Browse HDAC5 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF12203 Glutamine rich N terminal domain of histone deacetylase 4 PF00850 Histone deacetylase domain |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Involved in muscle maturation by repressing transcription of myocyte enhancer MEF2C. During muscle differentiation, it shuttles into the cytoplasm, allowing the expression of myocyte enhancer factors. Involved in the MTA1-mediated epigenetic regulation of ESR1 expression in breast cancer. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0001525 angiogenesis GO:0001667 ameboidal-type cell migration GO:0002040 sprouting angiogenesis GO:0002042 cell migration involved in sprouting angiogenesis GO:0002237 response to molecule of bacterial origin GO:0002521 leukocyte differentiation GO:0006338 chromatin remodeling GO:0006342 chromatin silencing GO:0006476 protein deacetylation GO:0010594 regulation of endothelial cell migration GO:0010596 negative regulation of endothelial cell migration GO:0010631 epithelial cell migration GO:0010632 regulation of epithelial cell migration GO:0010633 negative regulation of epithelial cell migration GO:0010830 regulation of myotube differentiation GO:0010832 negative regulation of myotube differentiation GO:0014823 response to activity GO:0014902 myotube differentiation GO:0016458 gene silencing GO:0016525 negative regulation of angiogenesis GO:0016570 histone modification GO:0016575 histone deacetylation GO:0030098 lymphocyte differentiation GO:0030183 B cell differentiation GO:0030336 negative regulation of cell migration GO:0032496 response to lipopolysaccharide GO:0032868 response to insulin GO:0032869 cellular response to insulin stimulus GO:0035601 protein deacylation GO:0040013 negative regulation of locomotion GO:0040029 regulation of gene expression, epigenetic GO:0042113 B cell activation GO:0042220 response to cocaine GO:0042493 response to drug GO:0042692 muscle cell differentiation GO:0043279 response to alkaloid GO:0043393 regulation of protein binding GO:0043434 response to peptide hormone GO:0043534 blood vessel endothelial cell migration GO:0043535 regulation of blood vessel endothelial cell migration GO:0043537 negative regulation of blood vessel endothelial cell migration GO:0043542 endothelial cell migration GO:0045765 regulation of angiogenesis GO:0045814 negative regulation of gene expression, epigenetic GO:0048514 blood vessel morphogenesis GO:0051090 regulation of sequence-specific DNA binding transcription factor activity GO:0051091 positive regulation of sequence-specific DNA binding transcription factor activity GO:0051098 regulation of binding GO:0051146 striated muscle cell differentiation GO:0051147 regulation of muscle cell differentiation GO:0051148 negative regulation of muscle cell differentiation GO:0051153 regulation of striated muscle cell differentiation GO:0051154 negative regulation of striated muscle cell differentiation GO:0051271 negative regulation of cellular component movement GO:0070932 histone H3 deacetylation GO:0071216 cellular response to biotic stimulus GO:0071219 cellular response to molecule of bacterial origin GO:0071222 cellular response to lipopolysaccharide GO:0071375 cellular response to peptide hormone stimulus GO:0071396 cellular response to lipid GO:0071417 cellular response to organonitrogen compound GO:0090049 regulation of cell migration involved in sprouting angiogenesis GO:0090051 negative regulation of cell migration involved in sprouting angiogenesis GO:0090130 tissue migration GO:0090132 epithelium migration GO:0098732 macromolecule deacylation GO:1901342 regulation of vasculature development GO:1901343 negative regulation of vasculature development GO:1901652 response to peptide GO:1901653 cellular response to peptide GO:1903670 regulation of sprouting angiogenesis GO:1903671 negative regulation of sprouting angiogenesis GO:2000146 negative regulation of cell motility GO:2000181 negative regulation of blood vessel morphogenesis |
| Molecular Function |
GO:0001025 RNA polymerase III transcription factor binding GO:0001047 core promoter binding GO:0003682 chromatin binding GO:0004407 histone deacetylase activity GO:0005080 protein kinase C binding GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0042826 histone deacetylase binding GO:0070491 repressing transcription factor binding |
| Cellular Component |
GO:0000118 histone deacetylase complex |
| KEGG | - |
| Reactome |
R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer |
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
| There is no record. |
|
| There is no record for HDAC5. |
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
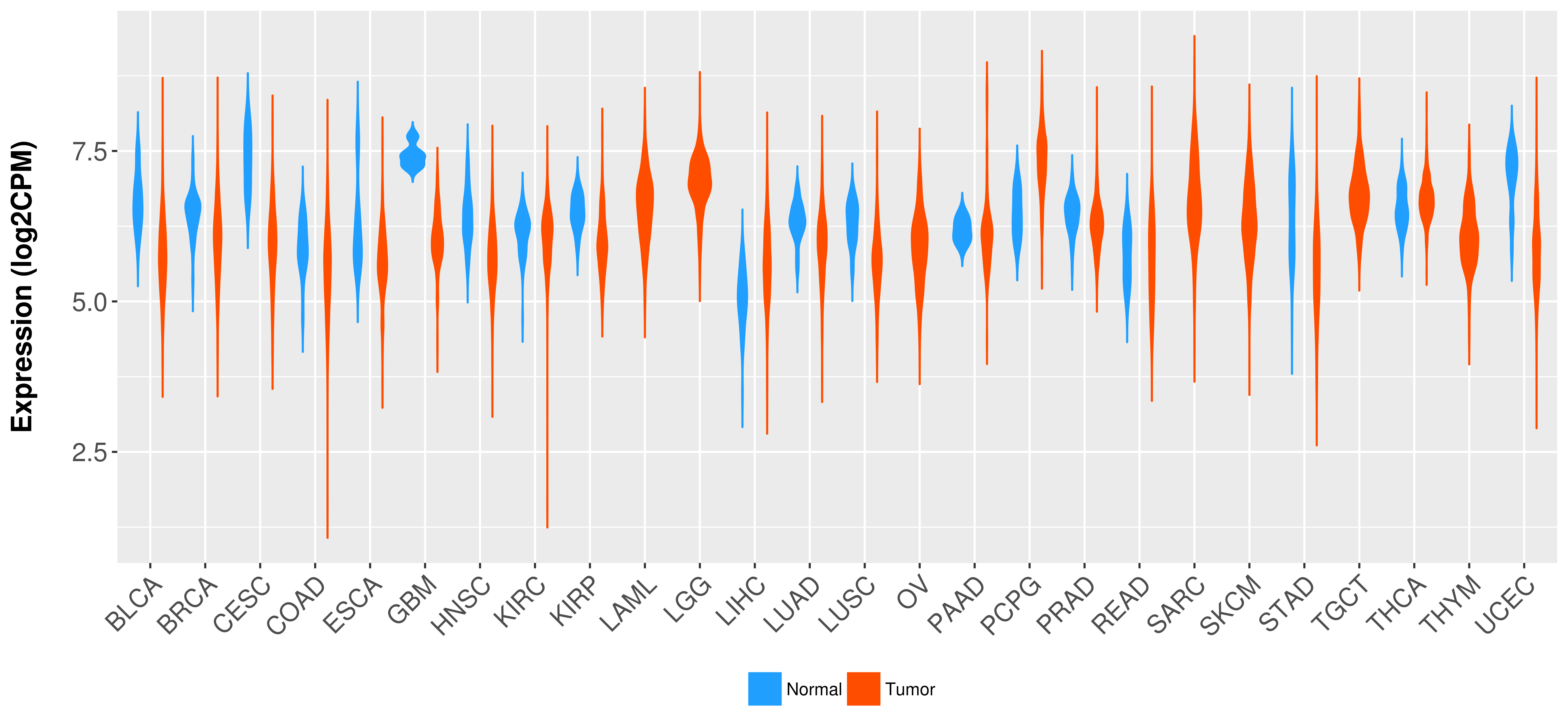
Differential expression analysis for cancers with more than 10 normal samples
|
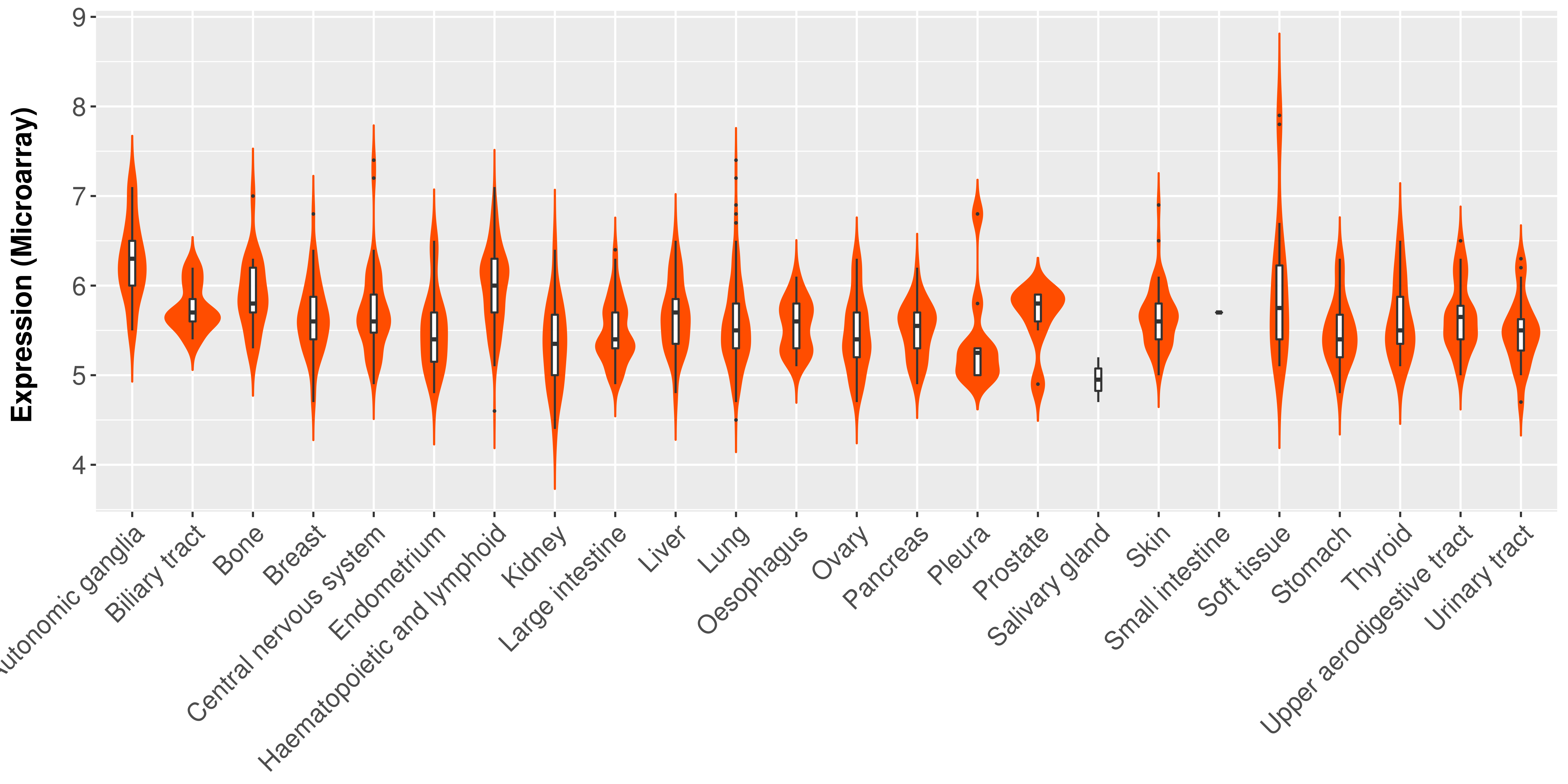
|
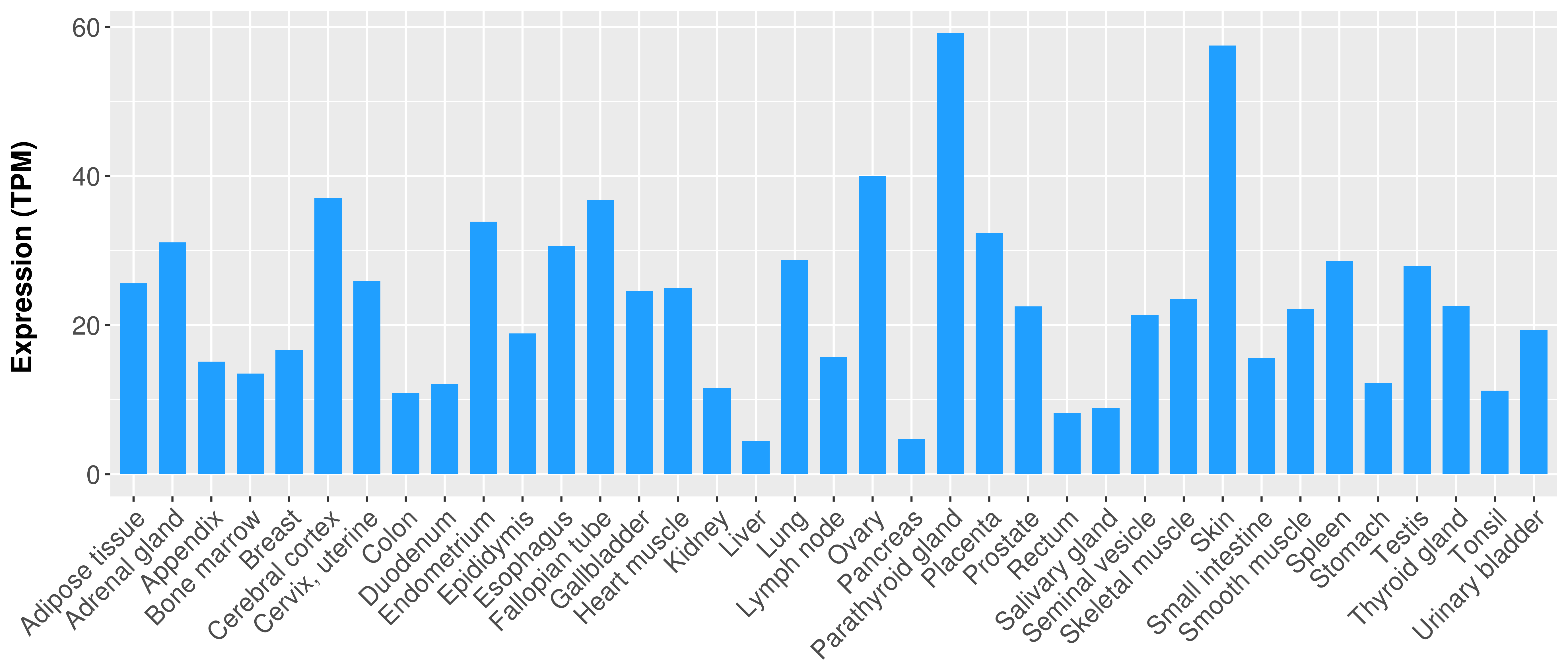
|
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
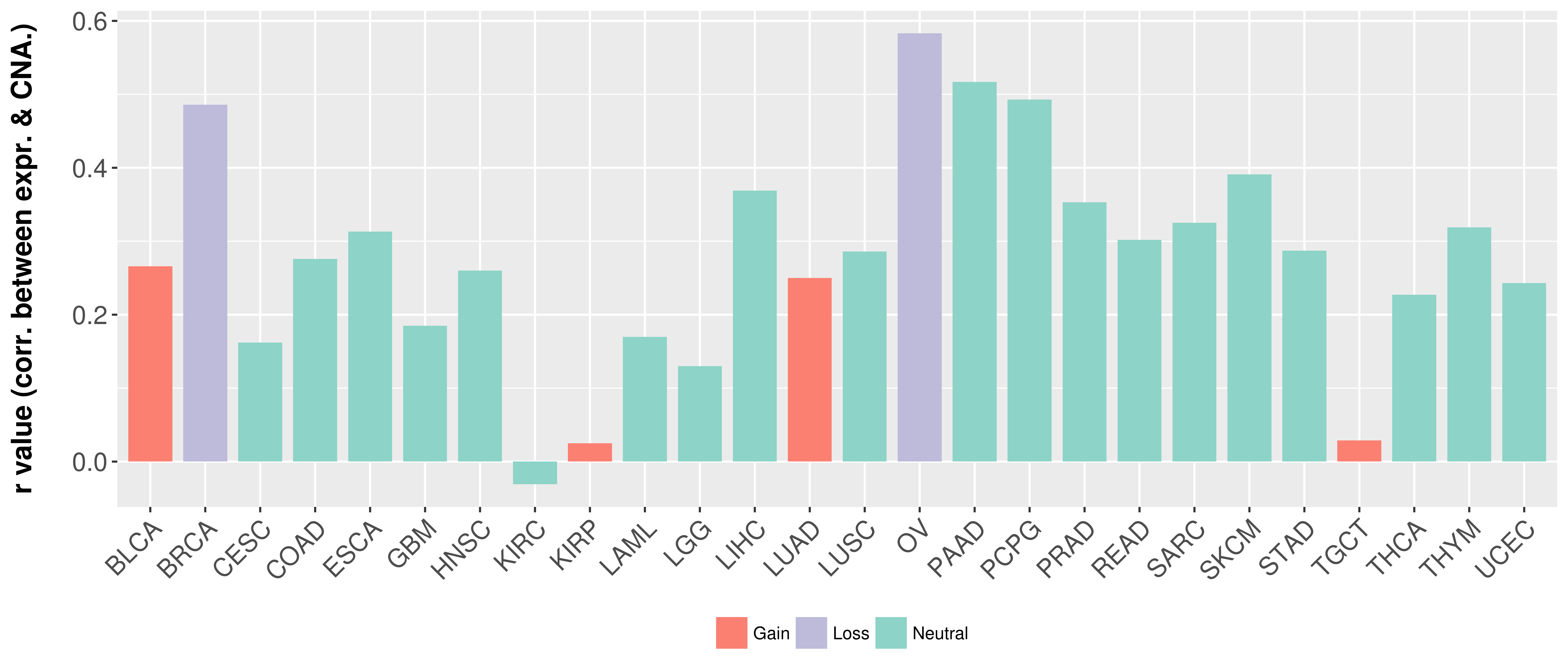
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
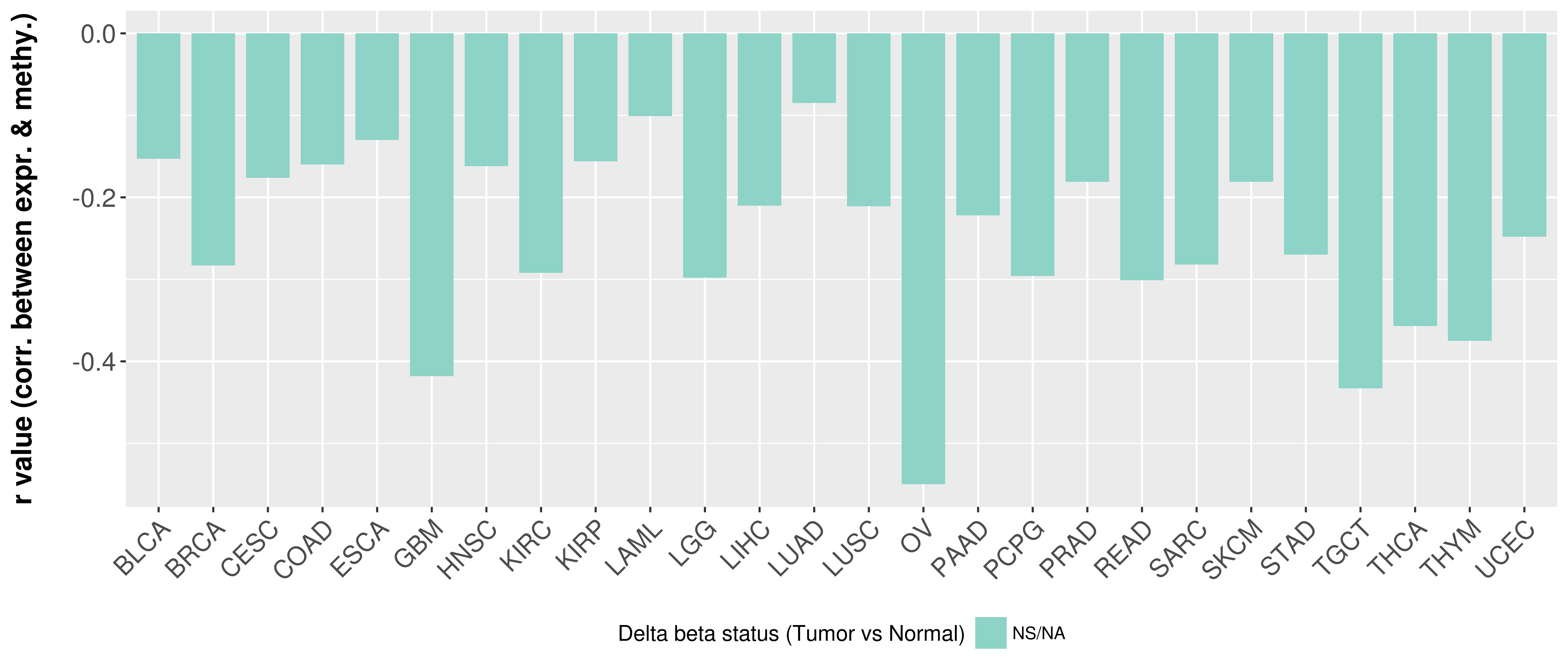
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |

|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
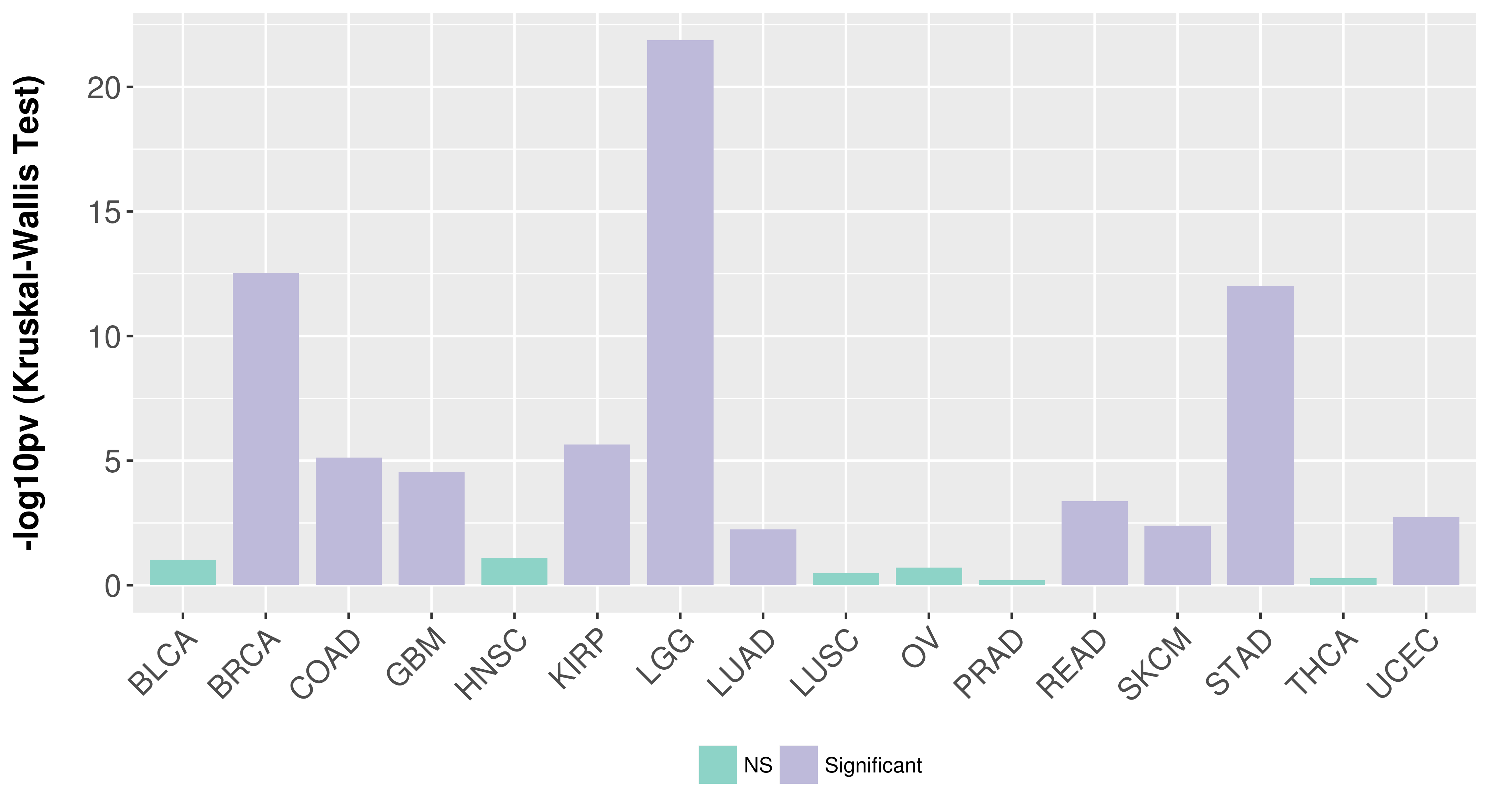
Association between expresson and subtype.
|
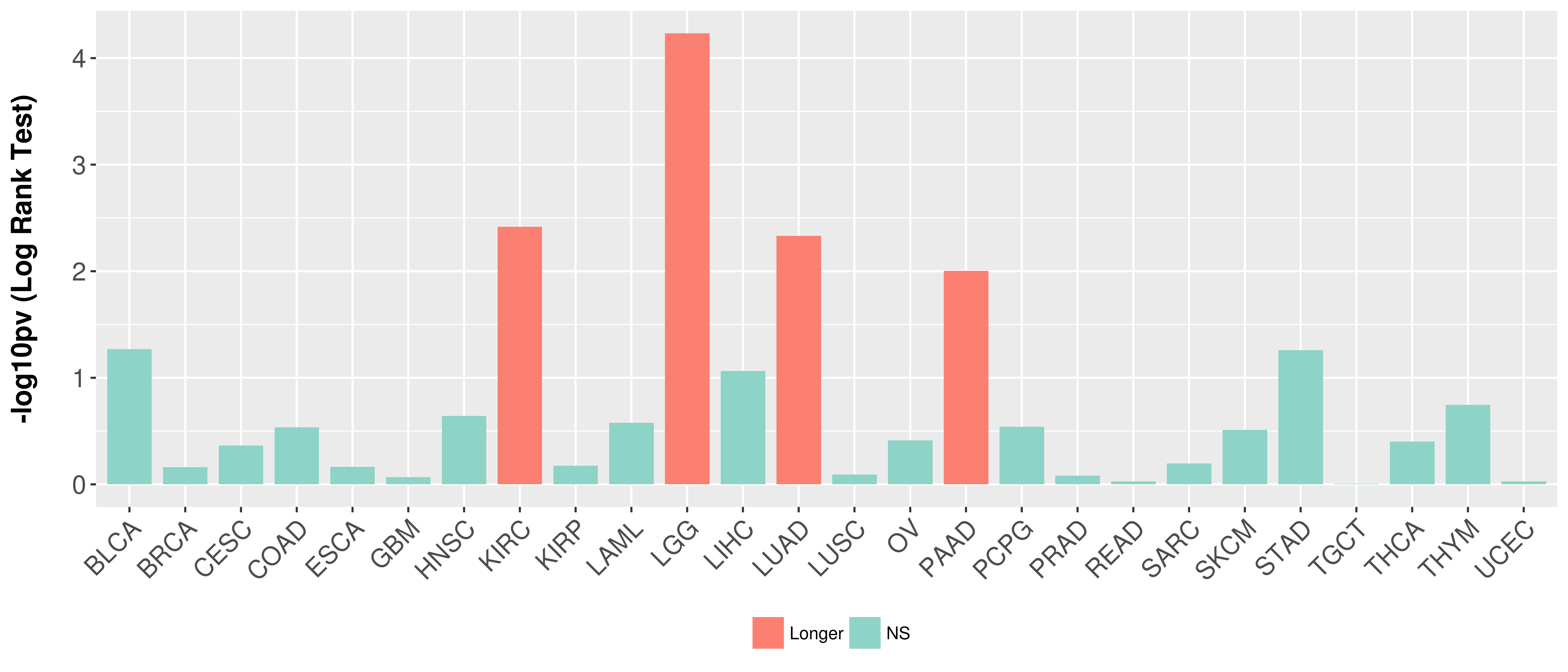
Overall survival analysis based on expression.
|
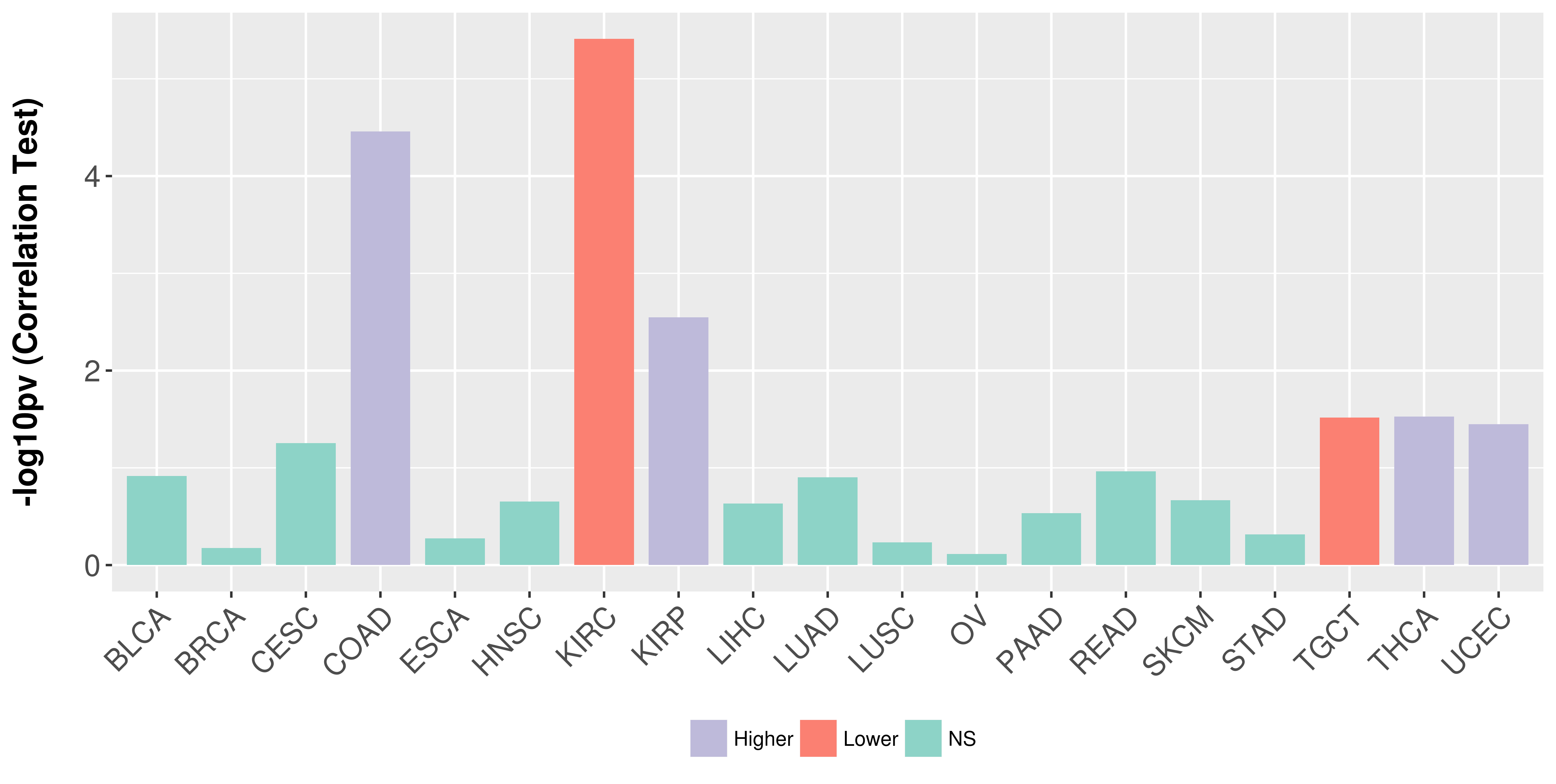
Association between expresson and stage.
|
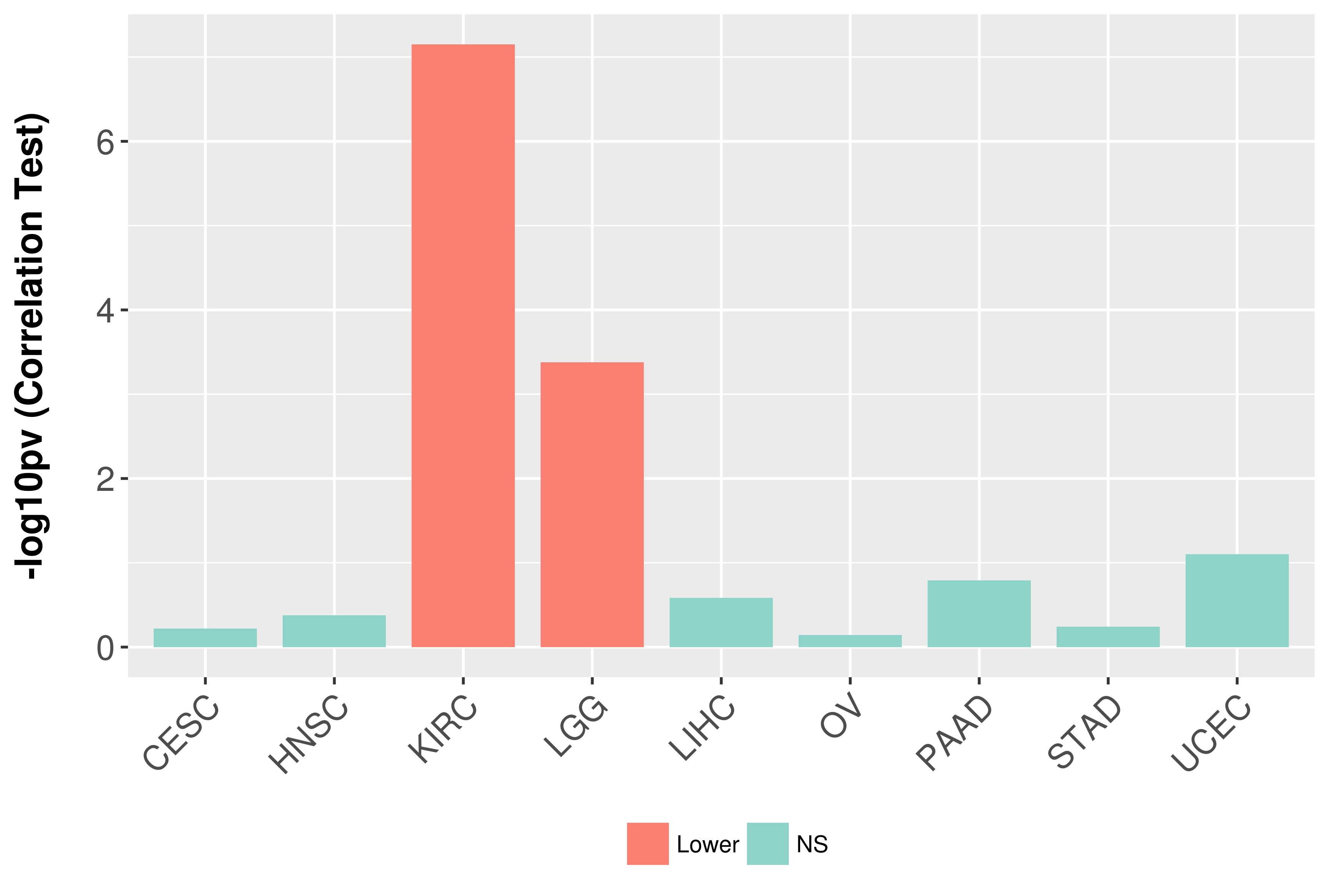
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
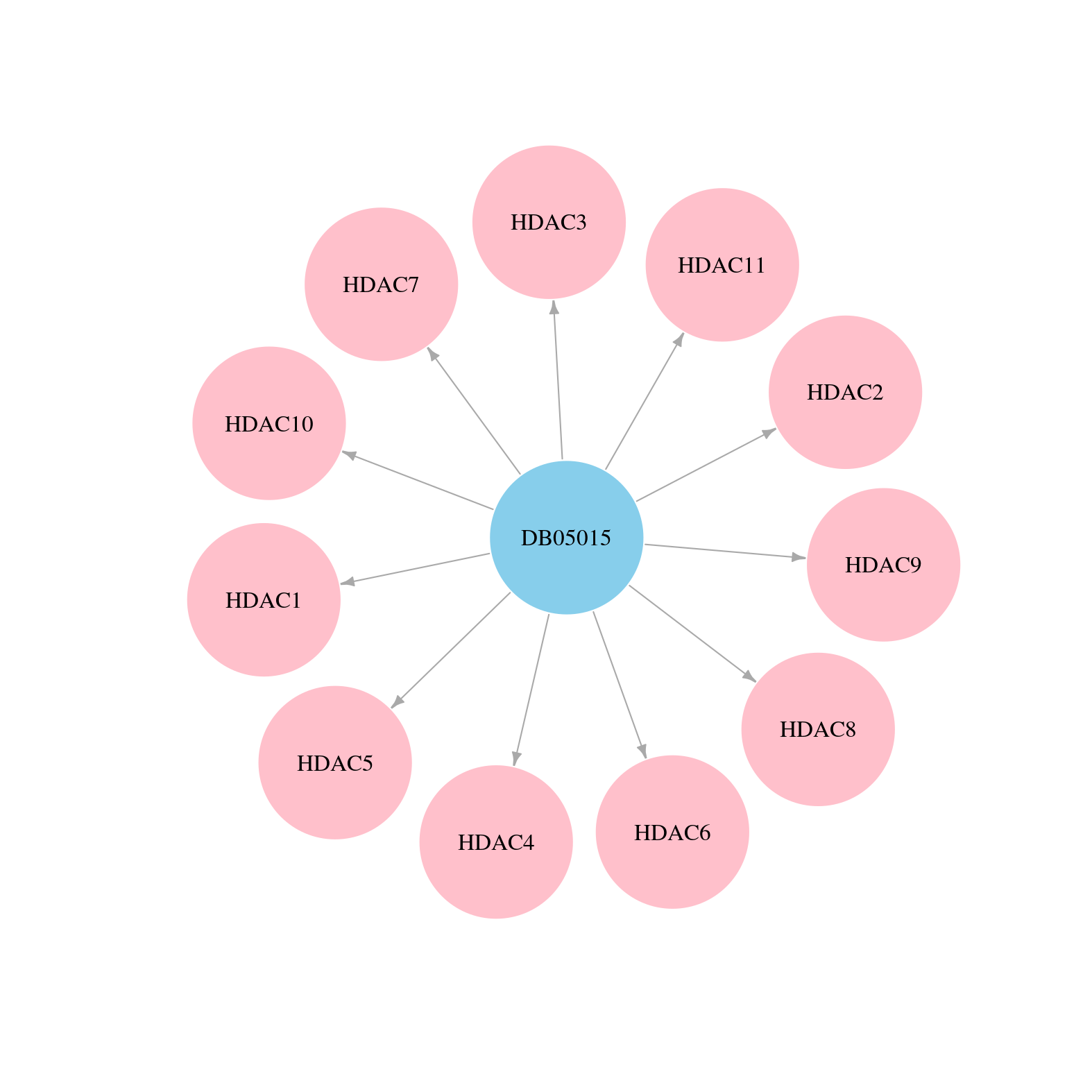
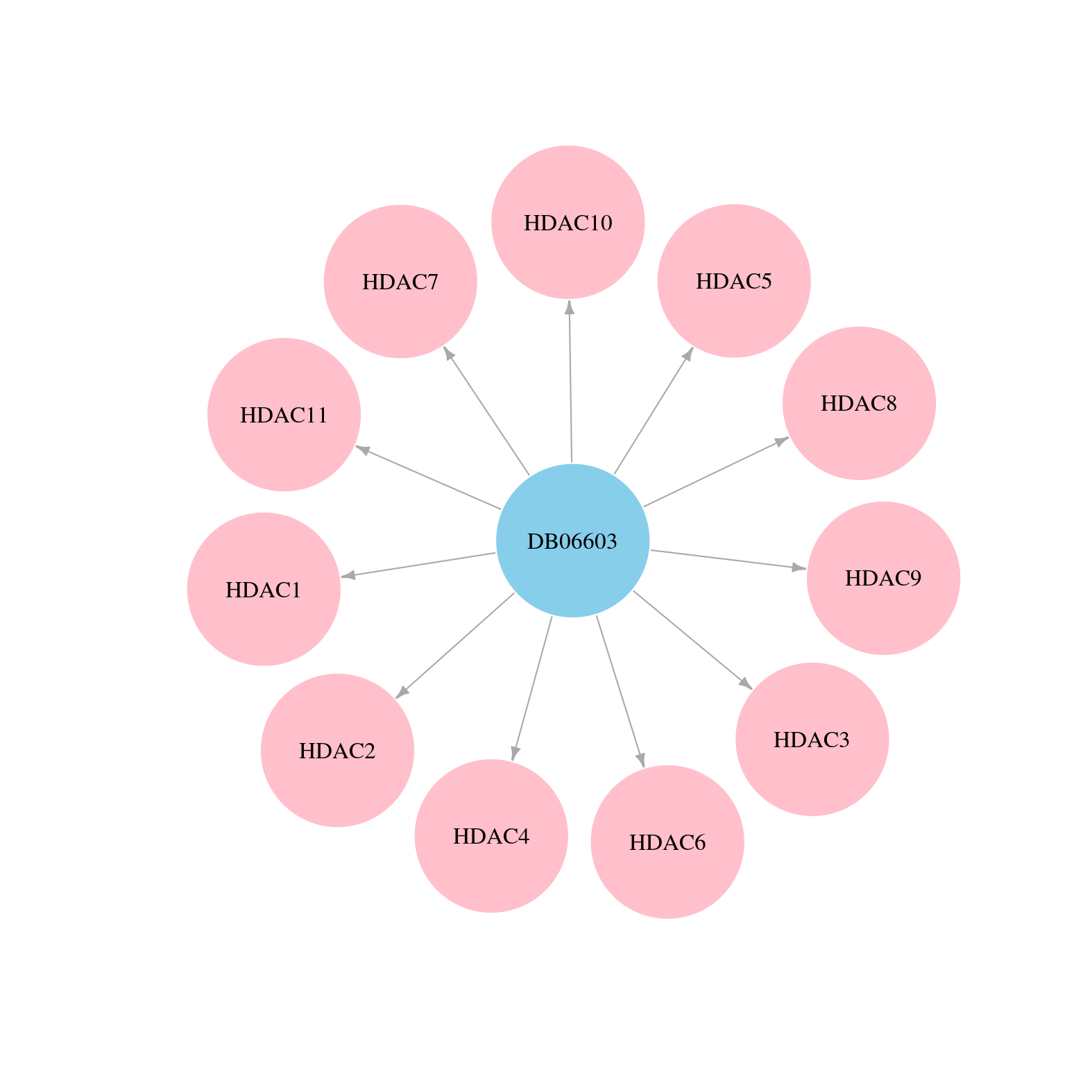
> Drugs from DrugBank database |
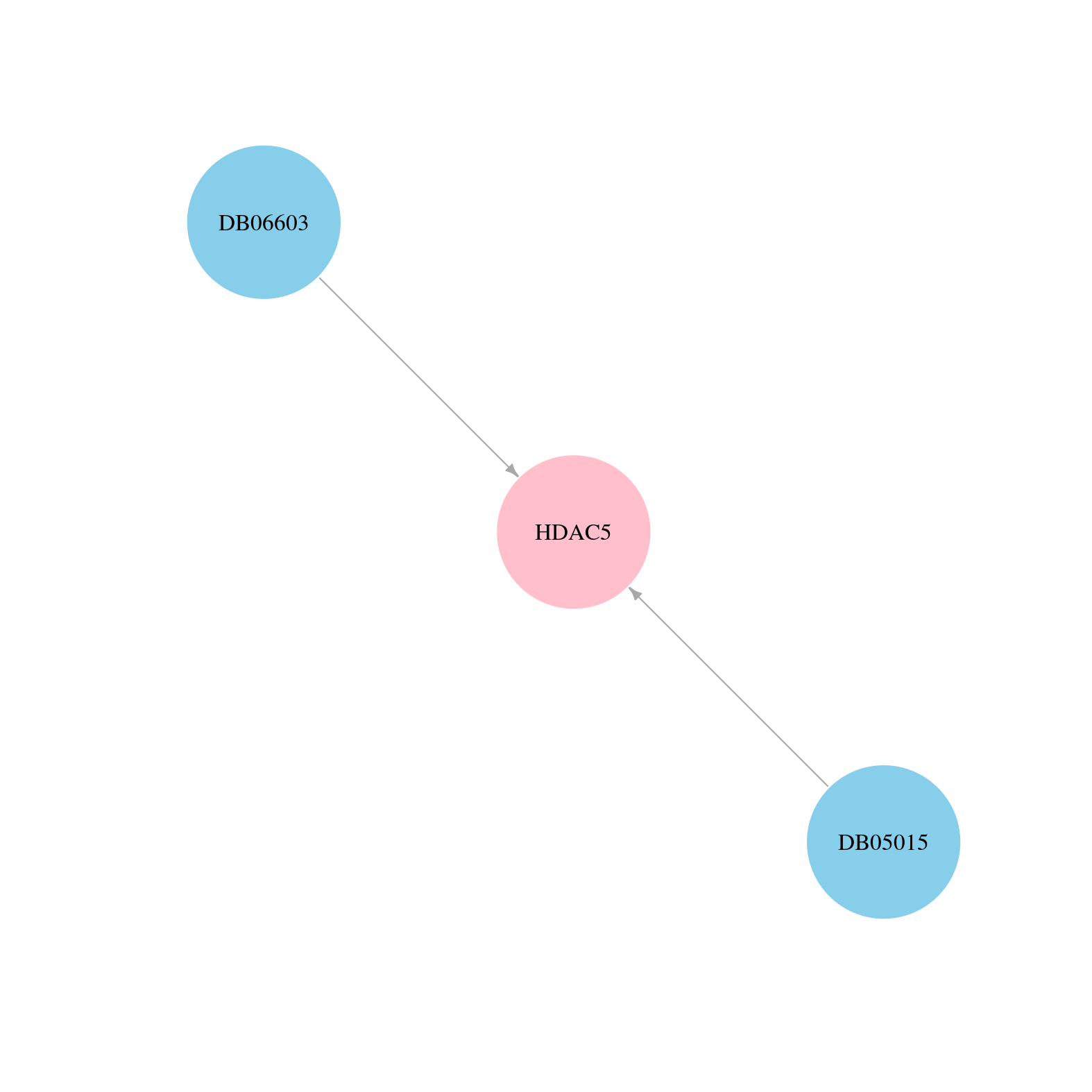
|
| Summary | |
|---|---|
| Symbol | HDAC5 |
| Name | histone deacetylase 5 |
| Aliases | KIAA0600; NY-CO-9; FLJ90614; HD5; antigen NY-CO-9 |
| Location | 17q21.31 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|