Browse HDAC3 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00850 Histone deacetylase domain |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4), and some other non-histone substrates. Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Participates in the BCL6 transcriptional repressor activity by deacetylating the H3 'Lys-27' (H3K27) on enhancer elements, antagonizing EP300 acetyltransferase activity and repressing proximal gene expression. Probably participates in the regulation of transcription through its binding to the zinc-finger transcription factor YY1; increases YY1 repression activity. Required to repress transcription of the POU1F1 transcription factor. Acts as a molecular chaperone for shuttling phosphorylated NR2C1 to PML bodies for sumoylation (PubMed:21444723, PubMed:23911289). Contributes, together with XBP1 isoform 1, to the activation of NFE2L2-mediated HMOX1 transcription factor gene expression in a PI(3)K/mTORC2/Akt-dependent signaling pathway leading to endothelial cell (EC) survival under disturbed flow/oxidative stress (PubMed:25190803). |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000226 microtubule cytoskeleton organization GO:0001933 negative regulation of protein phosphorylation GO:0006476 protein deacetylation GO:0006606 protein import into nucleus GO:0006913 nucleocytoplasmic transport GO:0007051 spindle organization GO:0007254 JNK cascade GO:0007623 circadian rhythm GO:0016570 histone modification GO:0016575 histone deacetylation GO:0017038 protein import GO:0031098 stress-activated protein kinase signaling cascade GO:0031647 regulation of protein stability GO:0031929 TOR signaling GO:0032006 regulation of TOR signaling GO:0032008 positive regulation of TOR signaling GO:0032386 regulation of intracellular transport GO:0032388 positive regulation of intracellular transport GO:0032872 regulation of stress-activated MAPK cascade GO:0032873 negative regulation of stress-activated MAPK cascade GO:0033157 regulation of intracellular protein transport GO:0034405 response to fluid shear stress GO:0034504 protein localization to nucleus GO:0035601 protein deacylation GO:0042306 regulation of protein import into nucleus GO:0042307 positive regulation of protein import into nucleus GO:0042326 negative regulation of phosphorylation GO:0042990 regulation of transcription factor import into nucleus GO:0042991 transcription factor import into nucleus GO:0042993 positive regulation of transcription factor import into nucleus GO:0043409 negative regulation of MAPK cascade GO:0044744 protein targeting to nucleus GO:0046328 regulation of JNK cascade GO:0046329 negative regulation of JNK cascade GO:0046822 regulation of nucleocytoplasmic transport GO:0046824 positive regulation of nucleocytoplasmic transport GO:0048511 rhythmic process GO:0051169 nuclear transport GO:0051170 nuclear import GO:0051222 positive regulation of protein transport GO:0051225 spindle assembly GO:0051403 stress-activated MAPK cascade GO:0070302 regulation of stress-activated protein kinase signaling cascade GO:0070303 negative regulation of stress-activated protein kinase signaling cascade GO:0070932 histone H3 deacetylation GO:0071498 cellular response to fluid shear stress GO:0090316 positive regulation of intracellular protein transport GO:0098732 macromolecule deacylation GO:1900180 regulation of protein localization to nucleus GO:1900182 positive regulation of protein localization to nucleus GO:1902532 negative regulation of intracellular signal transduction GO:1902593 single-organism nuclear import GO:1903533 regulation of protein targeting GO:1903829 positive regulation of cellular protein localization GO:1904589 regulation of protein import GO:1904591 positive regulation of protein import GO:1904951 positive regulation of establishment of protein localization |
| Molecular Function |
GO:0003682 chromatin binding GO:0003714 transcription corepressor activity GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0030332 cyclin binding GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0042826 histone deacetylase binding GO:0051059 NF-kappaB binding |
| Cellular Component |
GO:0000118 histone deacetylase complex GO:0005819 spindle GO:0005874 microtubule GO:0005876 spindle microtubule GO:0017053 transcriptional repressor complex |
| KEGG |
hsa04919 Thyroid hormone signaling pathway |
| Reactome |
R-HSA-5619507: Activation of HOX genes during differentiation R-HSA-5617472: Activation of anterior HOX genes in hindbrain development during early embryogenesis R-HSA-390471: Association of TriC/CCT with target proteins during biosynthesis R-HSA-1368108: BMAL1 R-HSA-390466: Chaperonin-mediated protein folding R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-400253: Circadian Clock R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-1266738: Developmental Biology R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-535734: Fatty acid, triacylglycerol, and ketone body metabolism R-HSA-3214815: HDACs deacetylate histones R-HSA-1430728: Metabolism R-HSA-556833: Metabolism of lipids and lipoproteins R-HSA-392499: Metabolism of proteins R-HSA-1592230: Mitochondrial biogenesis R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-1368071: NR1D1 (REV-ERBA) represses gene expression R-HSA-1852241: Organelle biogenesis and maintenance R-HSA-1989781: PPARA activates gene expression R-HSA-391251: Protein folding R-HSA-1368082: RORA activates gene expression R-HSA-400206: Regulation of lipid metabolism by Peroxisome proliferator-activated receptor alpha (PPARalpha) R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer R-HSA-166520: Signalling by NGF R-HSA-2151201: Transcriptional activation of mitochondrial biogenesis R-HSA-381340: Transcriptional regulation of white adipocyte differentiation R-HSA-193704: p75 NTR receptor-mediated signalling R-HSA-193670: p75NTR negatively regulates cell cycle via SC1 |
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for HDAC3. |
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
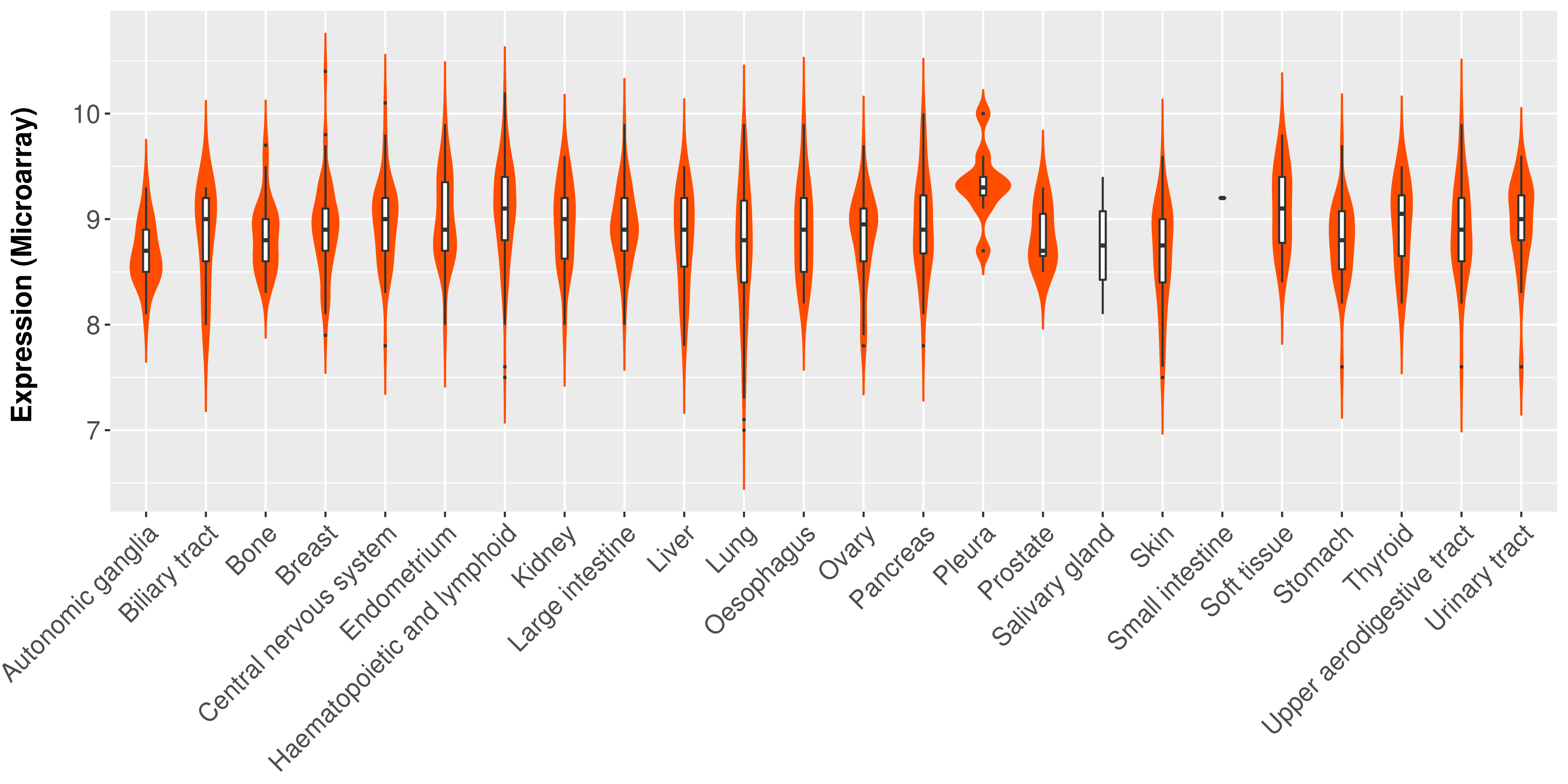
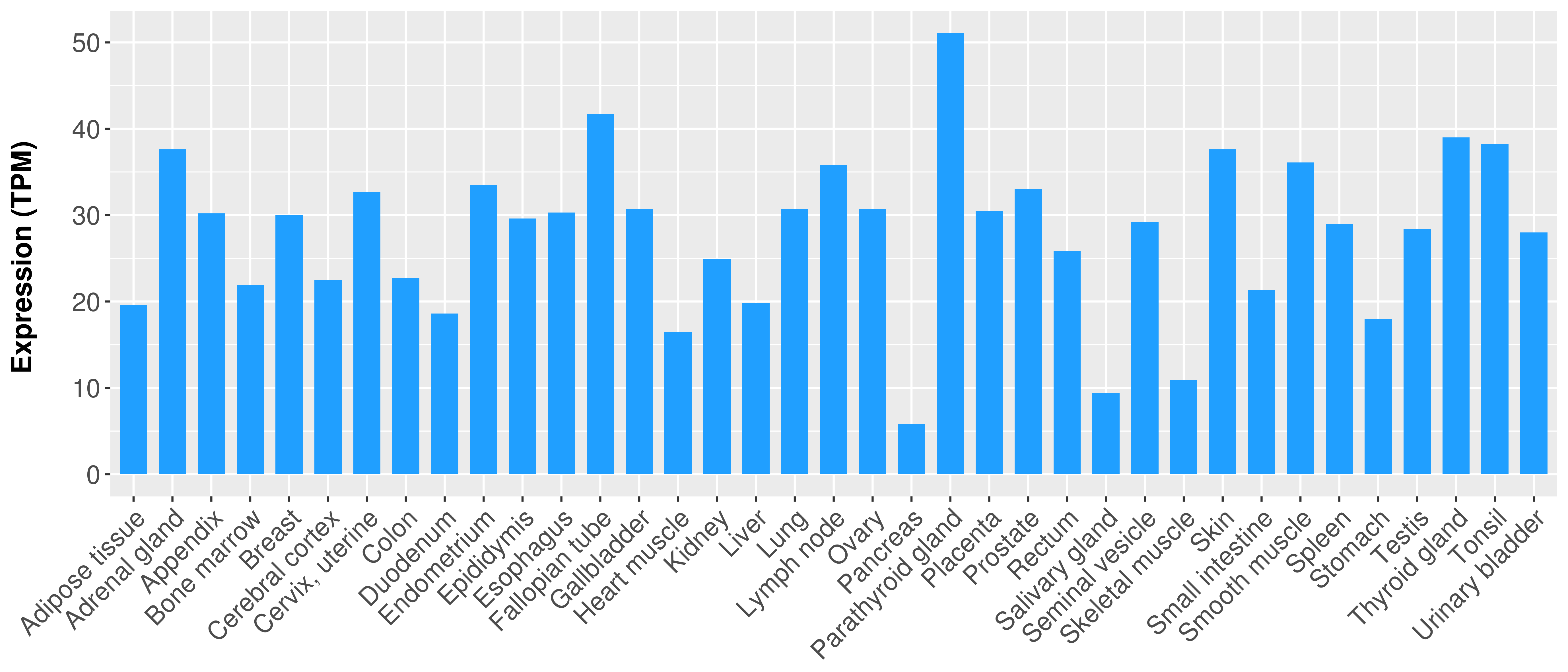
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
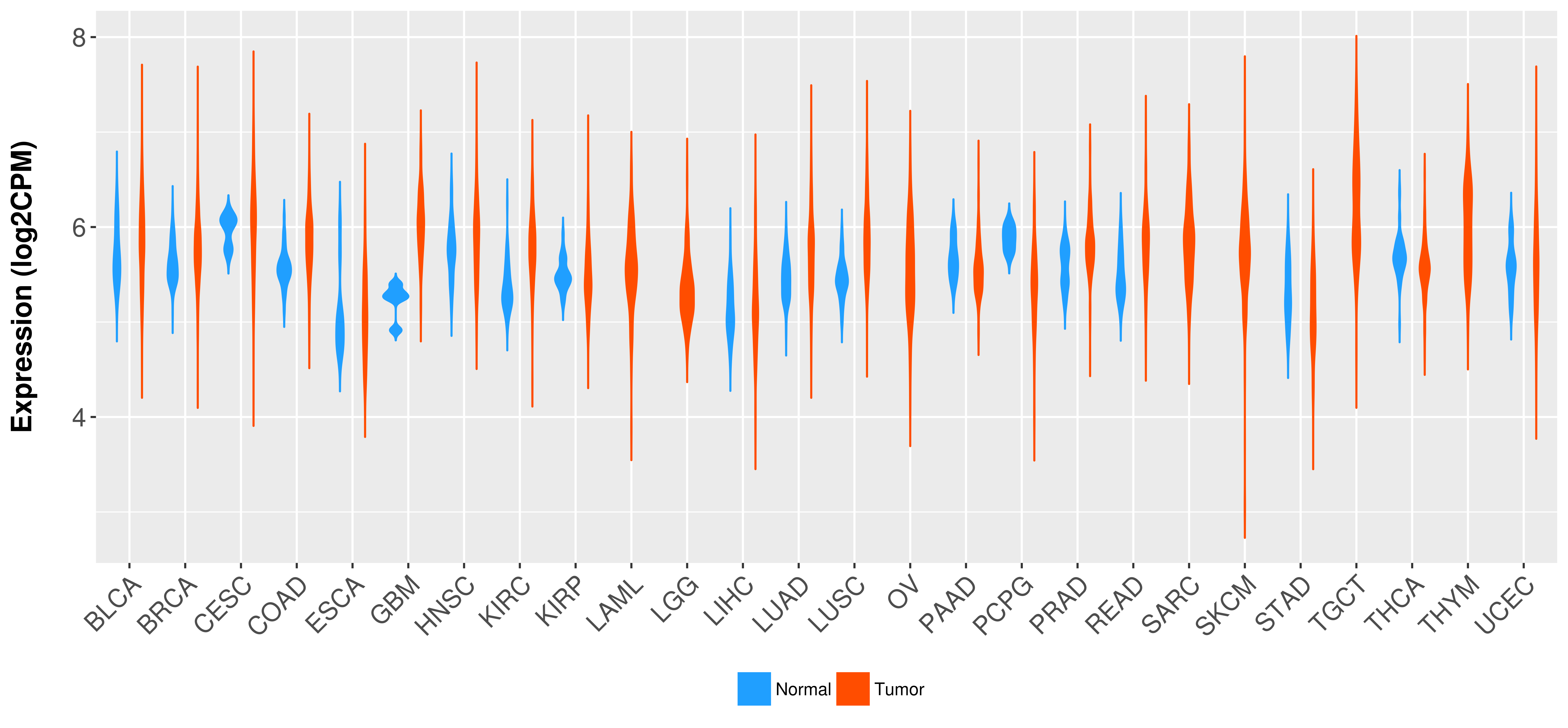
Differential expression analysis for cancers with more than 10 normal samples
|
|
|
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
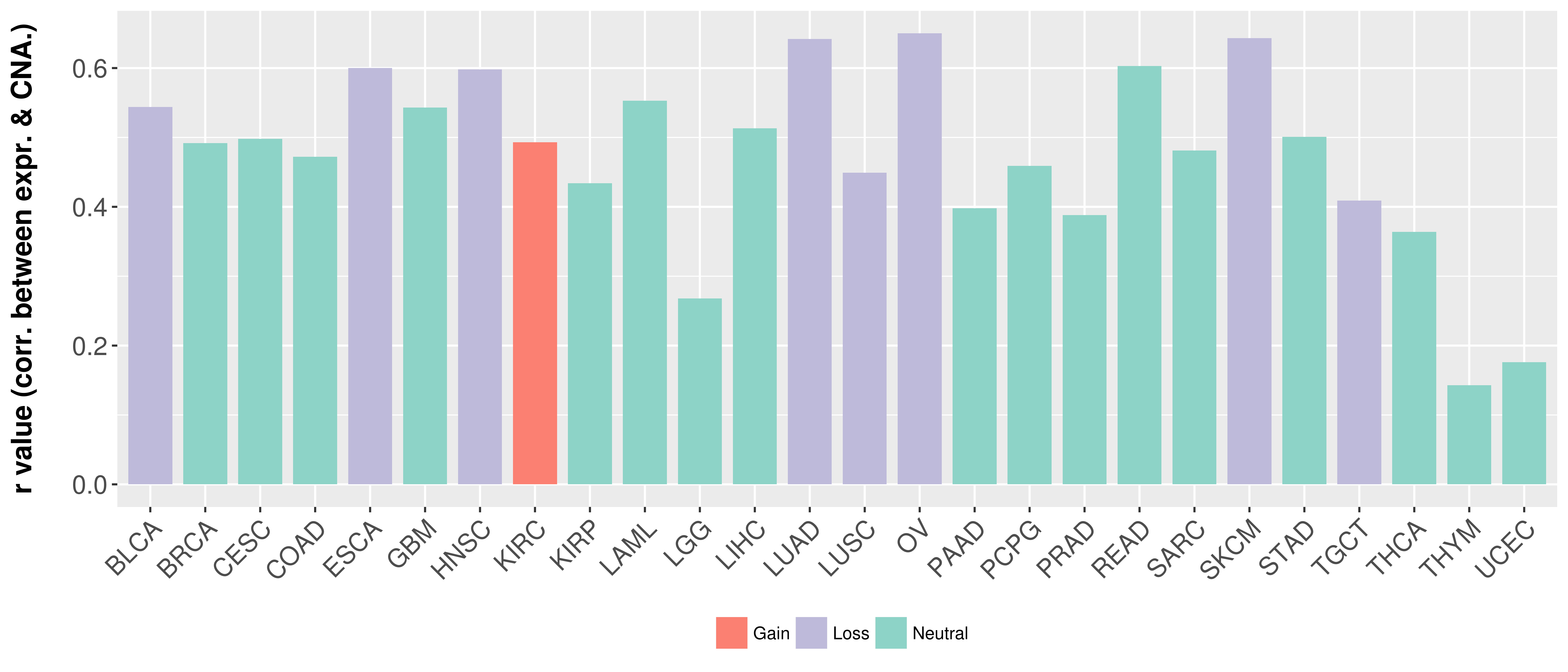
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
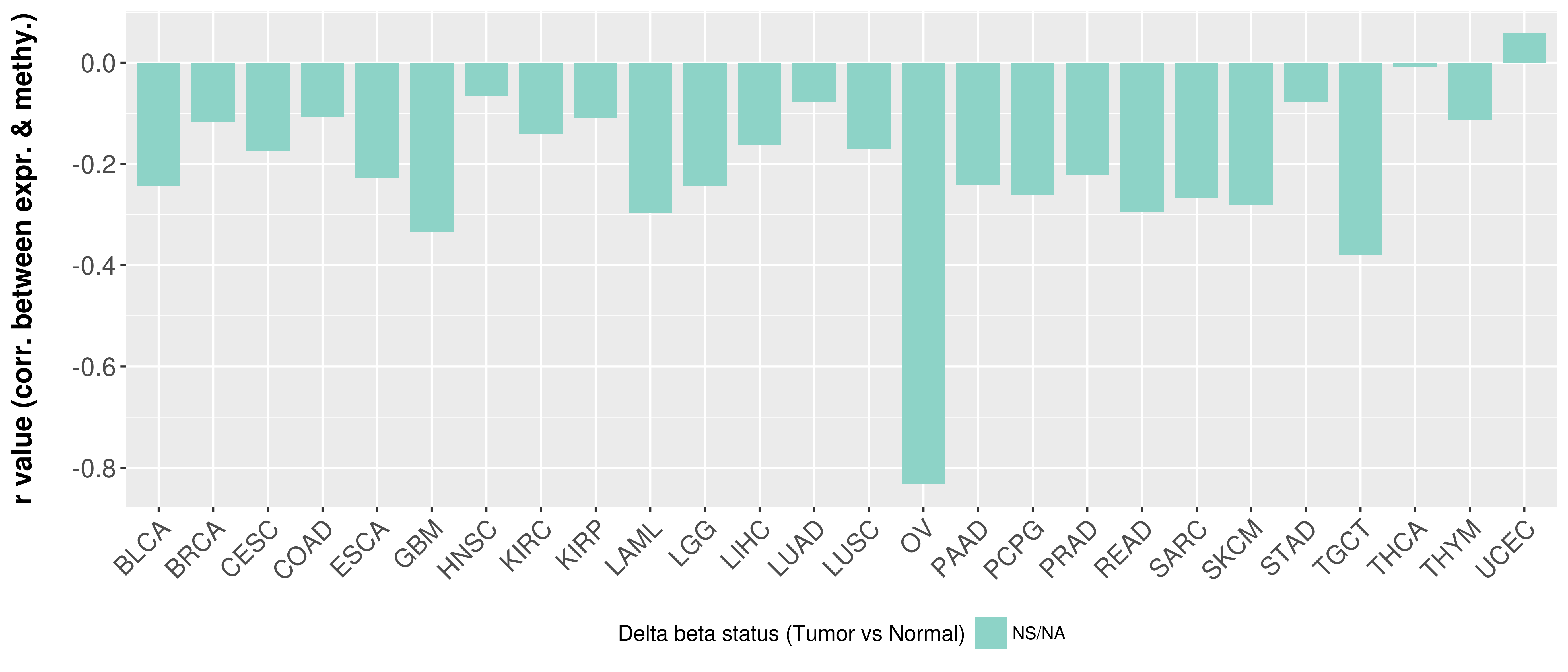
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
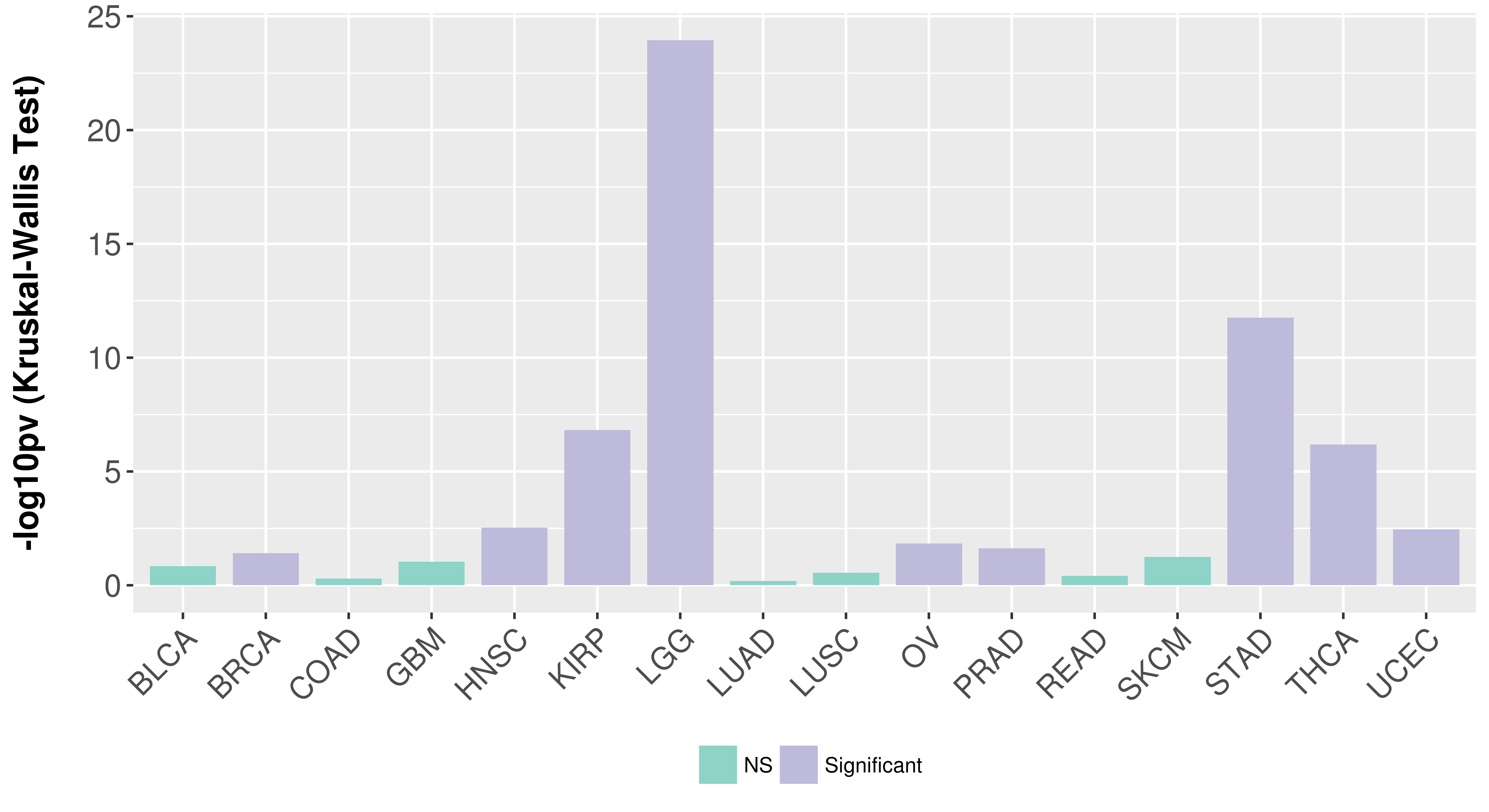
Association between expresson and subtype.
|
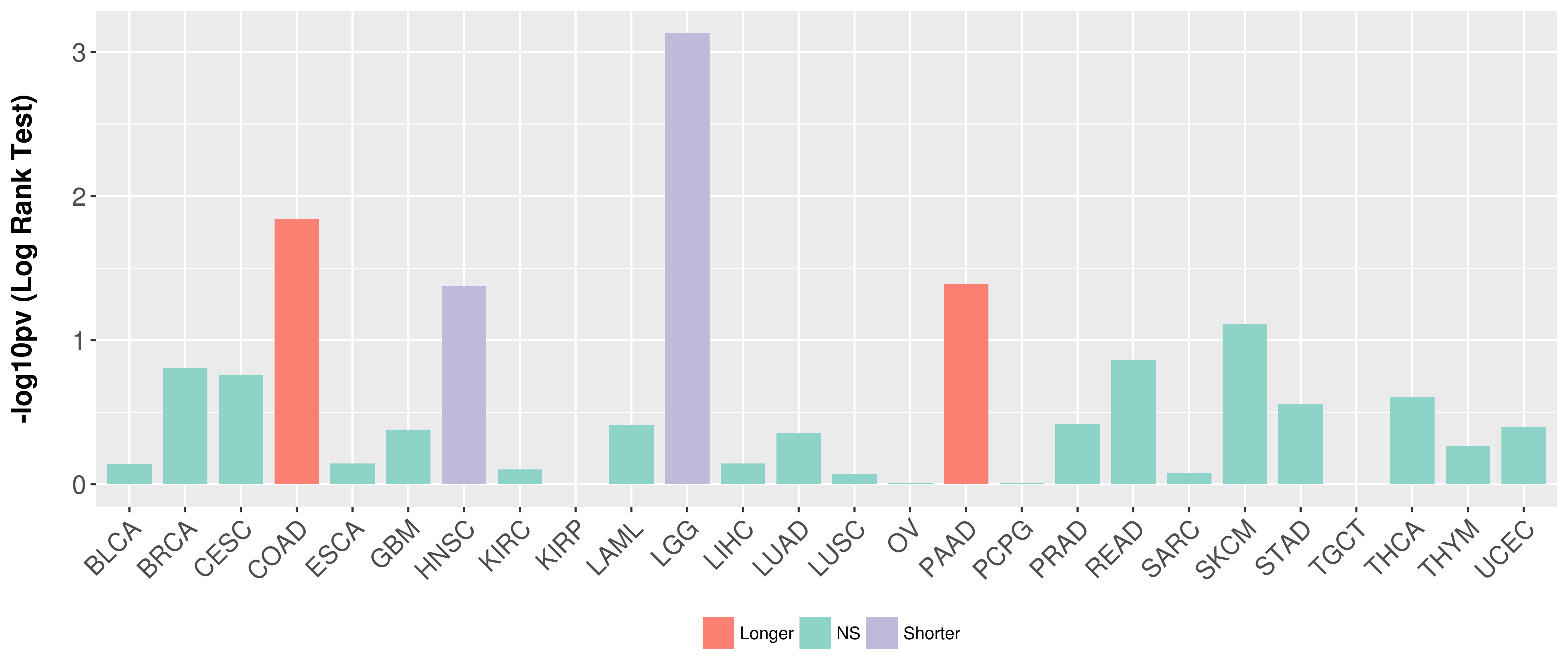
Overall survival analysis based on expression.
|
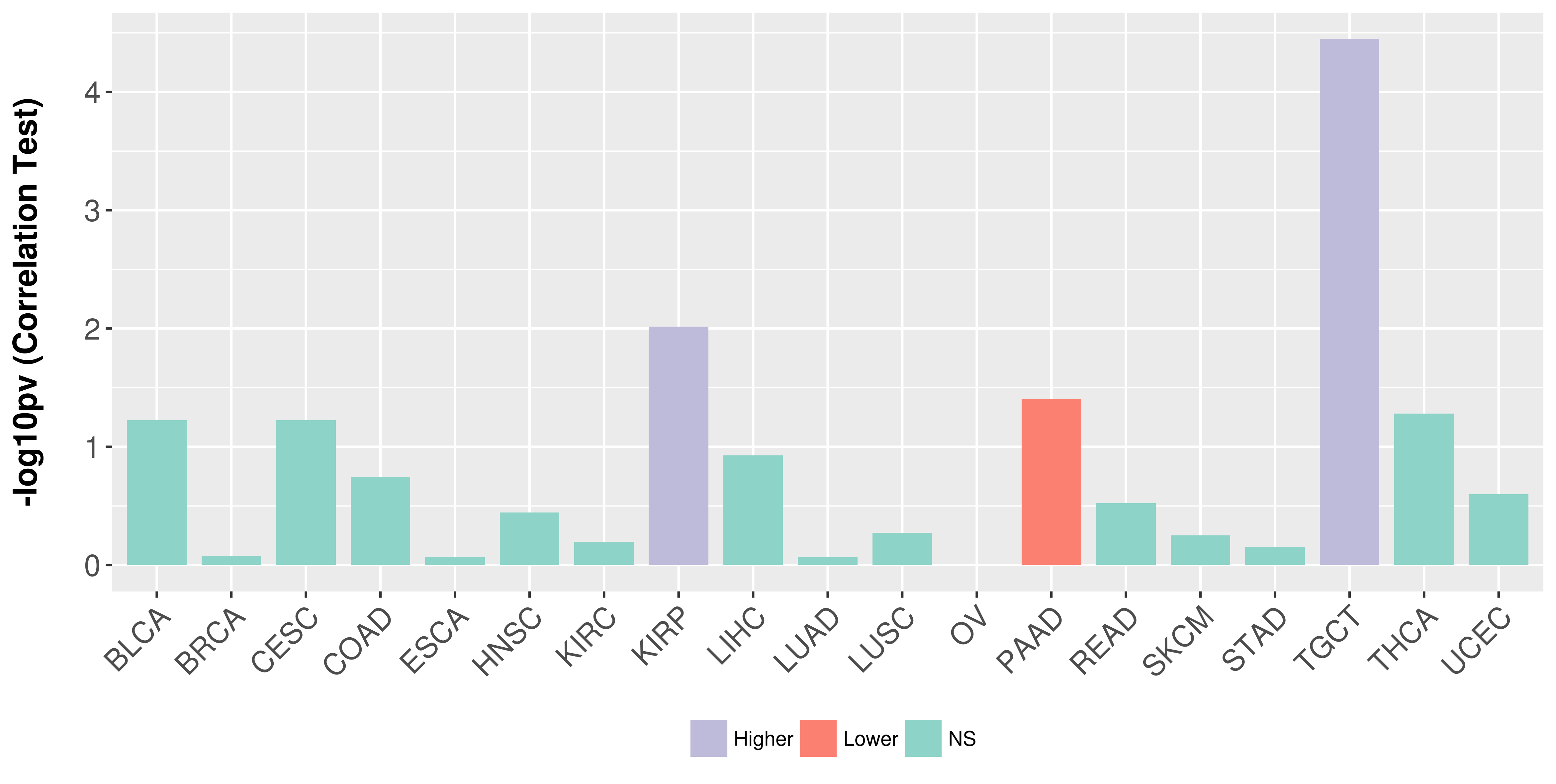
Association between expresson and stage.
|
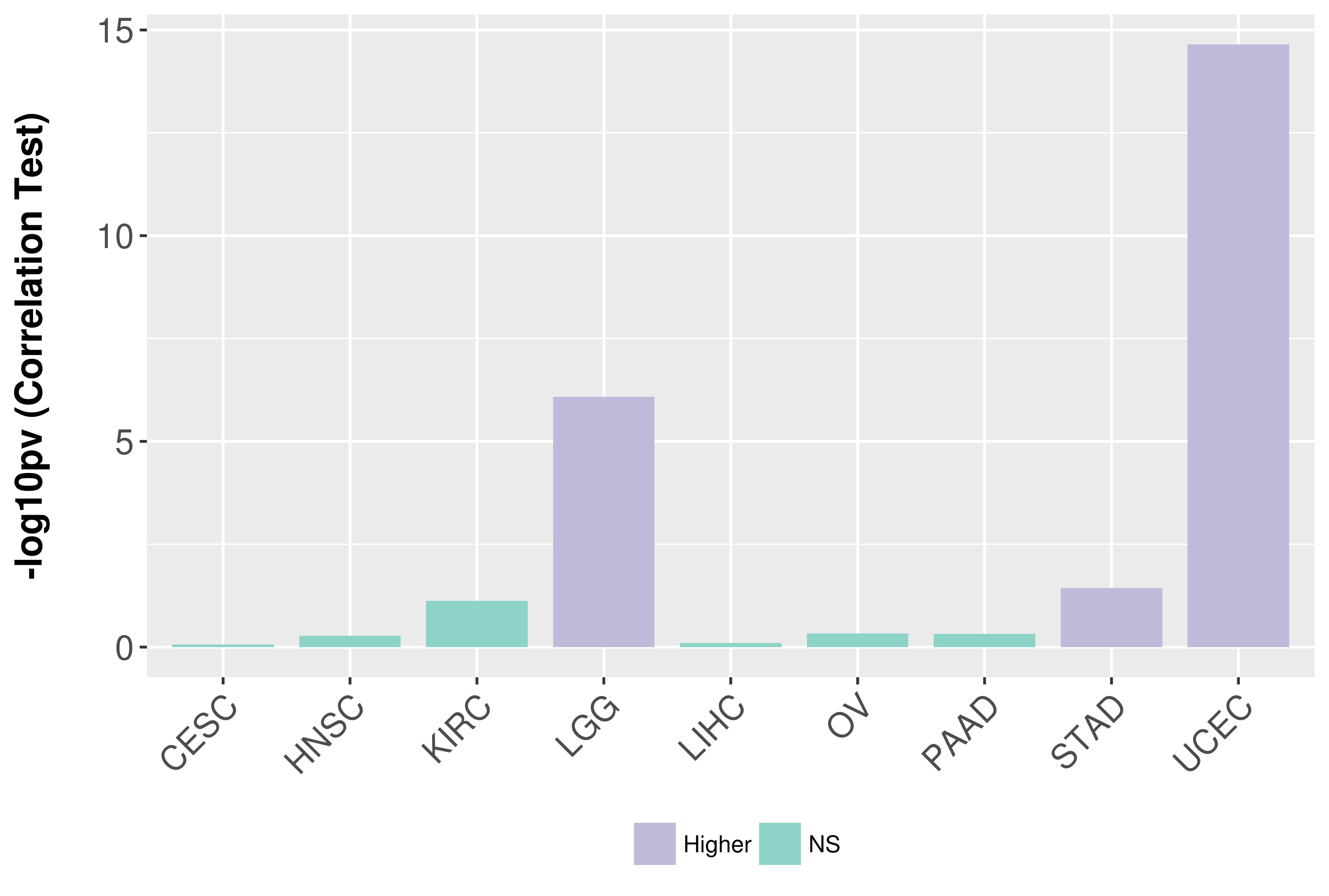
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
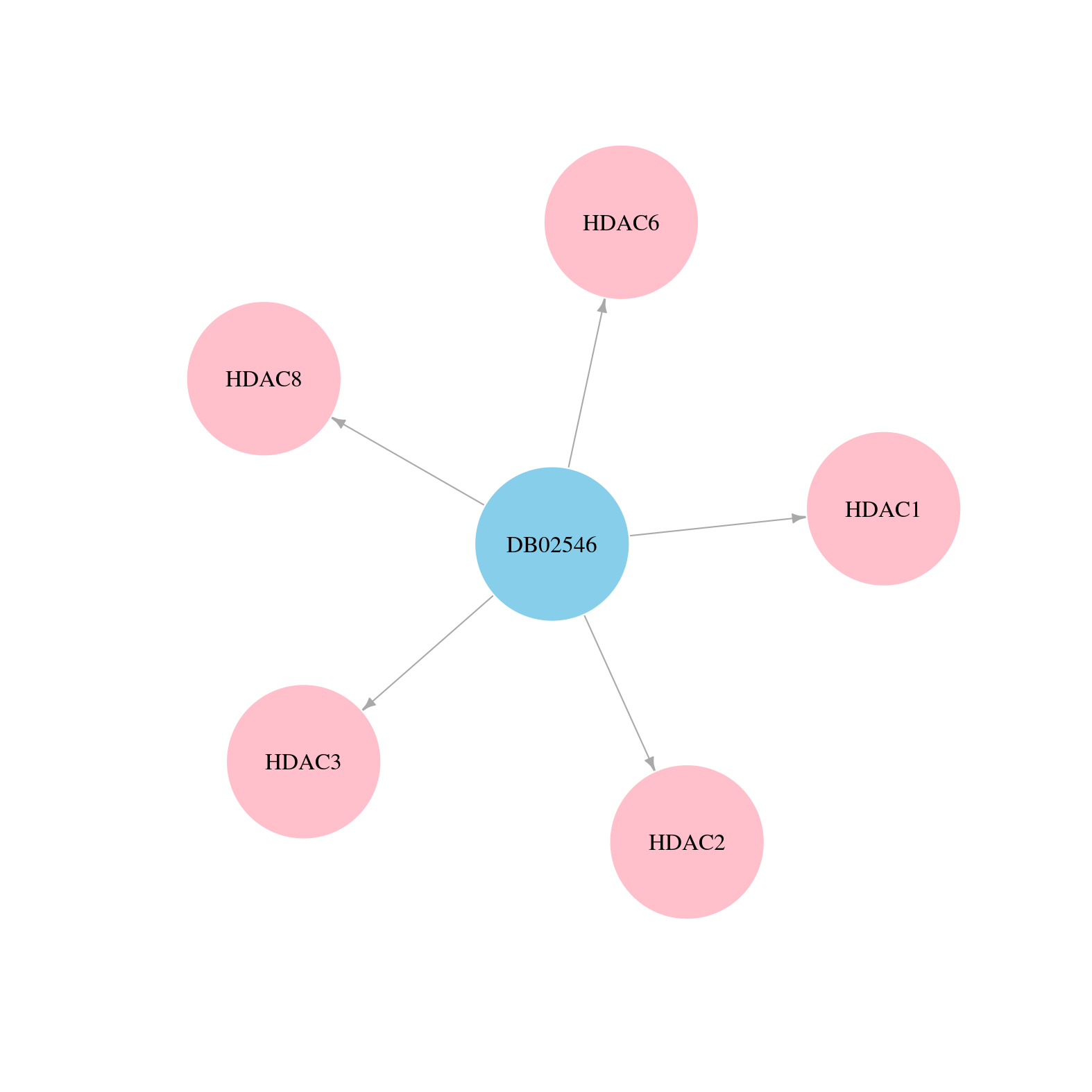
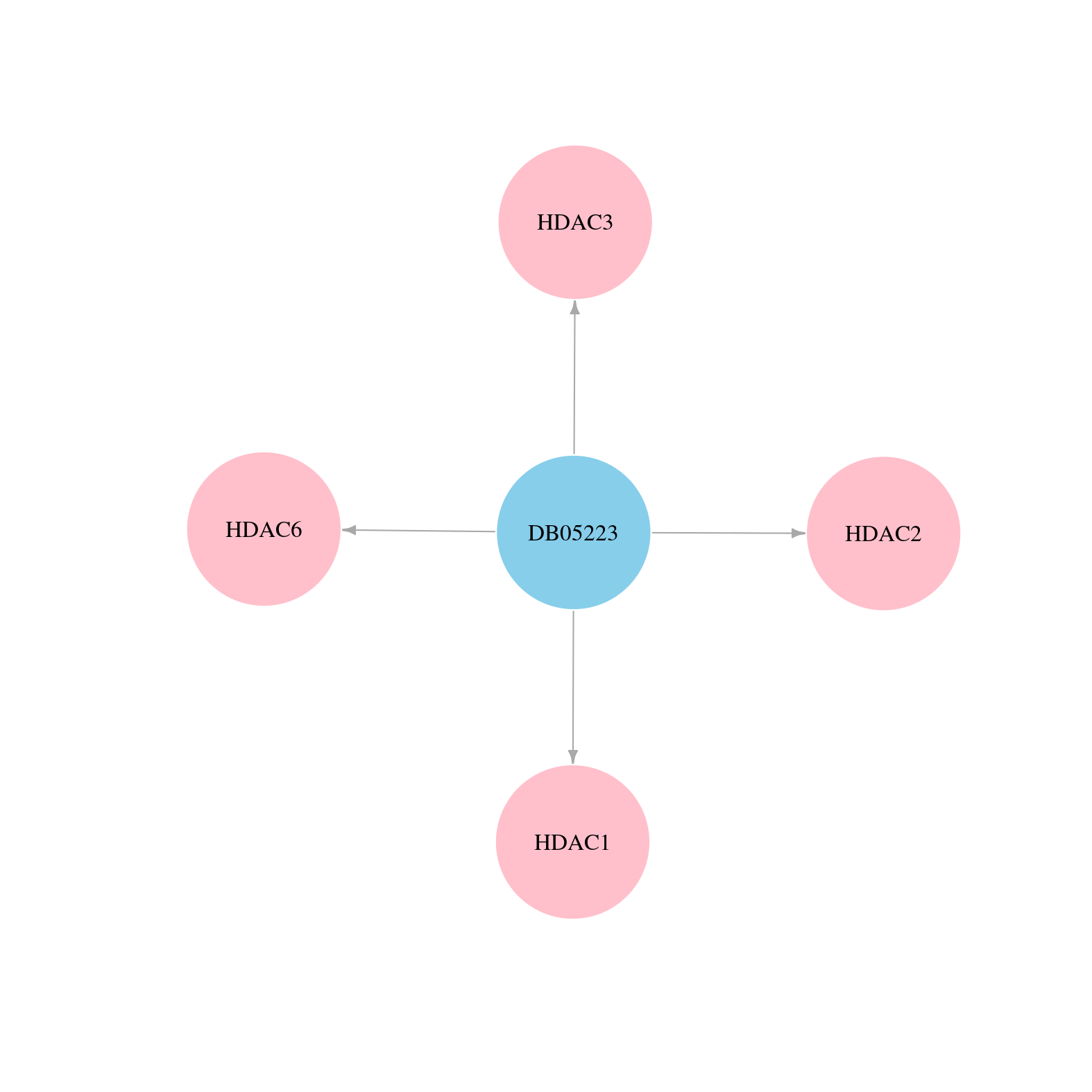
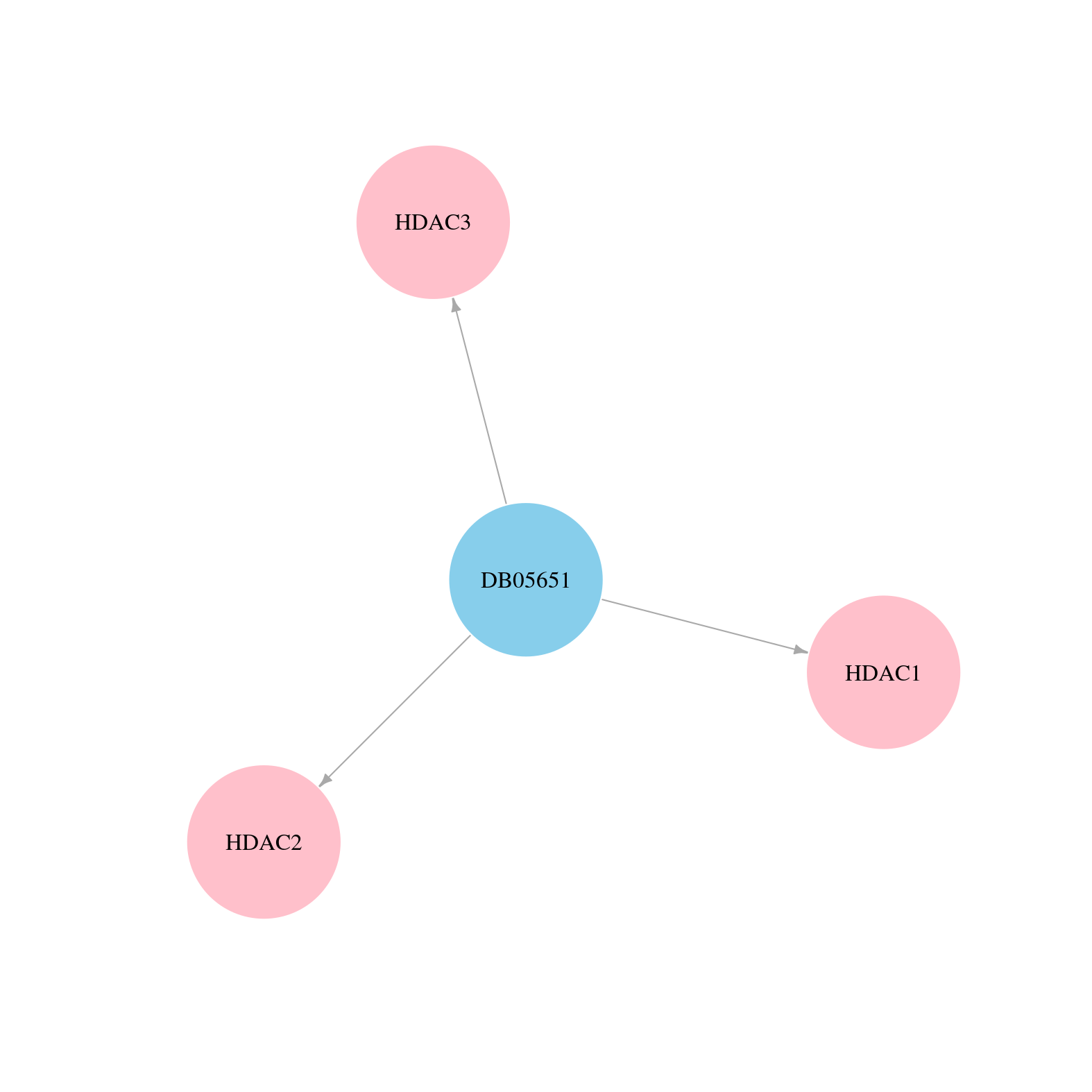
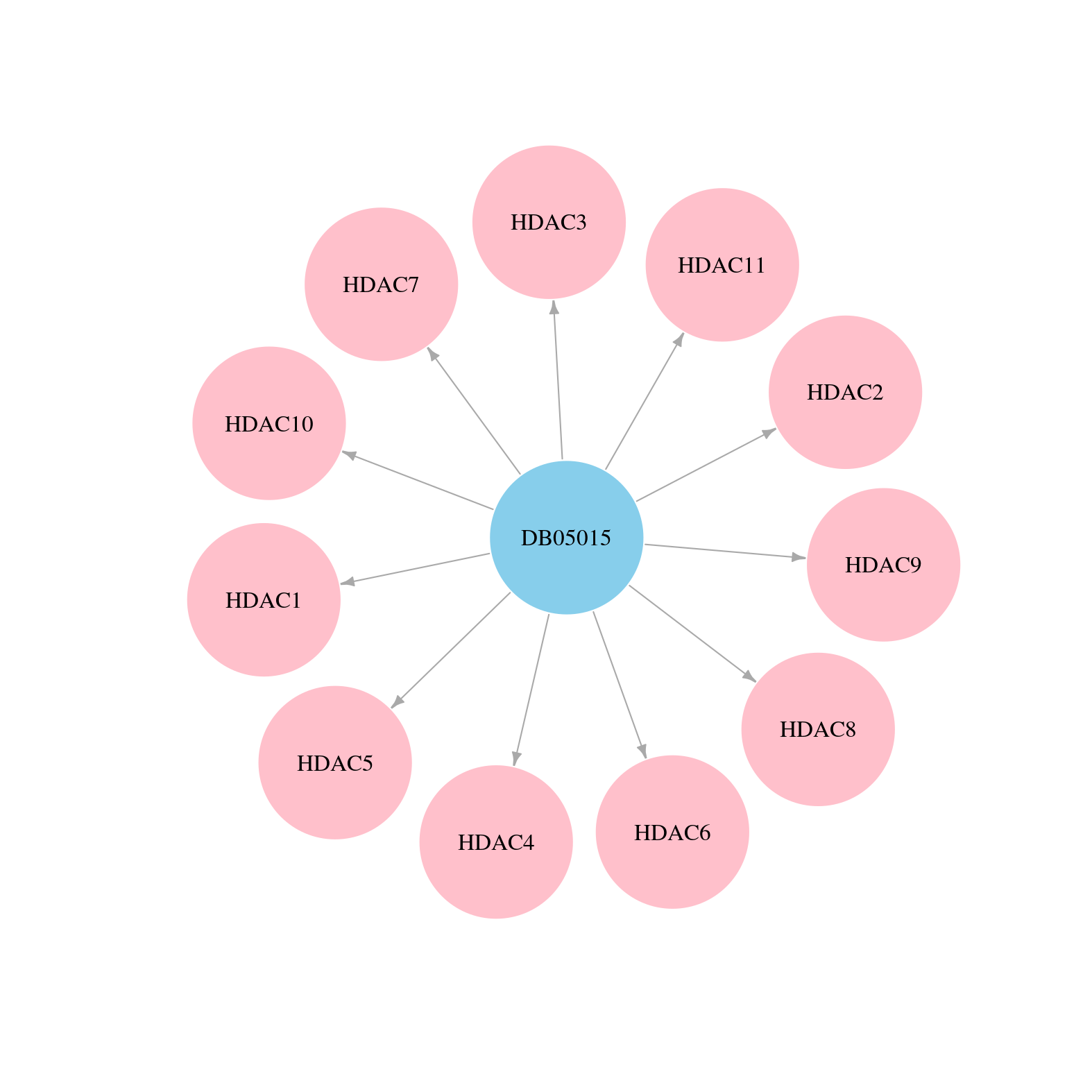
| Content |
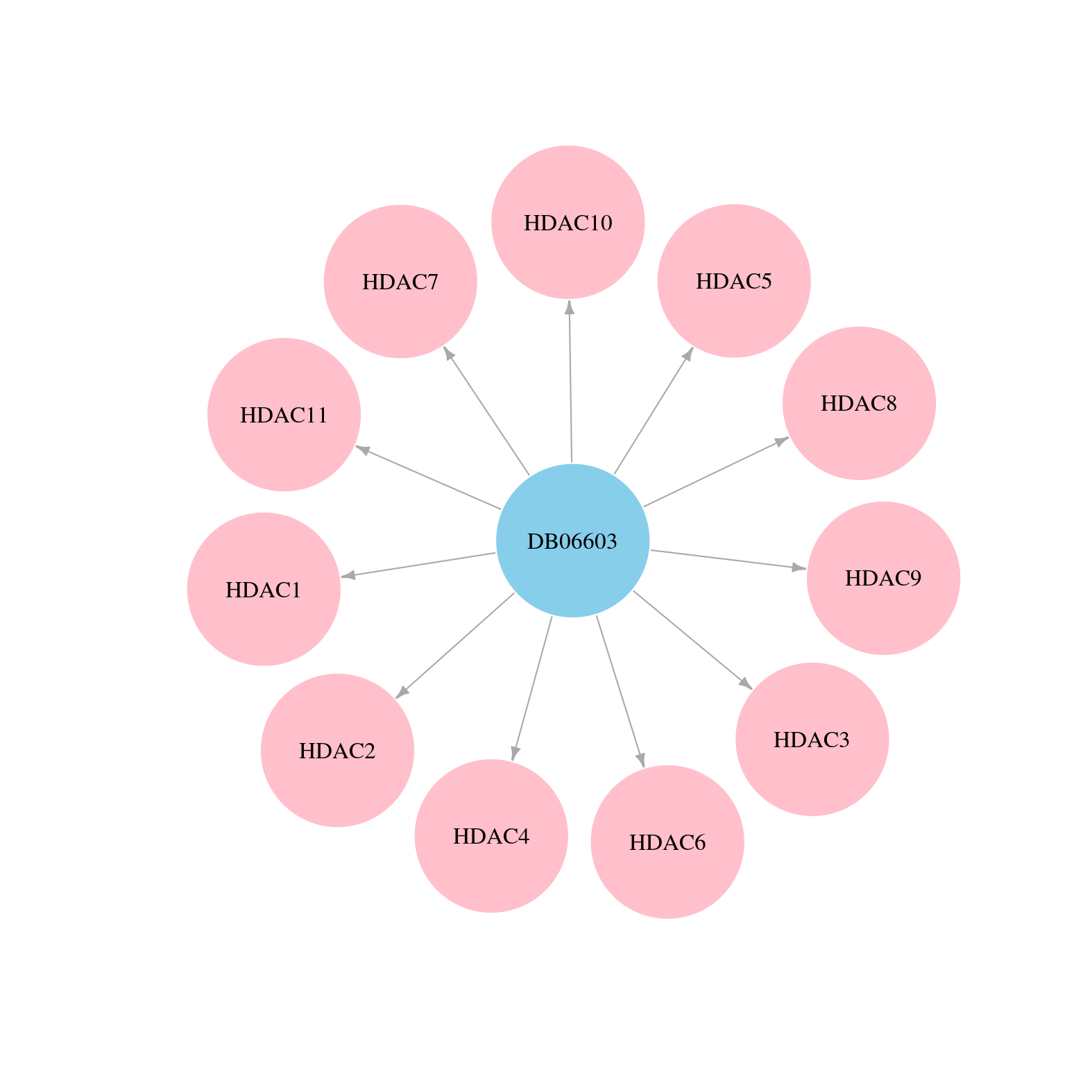
> Drugs from DrugBank database |
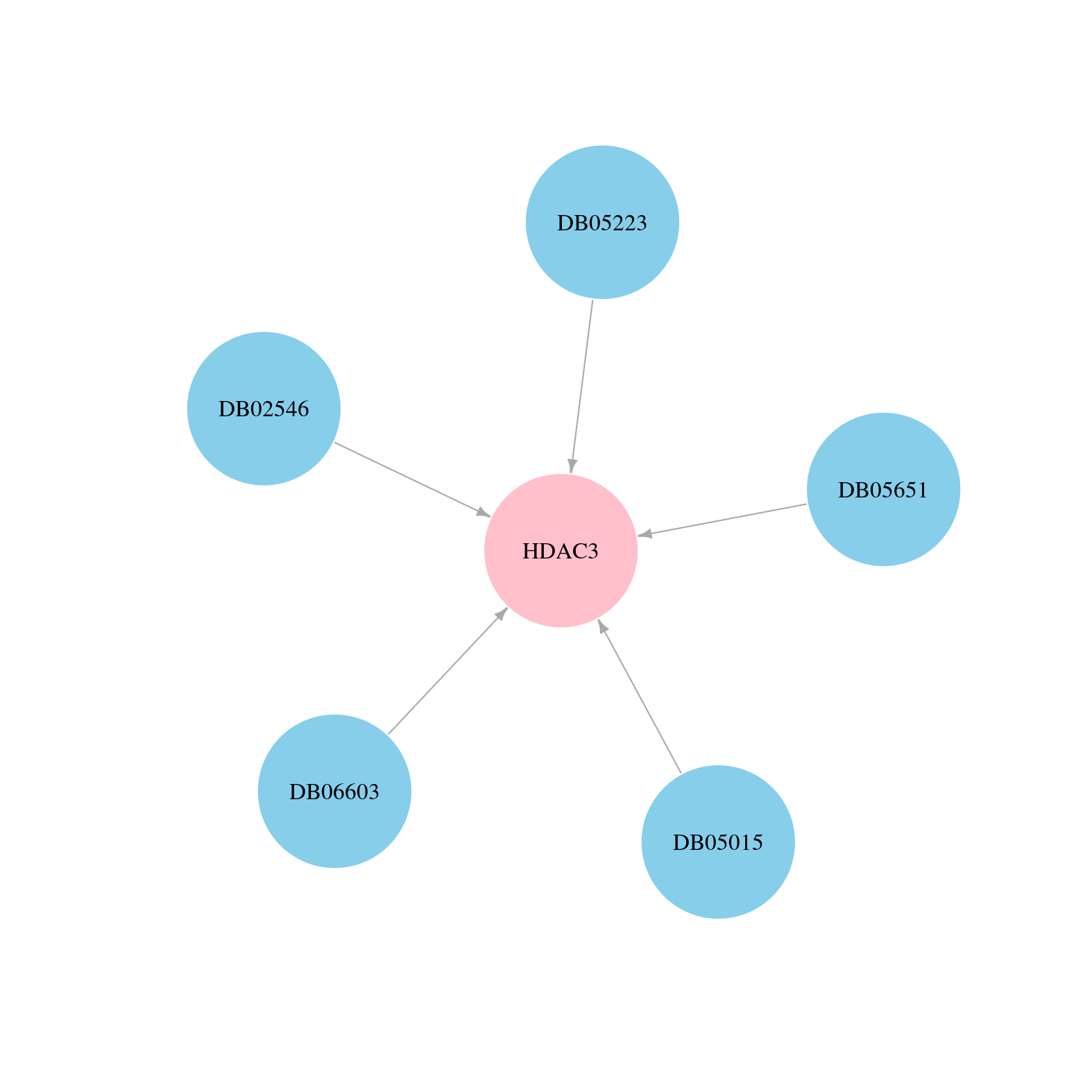
|
| Summary | |
|---|---|
| Symbol | HDAC3 |
| Name | histone deacetylase 3 |
| Aliases | HD3; RPD3-2; SMAP45 |
| Location | 5q31.3 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
| There is no record for HDAC3. |
|