA-1538133: G0 and Early G1 R-HSA-74160: Gene Expression R-HSA-212436: Generic Transcription Pathway R-HSA-3214815: HDACs deacetylate histones R-HSA-109582: Hemostasis R-HSA-453279: Mitotic G1-G1/S phases R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-5250941: Negative epigenetic regulation of rRNA expression R-HSA-427413: NoRC negatively regulates rRNA expression R-HSA-5250913: Positive epigenetic regulation of rRNA expression R-HSA-73854: RNA Polymerase I Promoter Clearance R-HSA-73864: RNA Polymerase I Transcription R-HSA-73762: RNA Polymerase I Transcription Initiation R-HSA-504046: RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription R-HSA-5633007: Regulation of TP53 Activity R-HSA-6804758: Regulation of TP53 Activity through Acetylation R-HSA-4641265: Repression of WNT target genes R-HSA-2173796: SMAD2/SMAD3 R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer R-HSA-170834: Signaling by TGF-beta Receptor Complex R-HSA-195721: Signaling by Wnt R-HSA-166520: Signalling by NGF R-HSA-201681: TCF dependent signaling in response to WNT R-HSA-3700989: Transcriptional Regulation by TP53 R-HSA-2173793: Transcriptional activity of SMAD2/SMAD3 R-HSA-193704: p75 NTR receptor-mediated signalling R-HSA-193670: p75NTR negatively regulates cell cycle via SC1 |
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
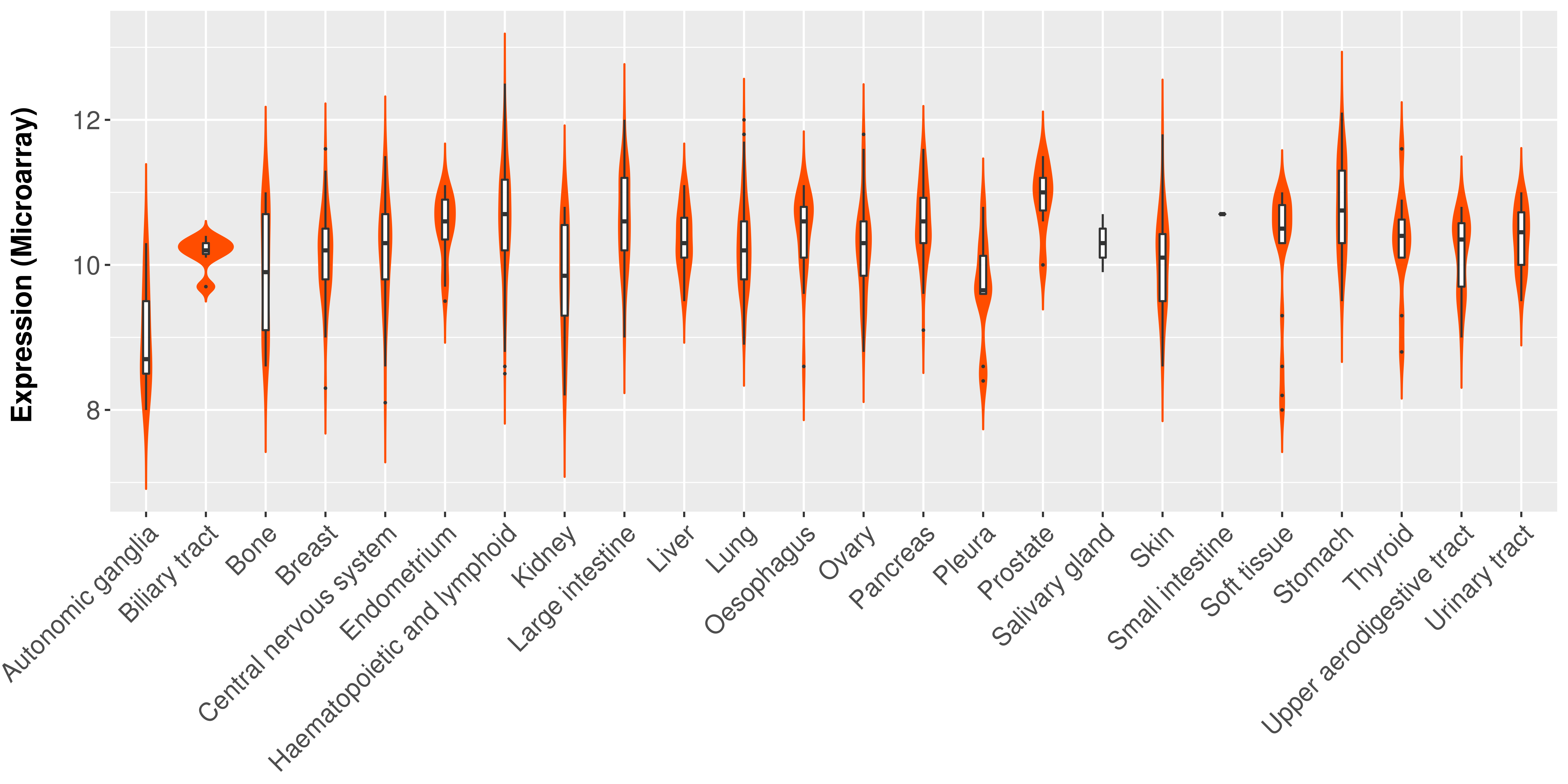
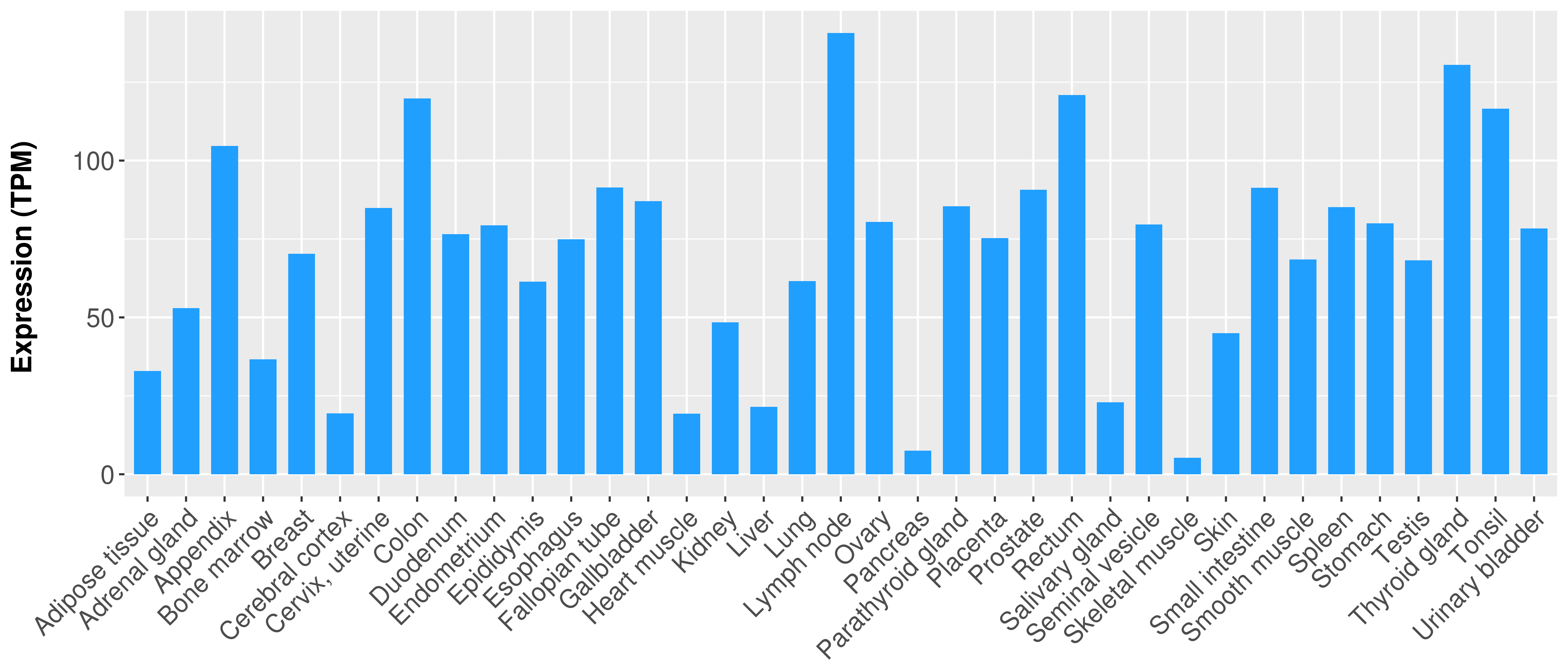
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
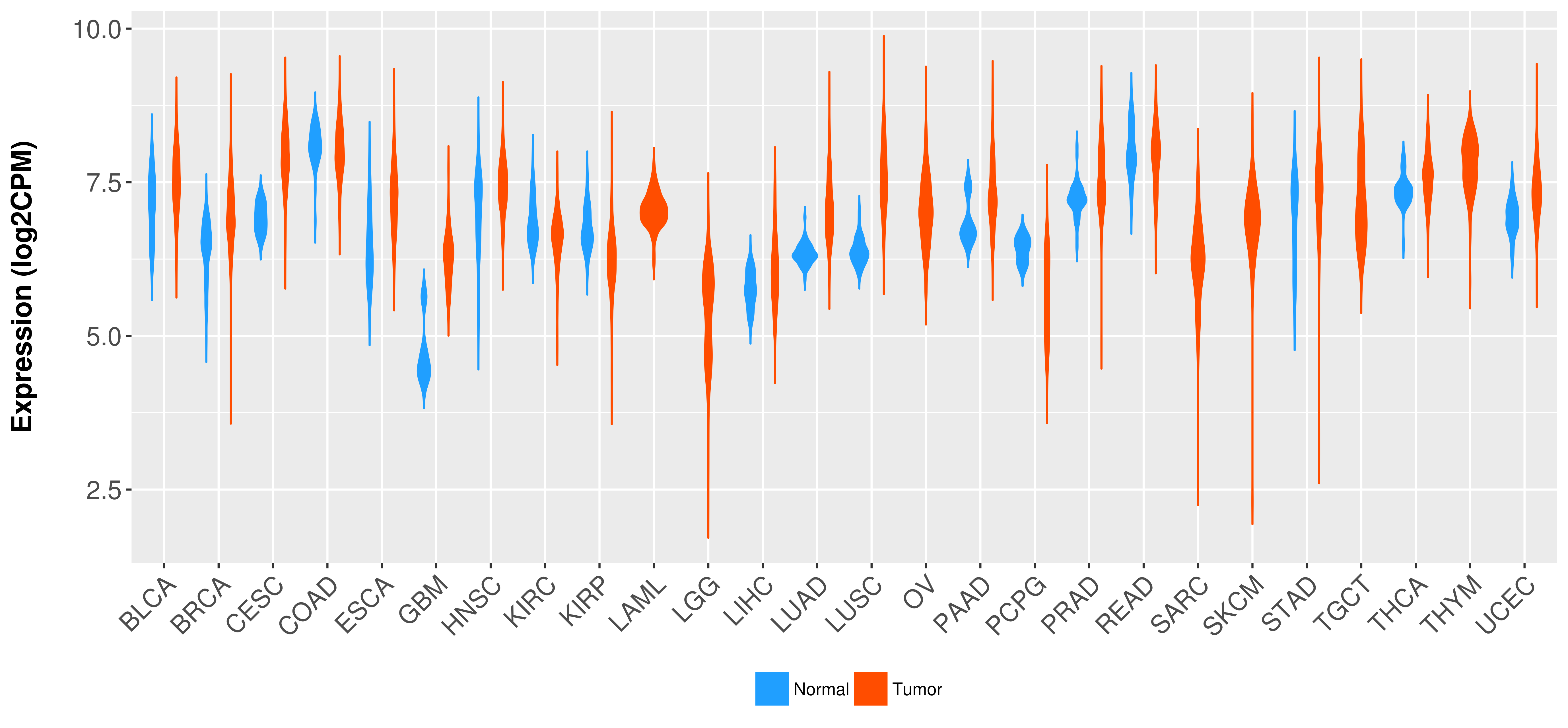
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
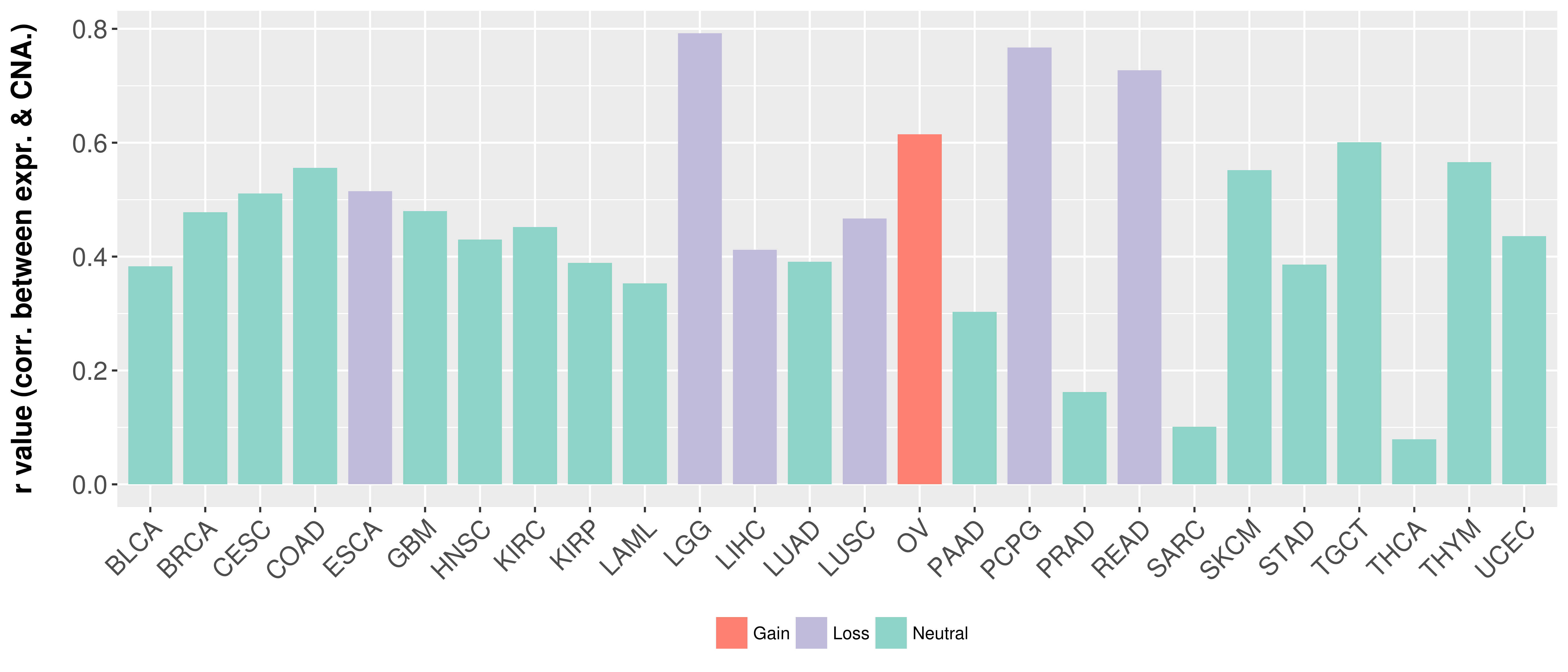
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
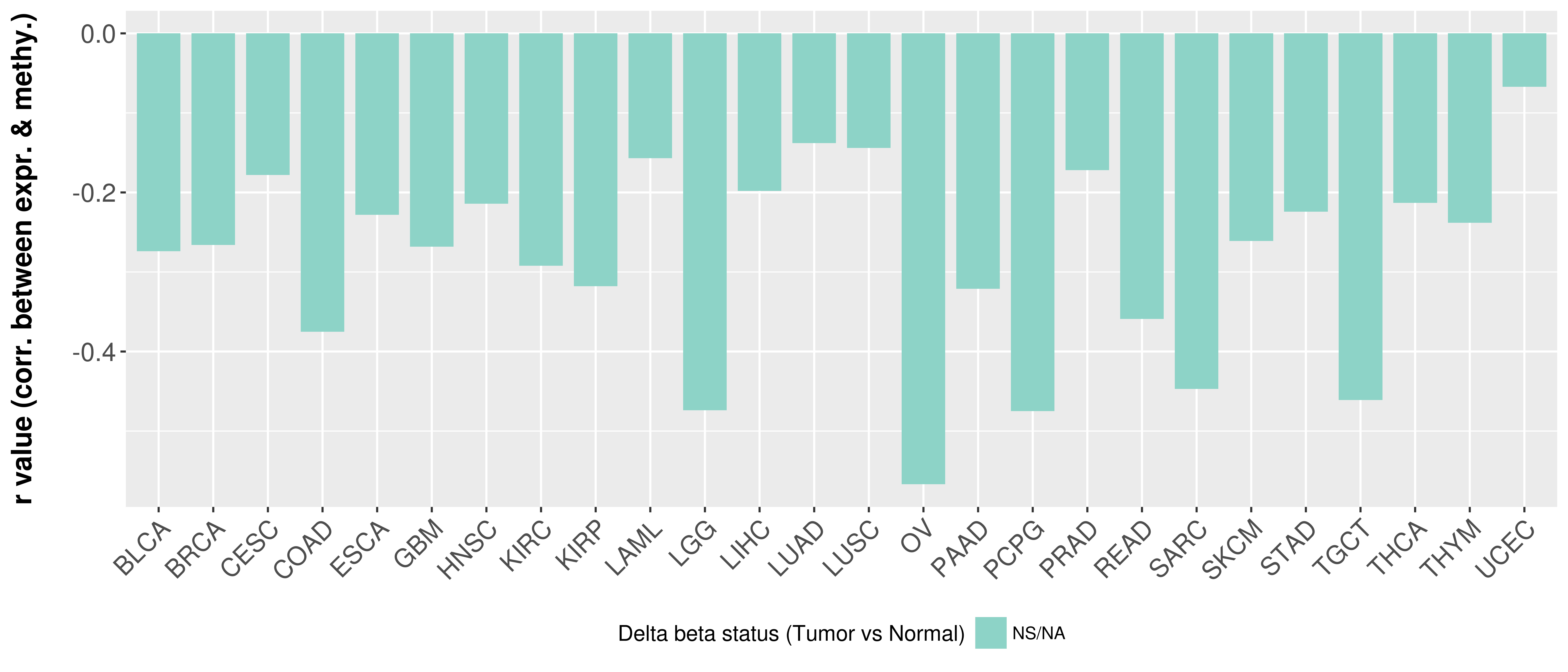
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
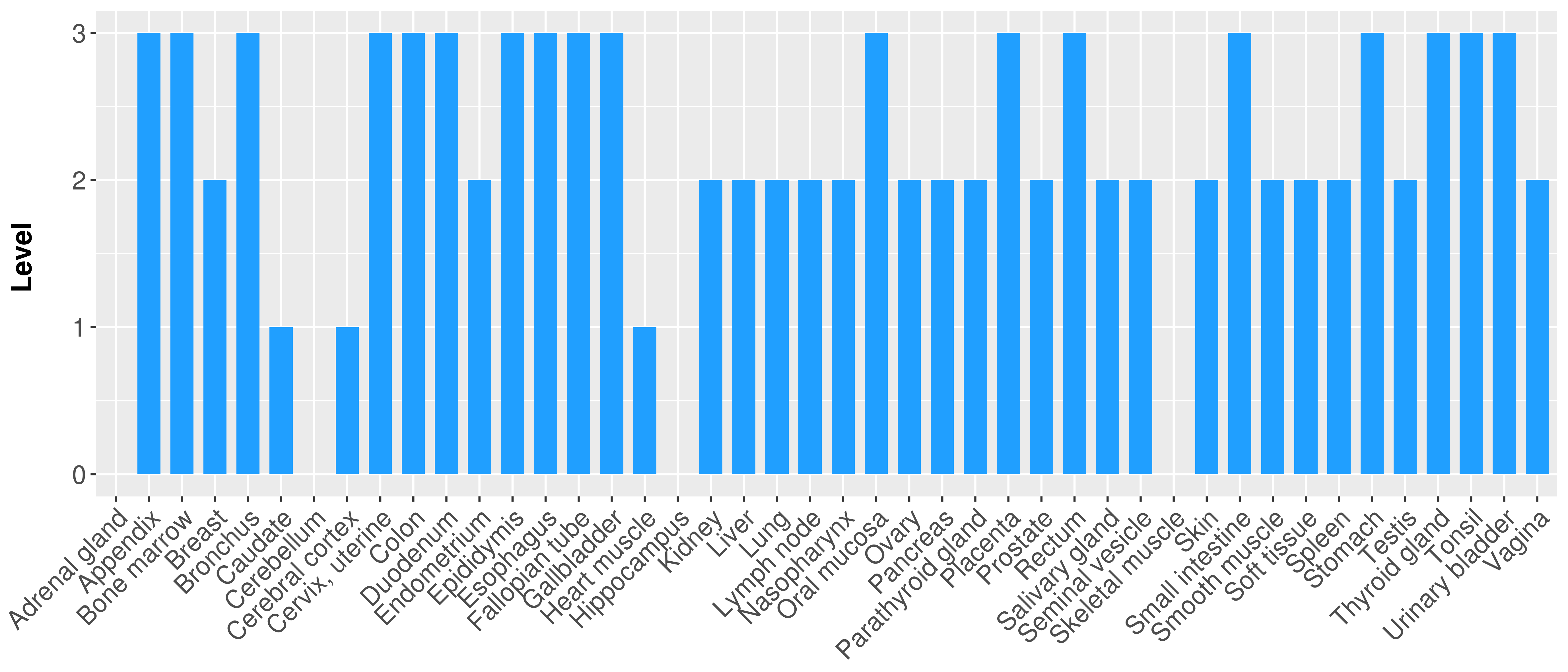
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
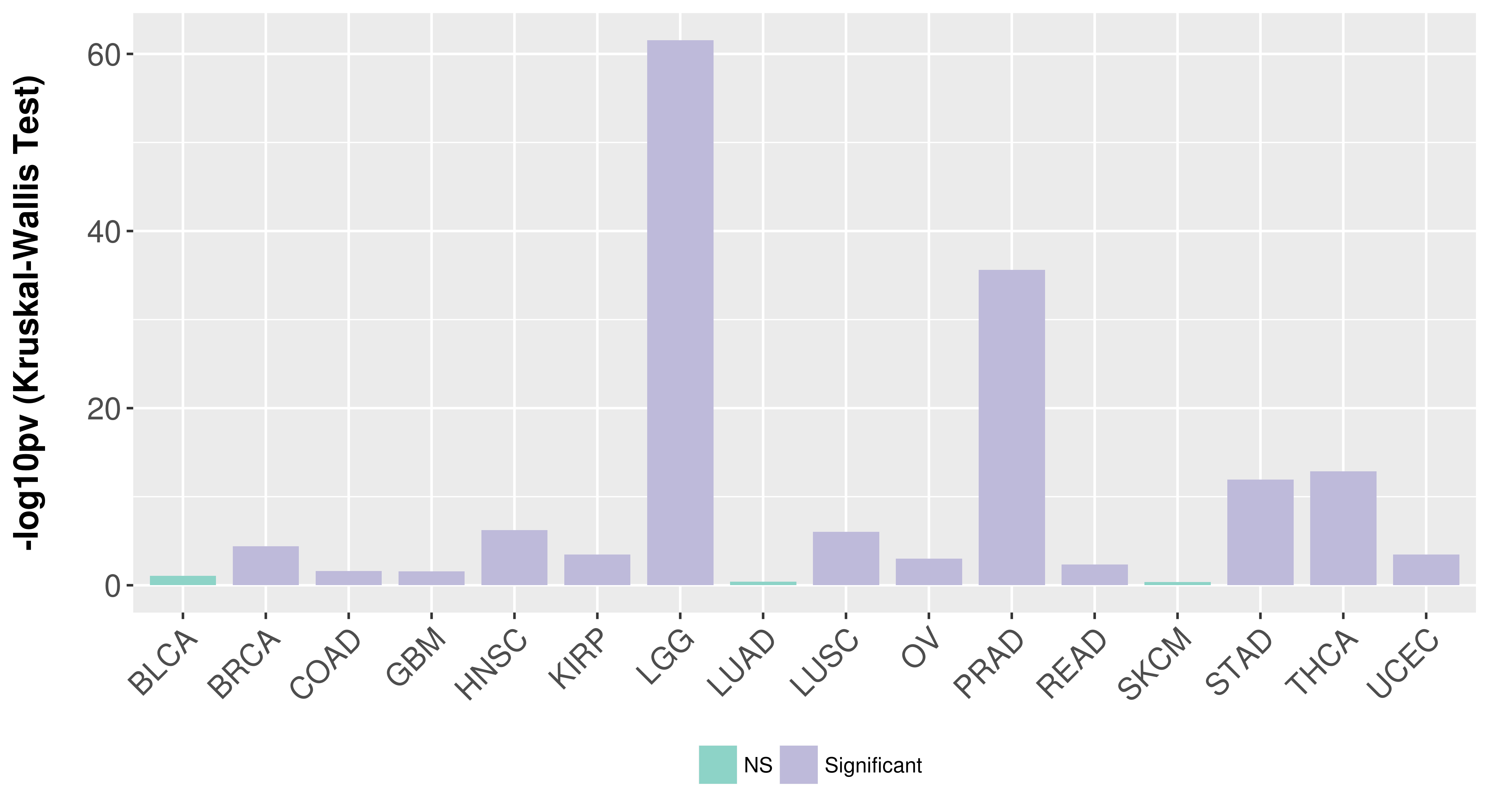
Association between expresson and subtype.
|
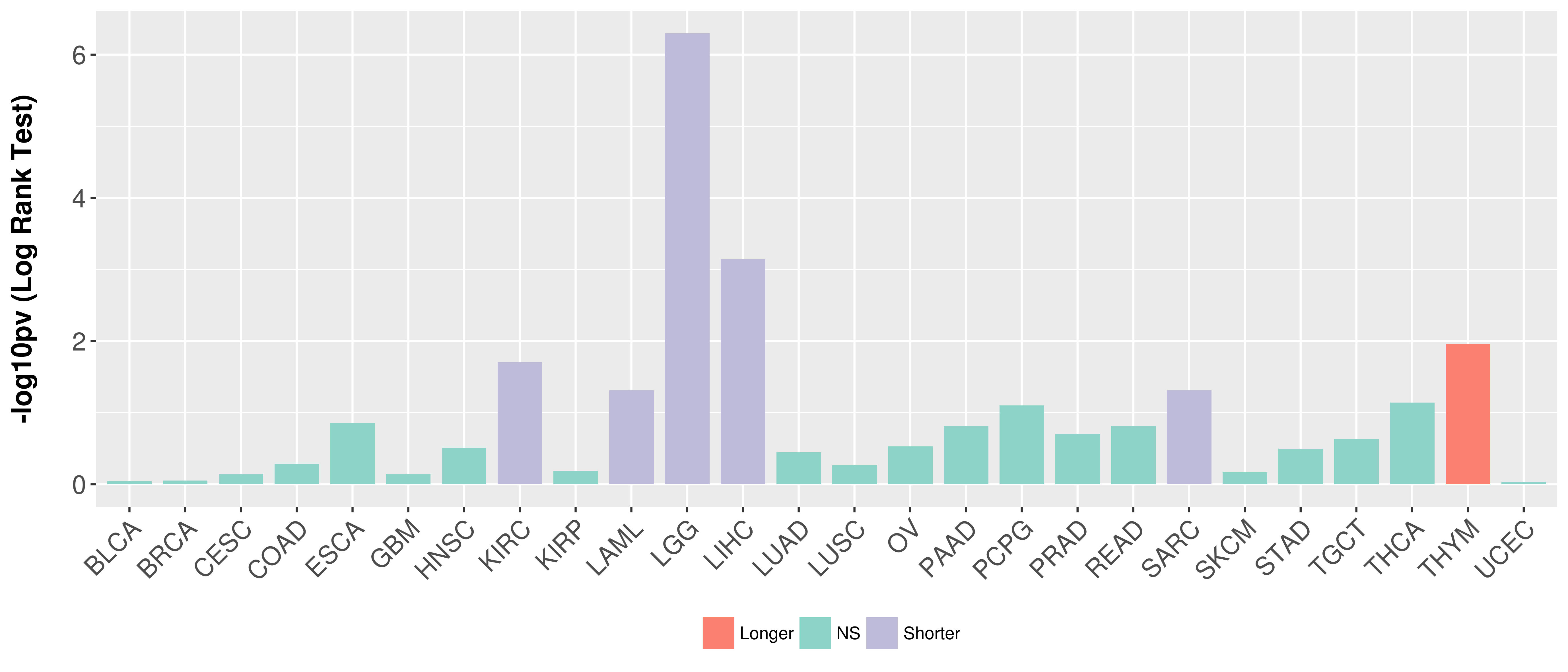
Overall survival analysis based on expression.
|
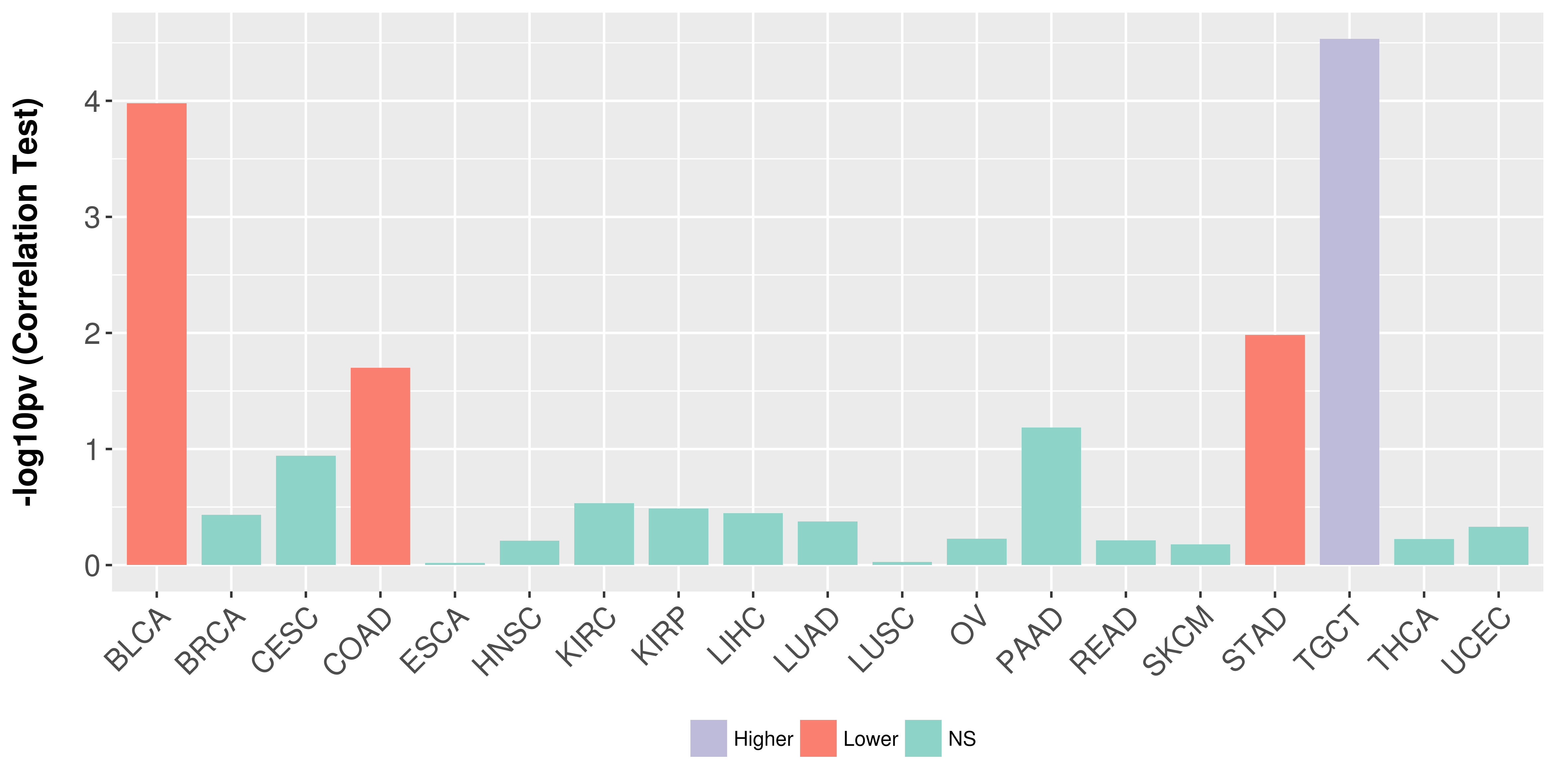
Association between expresson and stage.
|
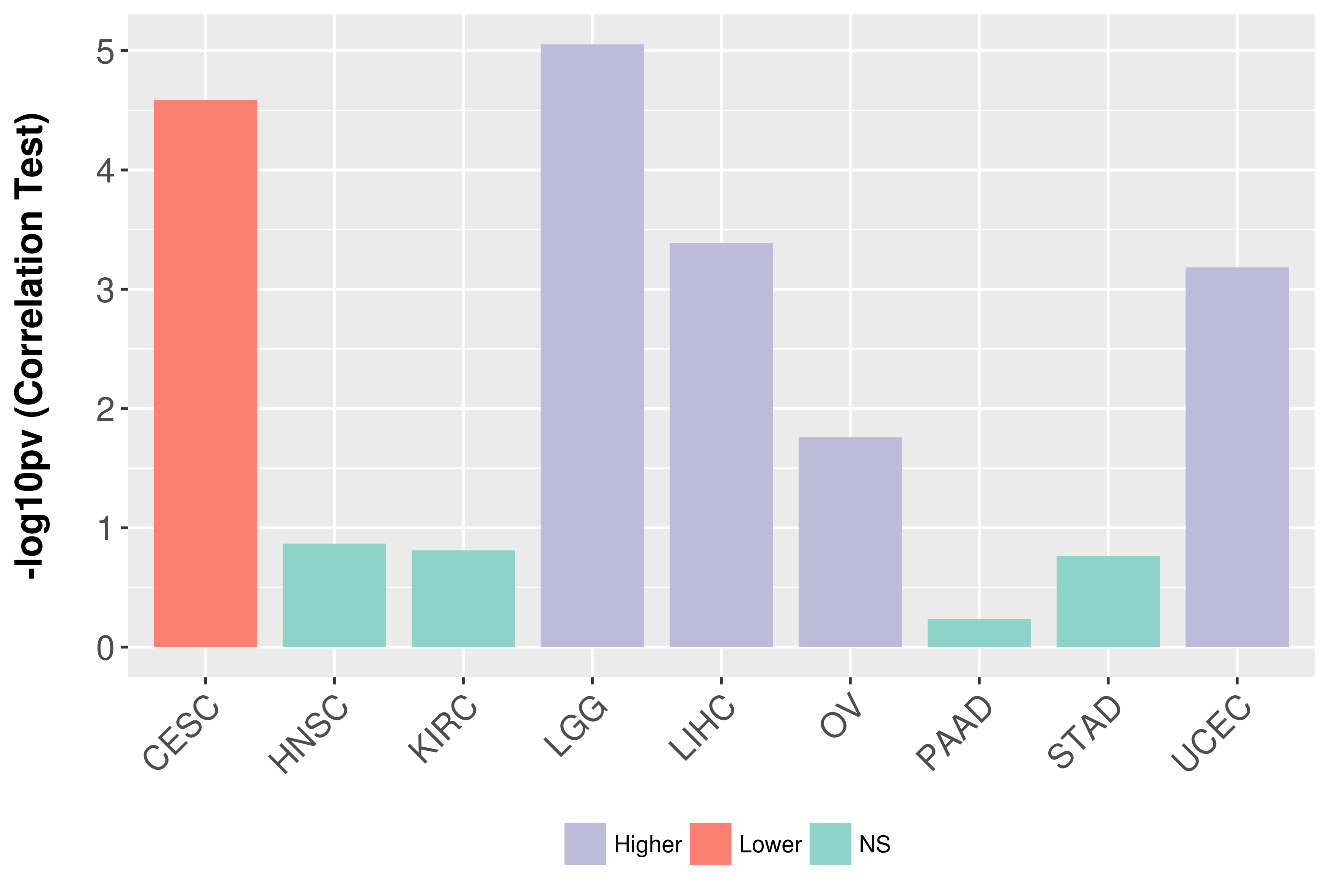
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
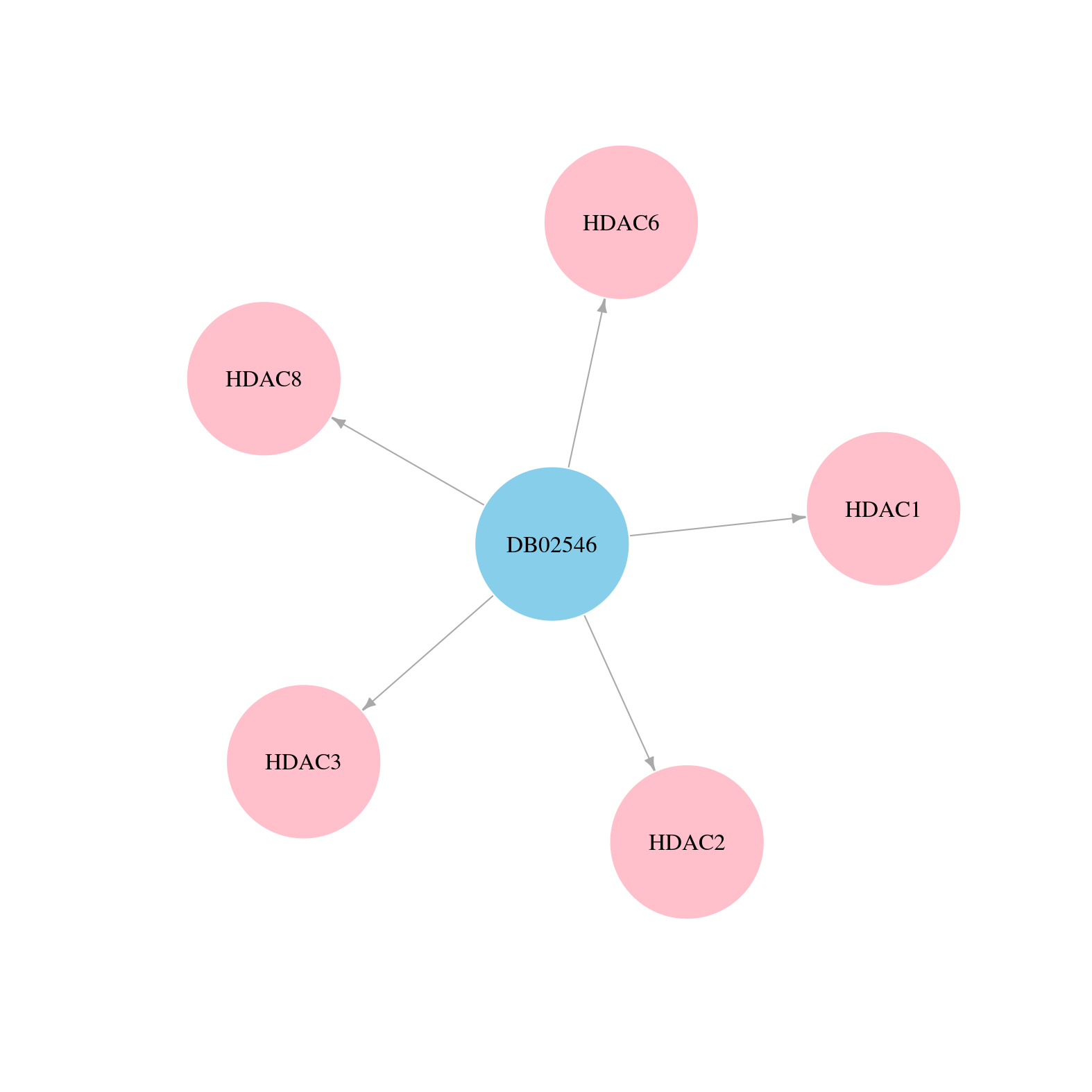
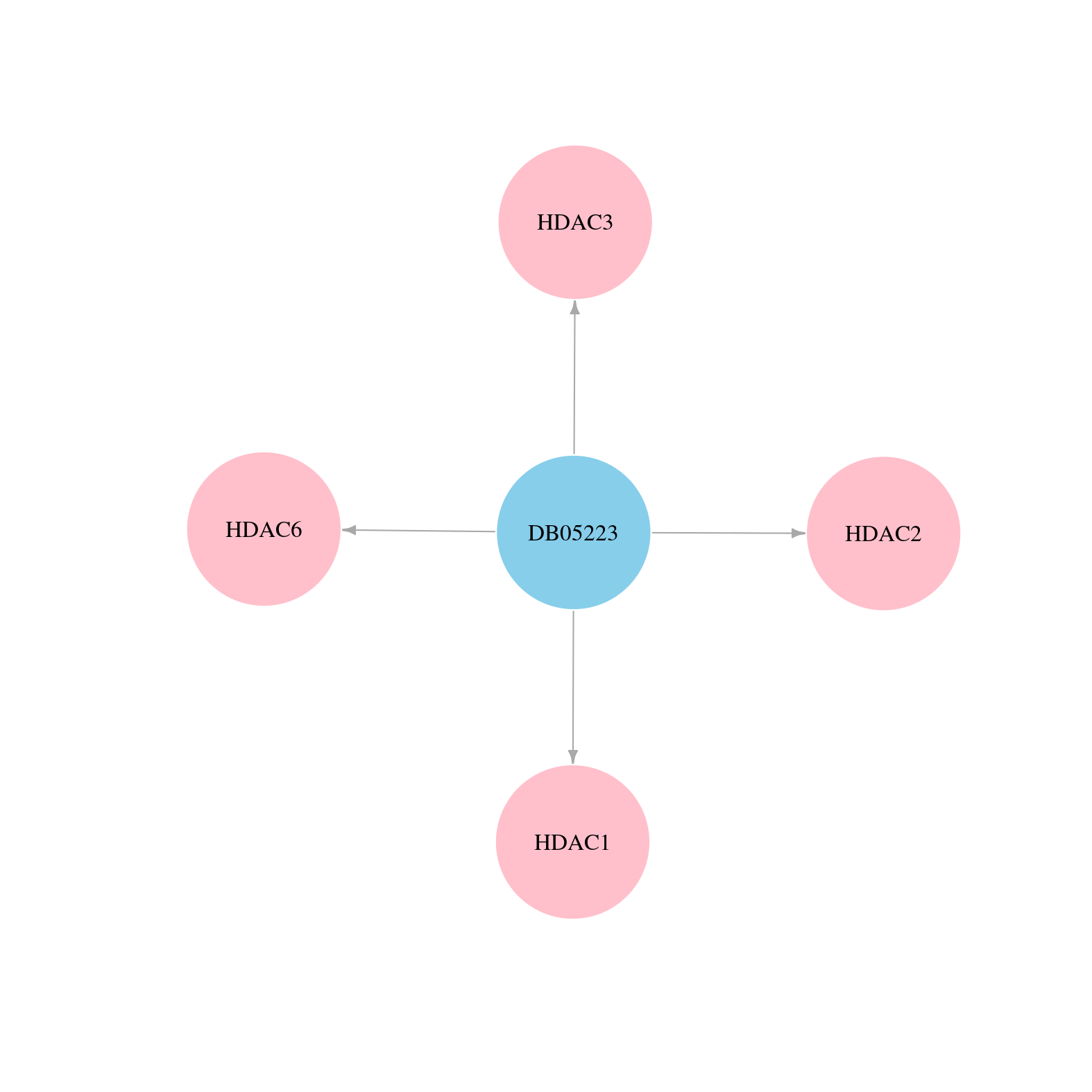
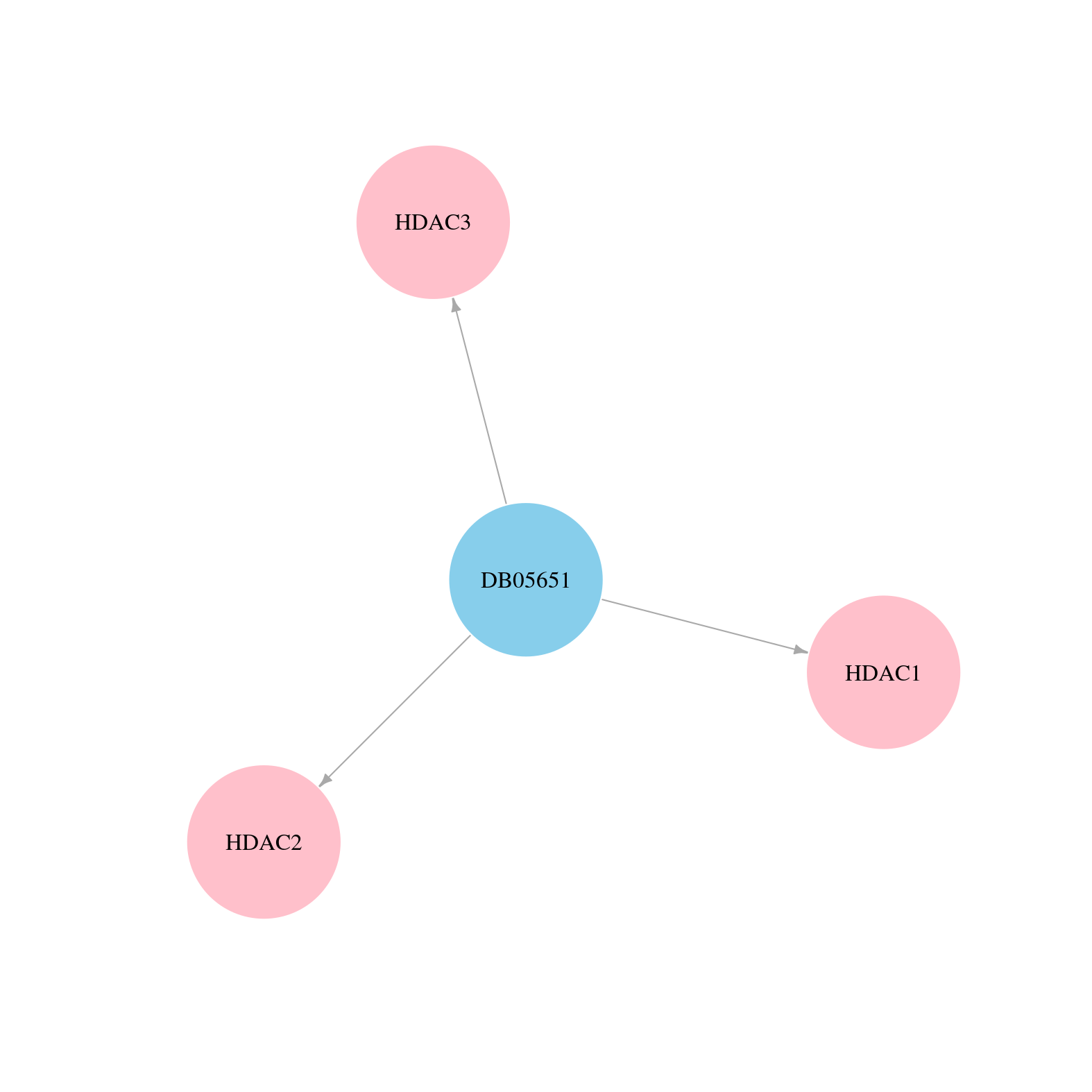
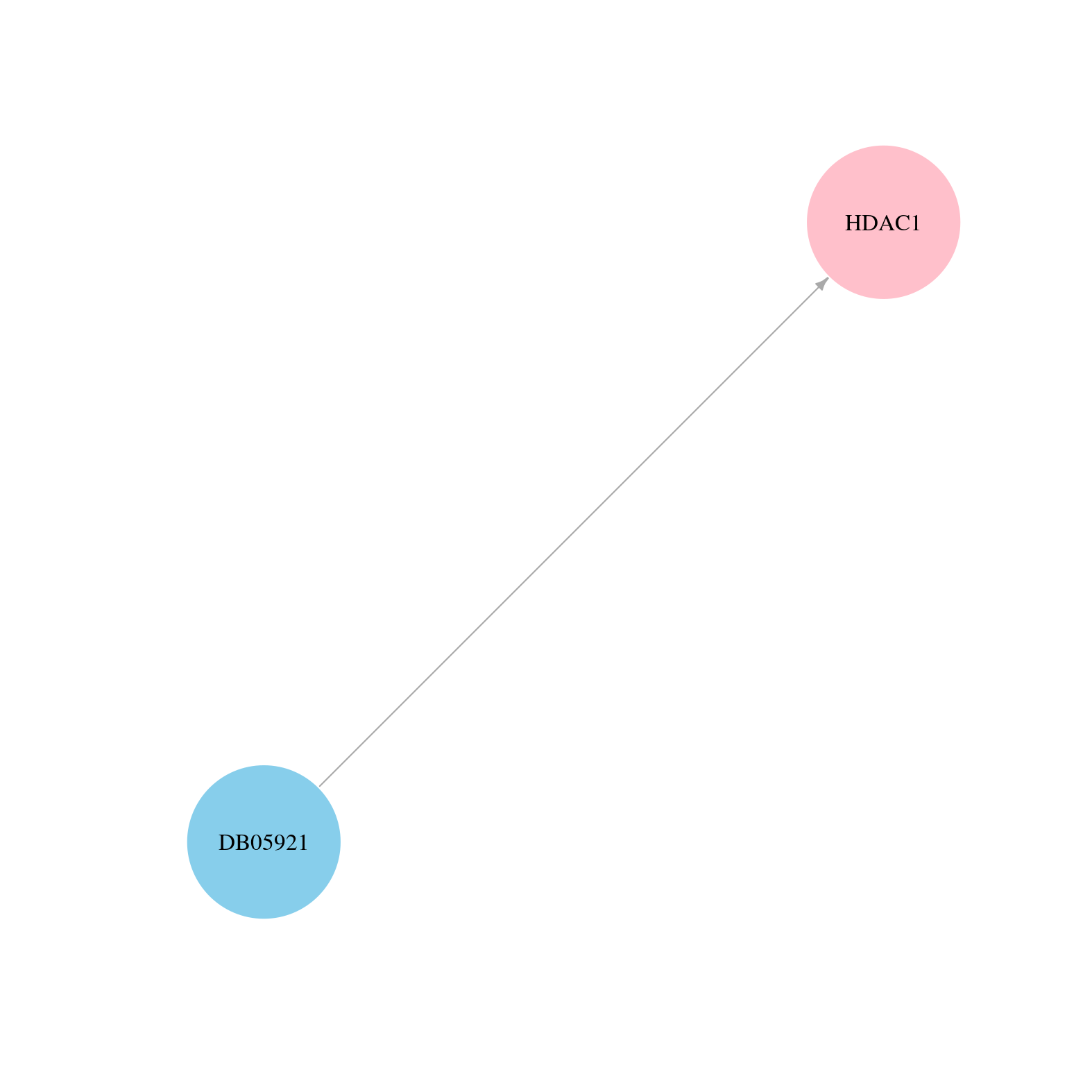
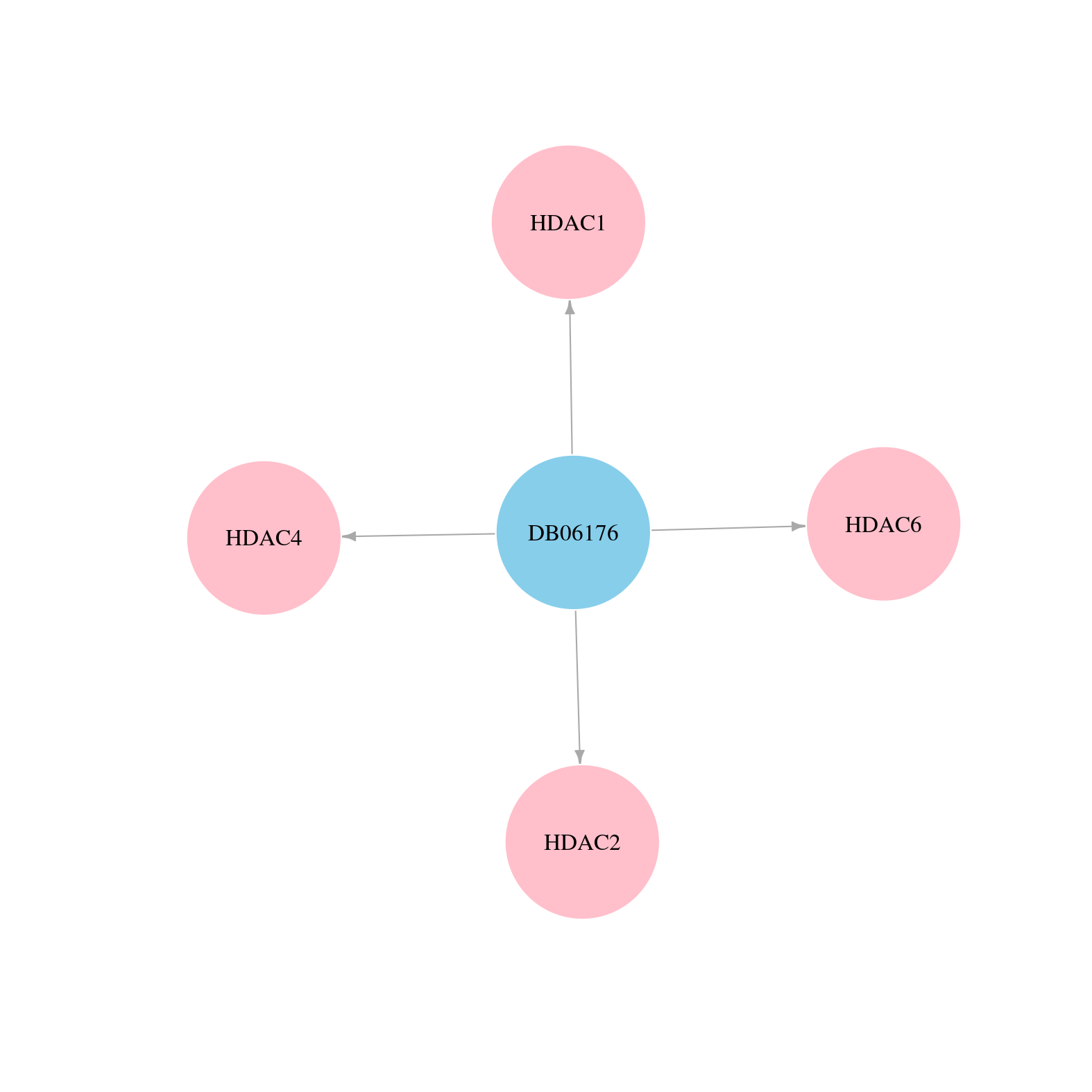
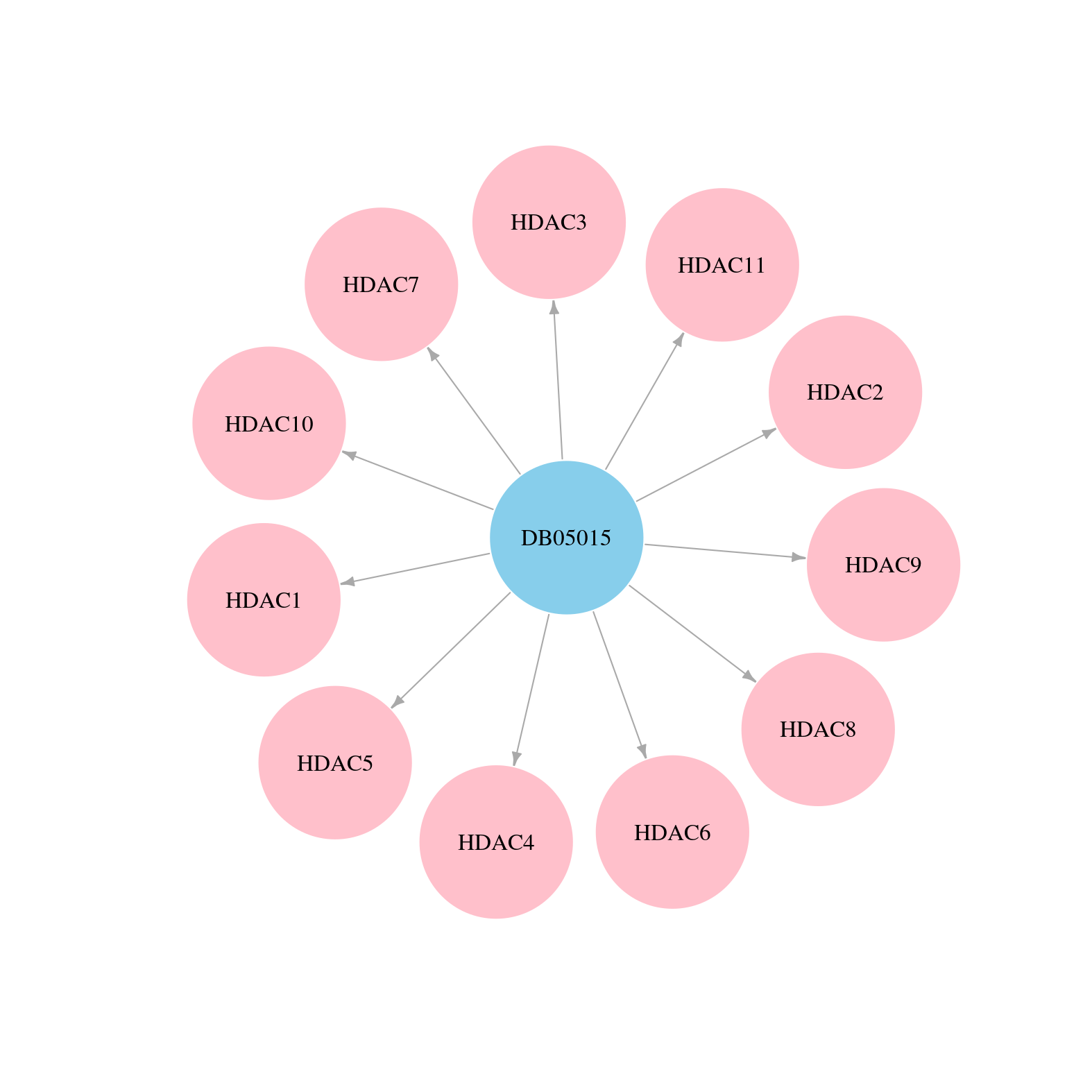
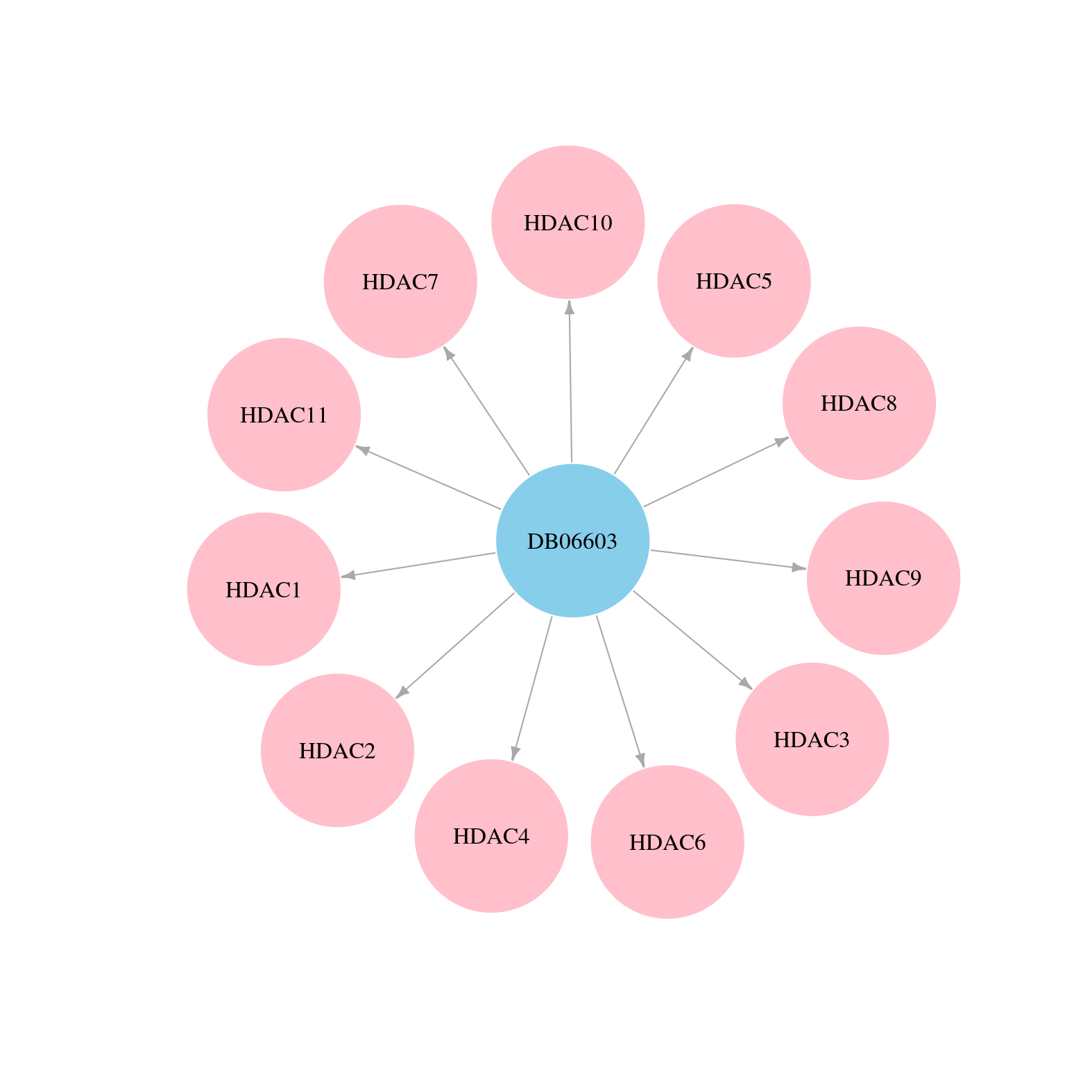
> Drugs from DrugBank database |
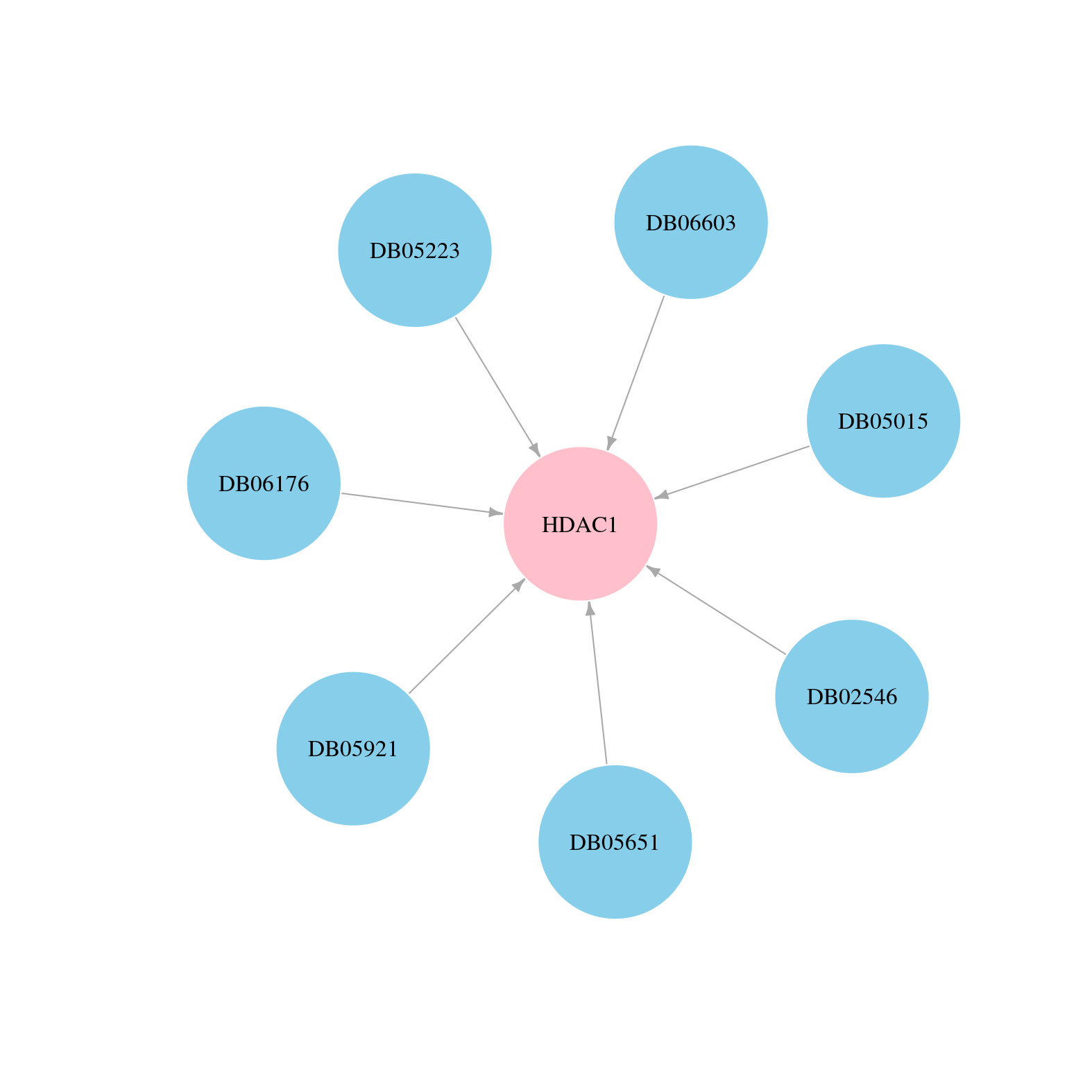
|
| Summary | |
|---|---|
| Symbol | HDAC1 |
| Name | histone deacetylase 1 |
| Aliases | GON-10; RPD3L1; reduced potassium dependency, yeast homolog-like 1 |
| Location | 1p35.2-p35.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|