Browse HDAC8 in pancancer
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Domain, Function and Classification > Gene Ontology > KEGG and Reactome Pathway |
| Domain |
PF00850 Histone deacetylase domain |
||||||||||
| Function |
Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Also involved in the deacetylation of cohesin complex protein SMC3 regulating release of cohesin complexes from chromatin. May play a role in smooth muscle cell contractility. |
||||||||||
| Classification |
|
||||||||||
| Biological Process |
GO:0000723 telomere maintenance GO:0000819 sister chromatid segregation GO:0006333 chromatin assembly or disassembly GO:0006476 protein deacetylation GO:0007059 chromosome segregation GO:0007062 sister chromatid cohesion GO:0007063 regulation of sister chromatid cohesion GO:0016570 histone modification GO:0016575 histone deacetylation GO:0031396 regulation of protein ubiquitination GO:0031397 negative regulation of protein ubiquitination GO:0031647 regulation of protein stability GO:0032200 telomere organization GO:0032204 regulation of telomere maintenance GO:0032844 regulation of homeostatic process GO:0033044 regulation of chromosome organization GO:0033045 regulation of sister chromatid segregation GO:0034085 establishment of sister chromatid cohesion GO:0034502 protein localization to chromosome GO:0035601 protein deacylation GO:0043388 positive regulation of DNA binding GO:0051052 regulation of DNA metabolic process GO:0051098 regulation of binding GO:0051099 positive regulation of binding GO:0051101 regulation of DNA binding GO:0051983 regulation of chromosome segregation GO:0060249 anatomical structure homeostasis GO:0070932 histone H3 deacetylation GO:0071168 protein localization to chromatin GO:0071921 cohesin loading GO:0071922 regulation of cohesin loading GO:0098732 macromolecule deacylation GO:0098813 nuclear chromosome segregation GO:1903320 regulation of protein modification by small protein conjugation or removal GO:1903321 negative regulation of protein modification by small protein conjugation or removal |
| Molecular Function |
GO:0004407 histone deacetylase activity GO:0008134 transcription factor binding GO:0016810 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds GO:0016811 hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides GO:0017136 NAD-dependent histone deacetylase activity GO:0019213 deacetylase activity GO:0030544 Hsp70 protein binding GO:0031072 heat shock protein binding GO:0031078 histone deacetylase activity (H3-K14 specific) GO:0032041 NAD-dependent histone deacetylase activity (H3-K14 specific) GO:0033558 protein deacetylase activity GO:0034979 NAD-dependent protein deacetylase activity GO:0051879 Hsp90 protein binding |
| Cellular Component |
GO:0000118 histone deacetylase complex |
| KEGG | - |
| Reactome |
R-HSA-1640170: Cell Cycle R-HSA-69278: Cell Cycle, Mitotic R-HSA-3247509: Chromatin modifying enzymes R-HSA-4839726: Chromatin organization R-HSA-2894862: Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants R-HSA-2644606: Constitutive Signaling by NOTCH1 PEST Domain Mutants R-HSA-1643685: Disease R-HSA-5663202: Diseases of signal transduction R-HSA-3214815: HDACs deacetylate histones R-HSA-68886: M Phase R-HSA-68882: Mitotic Anaphase R-HSA-2555396: Mitotic Metaphase and Anaphase R-HSA-68877: Mitotic Prometaphase R-HSA-2122947: NOTCH1 Intracellular Domain Regulates Transcription R-HSA-2500257: Resolution of Sister Chromatid Cohesion R-HSA-2467813: Separation of Sister Chromatids R-HSA-162582: Signal Transduction R-HSA-157118: Signaling by NOTCH R-HSA-1980143: Signaling by NOTCH1 R-HSA-2894858: Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer R-HSA-2644602: Signaling by NOTCH1 PEST Domain Mutants in Cancer R-HSA-2644603: Signaling by NOTCH1 in Cancer |
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Mutation landscape in primary tumor tissue from TCGA > Mutation landscape in cancer cell line from CCLE > All mutations from COSMIC database V81 > Variations from text mining |
|
| There is no record for HDAC8. |
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Post-translational modification (PTM) |
|
Filter By:
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Expression analysis in primary tumor tissue from TCGA > Expression level in cancer cell line from CCLE > Expression level in human normal tissue from HPA > Text mining based expression change |
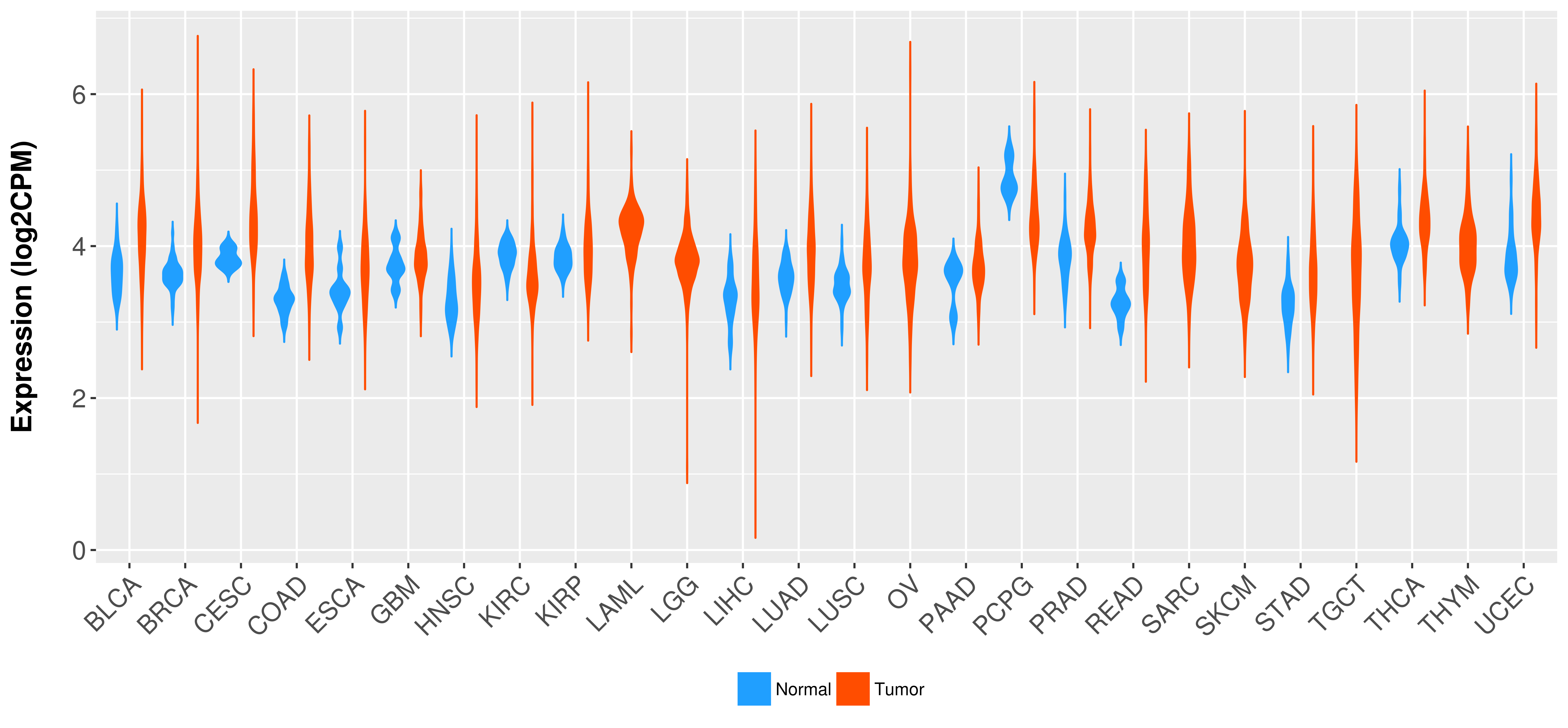
Differential expression analysis for cancers with more than 10 normal samples
|
|
There is no record. |
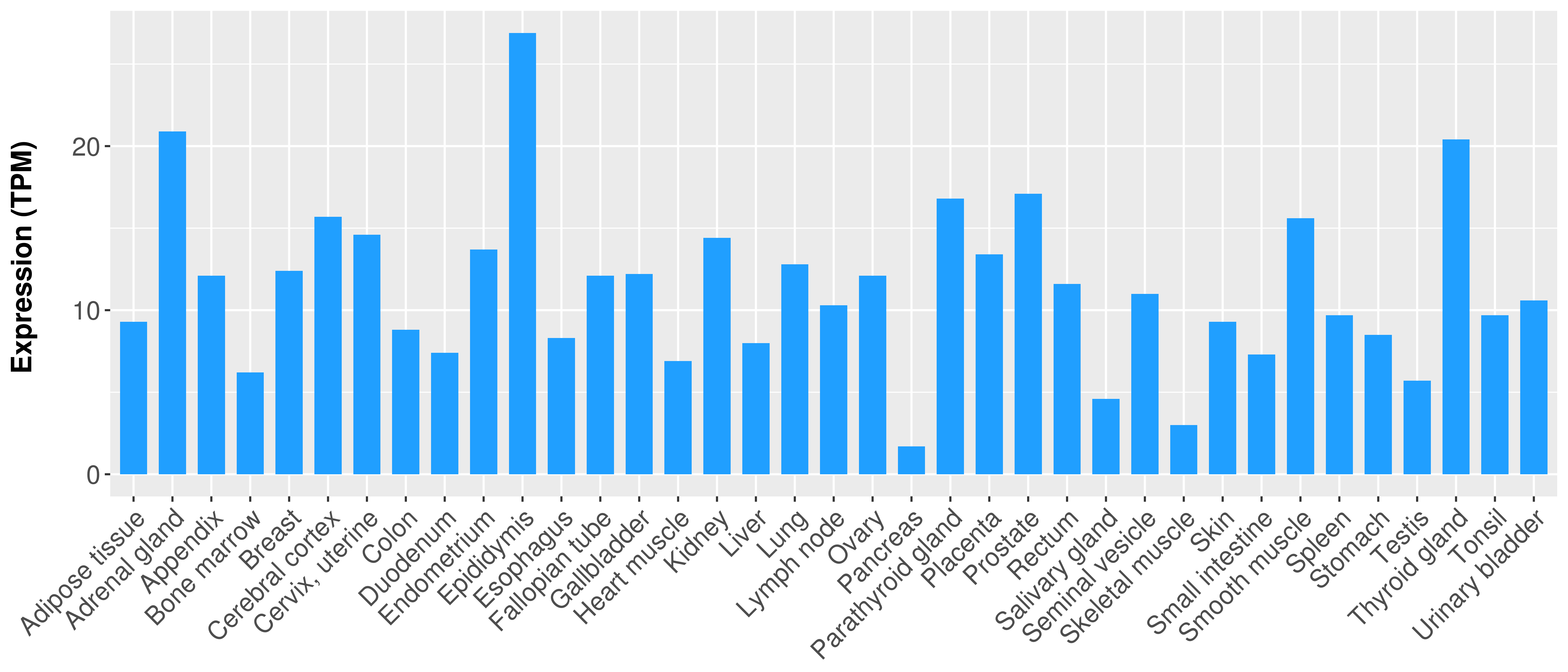
|
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Somatic copy number alteration in primary tomur tissue |
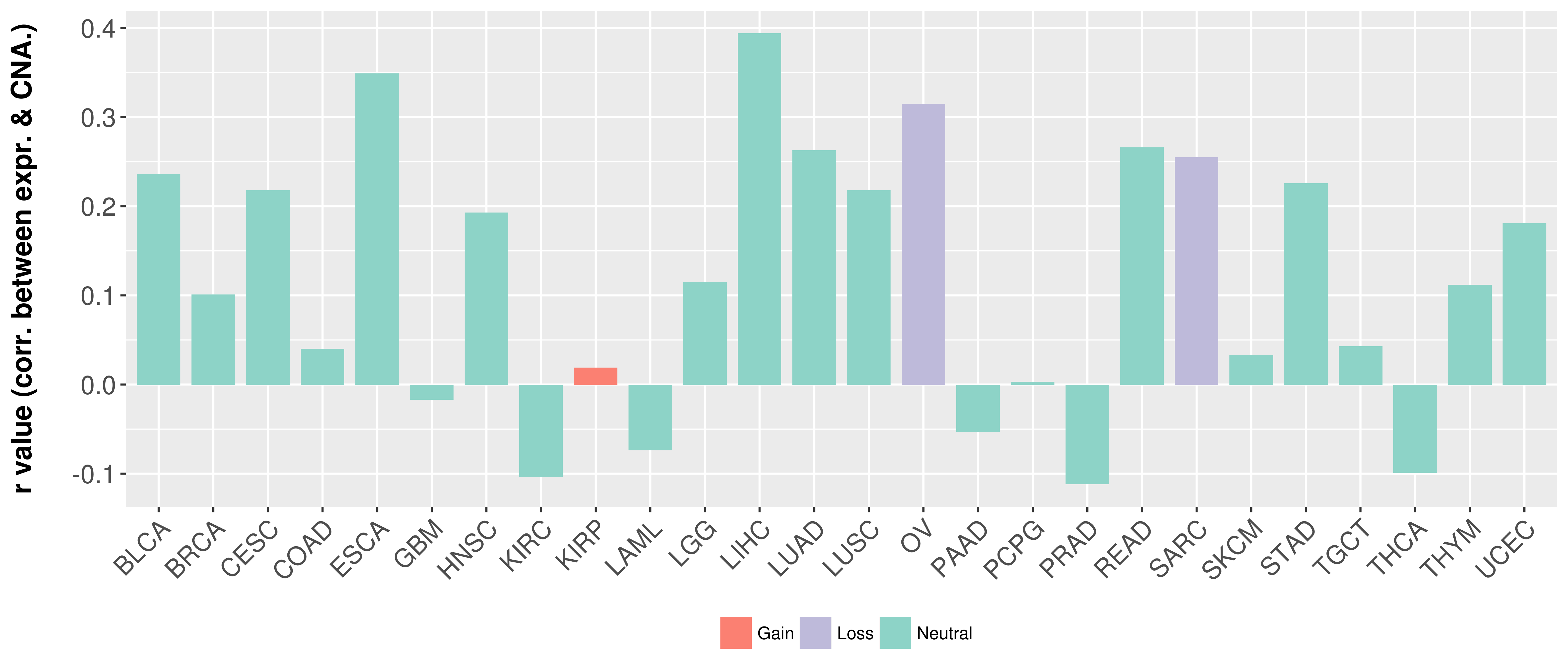
Correlation between expression and SCNA as well as percentage of patients in different status.
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Methylation level in the promoter region of CR |
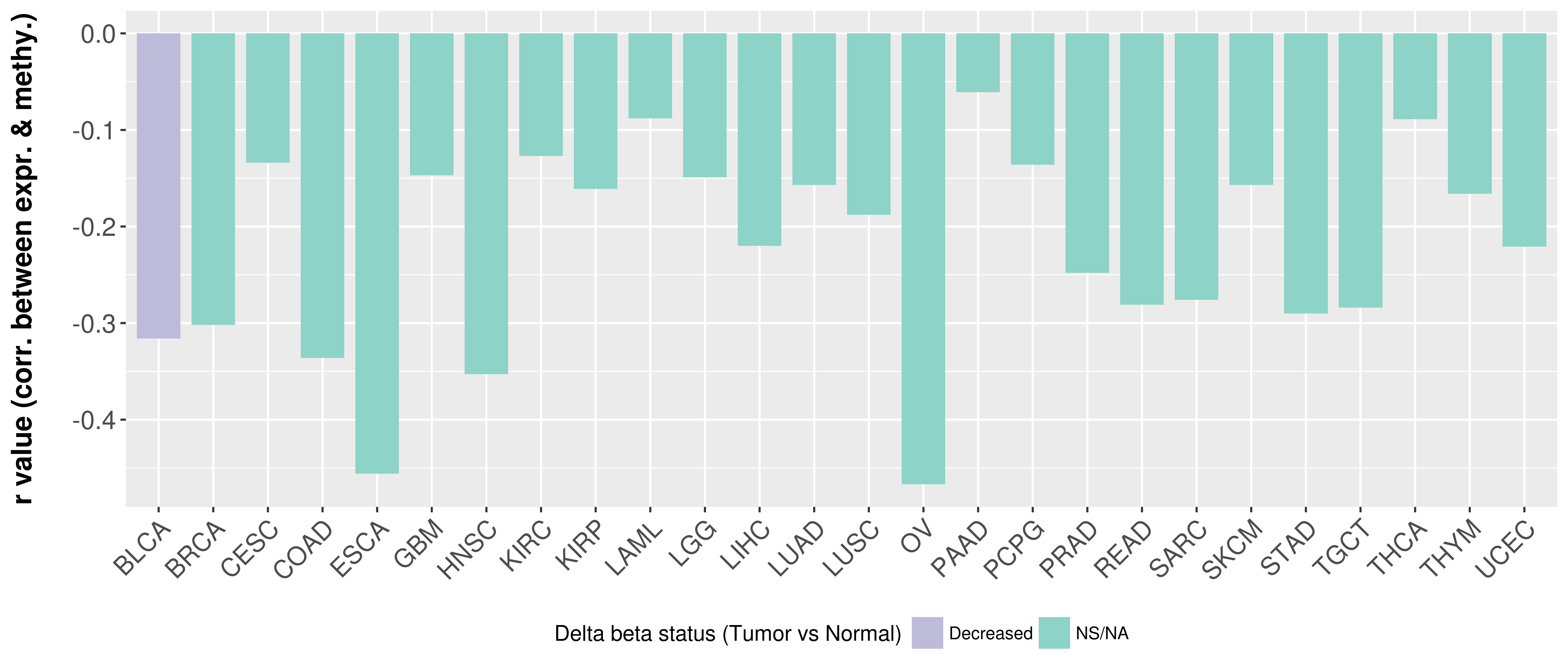
Correlation between expression and methylation as well as differential methylation analysis.
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Primary tumor tissue from TCGA > Normal tumor tissue from HPA |
| There is no record. |
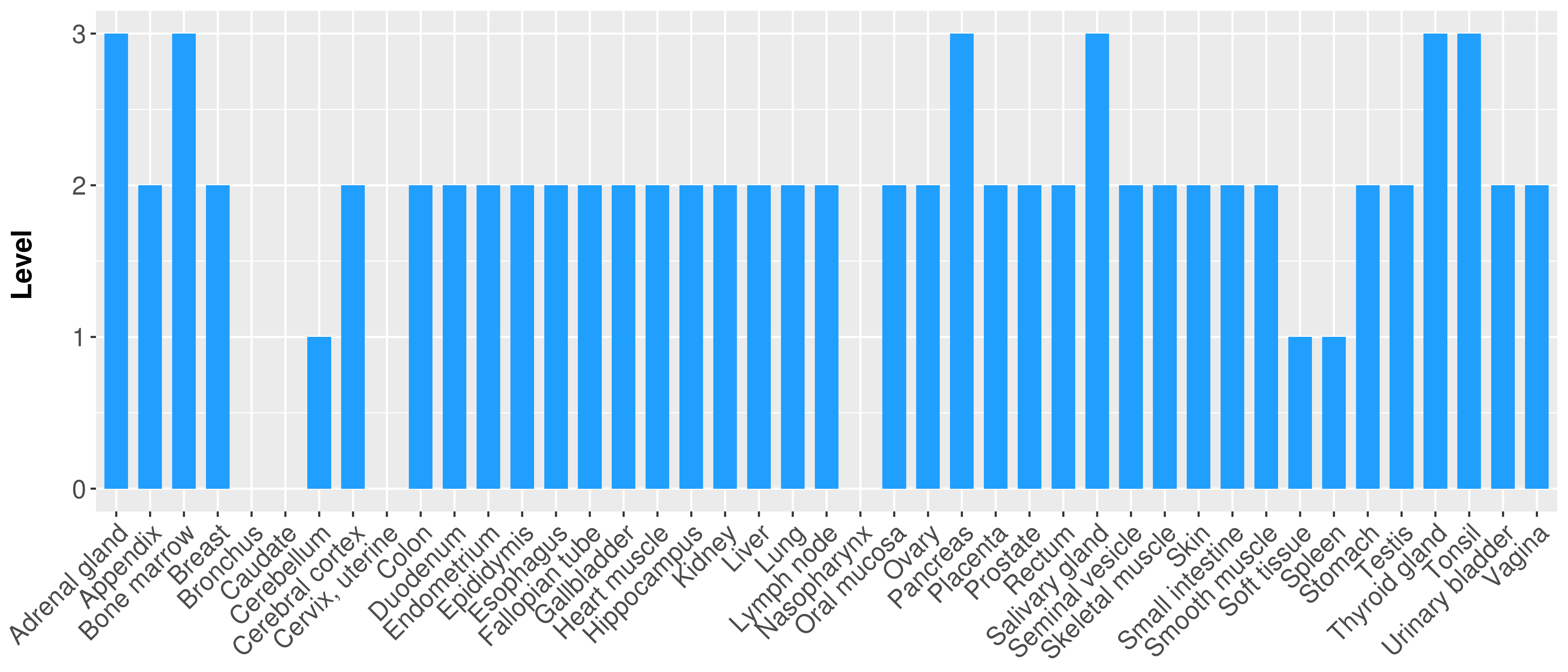
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
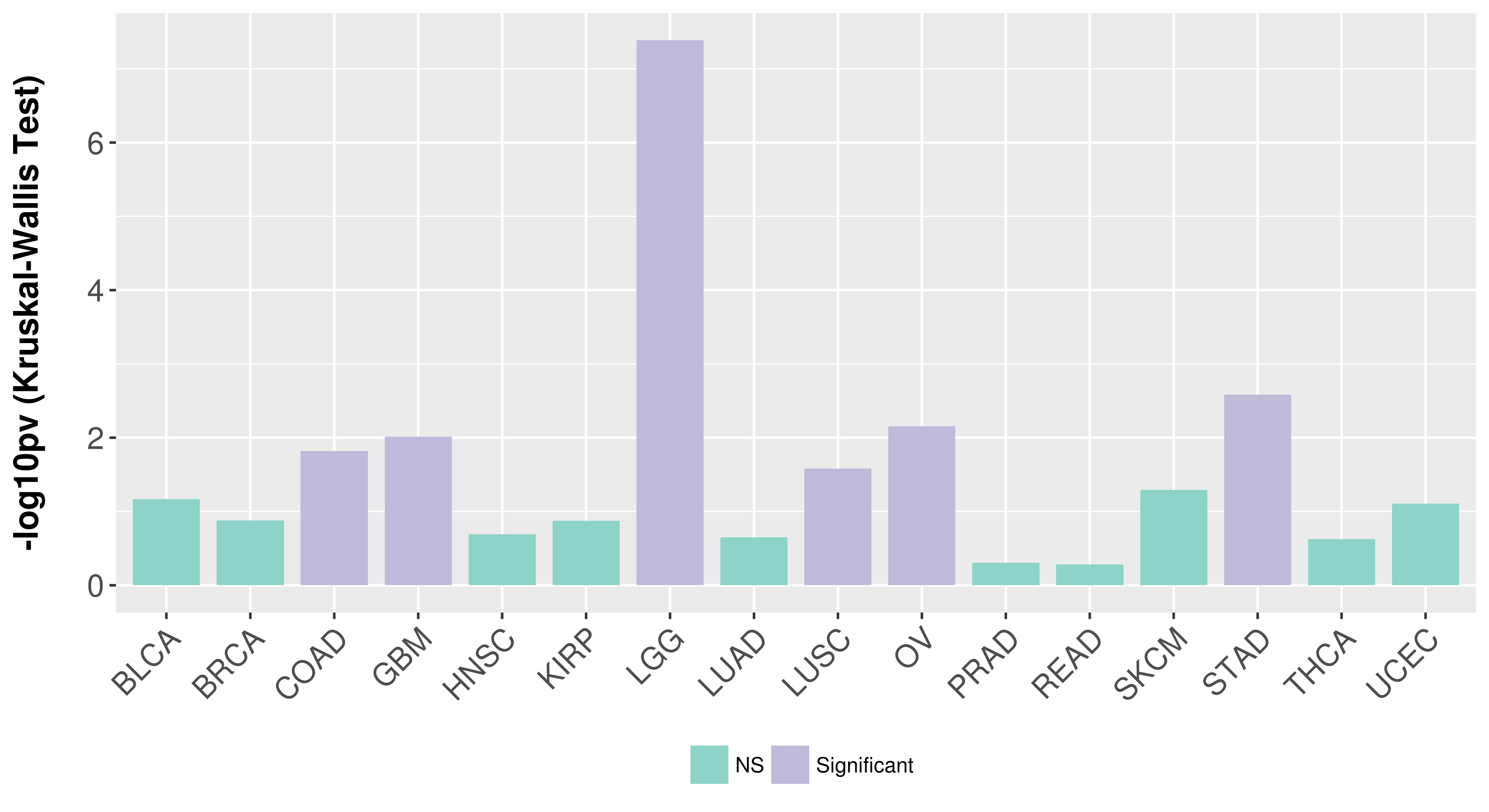
> Association between expresson and subtype > Overall survival analysis based on expression > Association between expresson and stage > Association between expresson and grade |
Association between expresson and subtype.
|
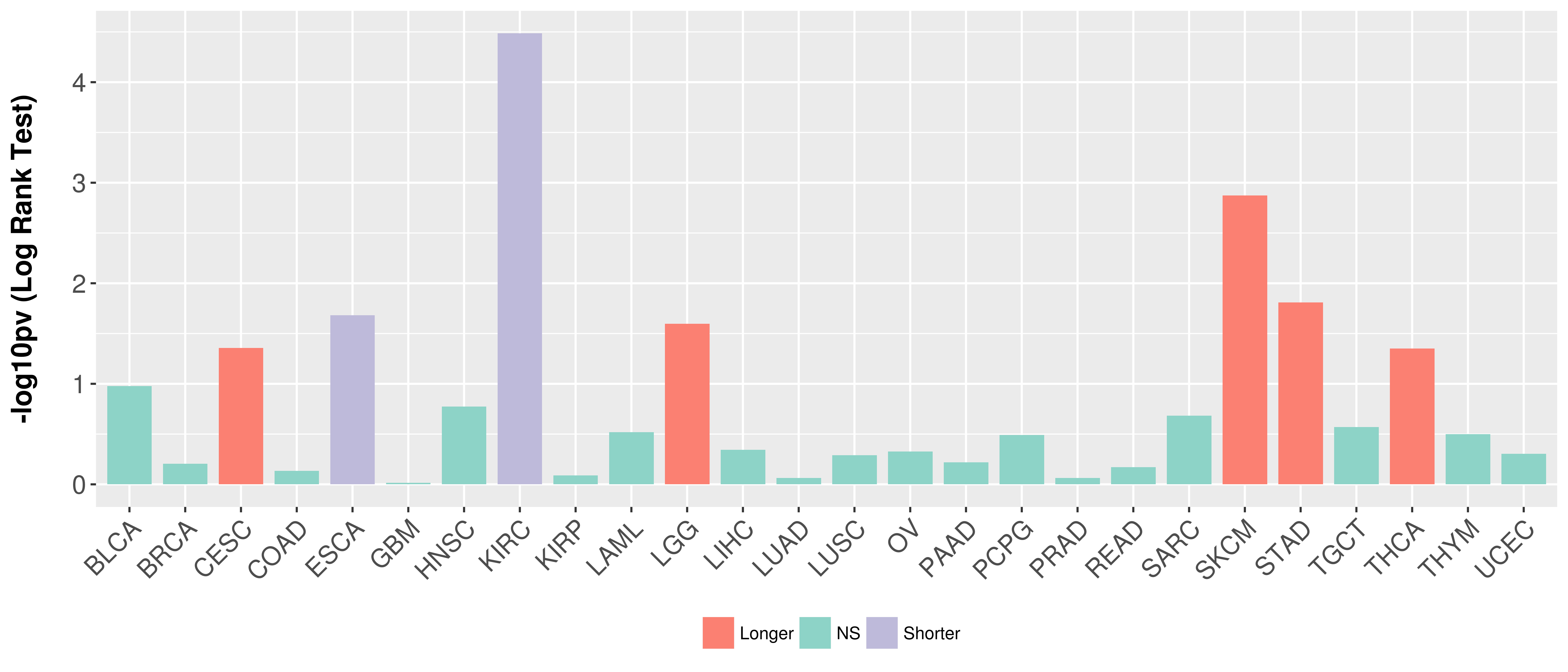
Overall survival analysis based on expression.
|
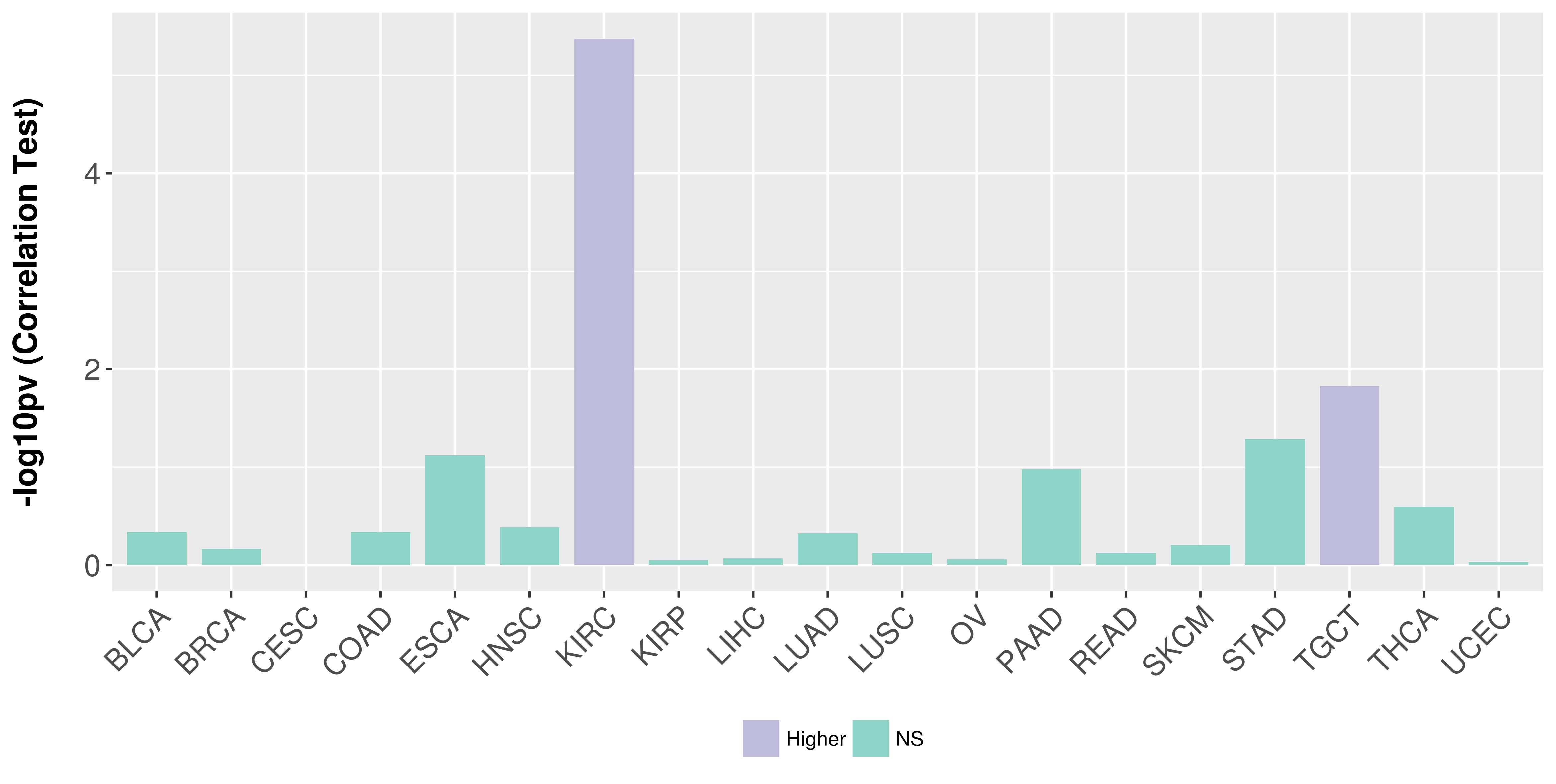
Association between expresson and stage.
|
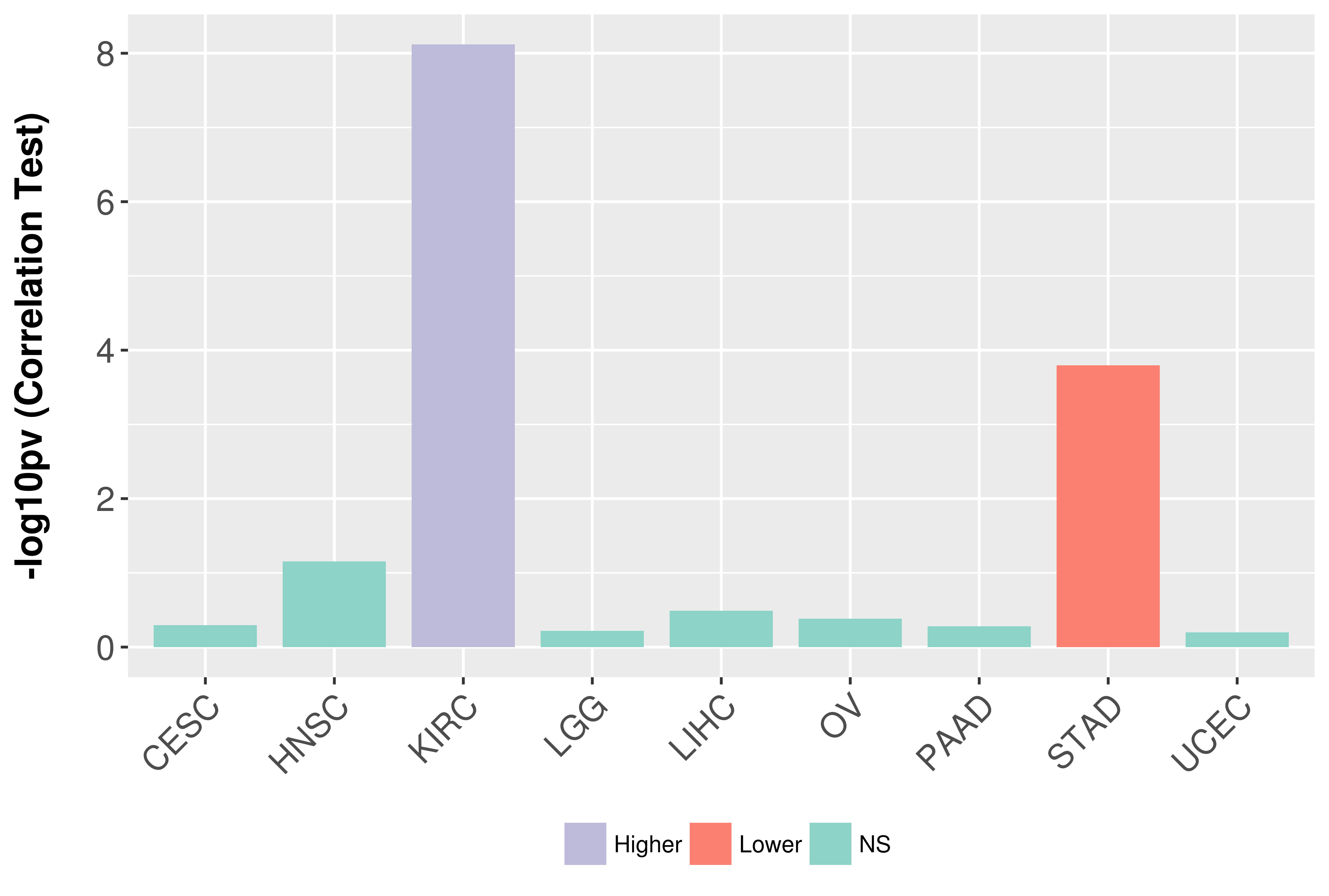
Association between expresson and grade.
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
> Targets inferred by reverse engineering method > Targets identified by ChIP-seq data |
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
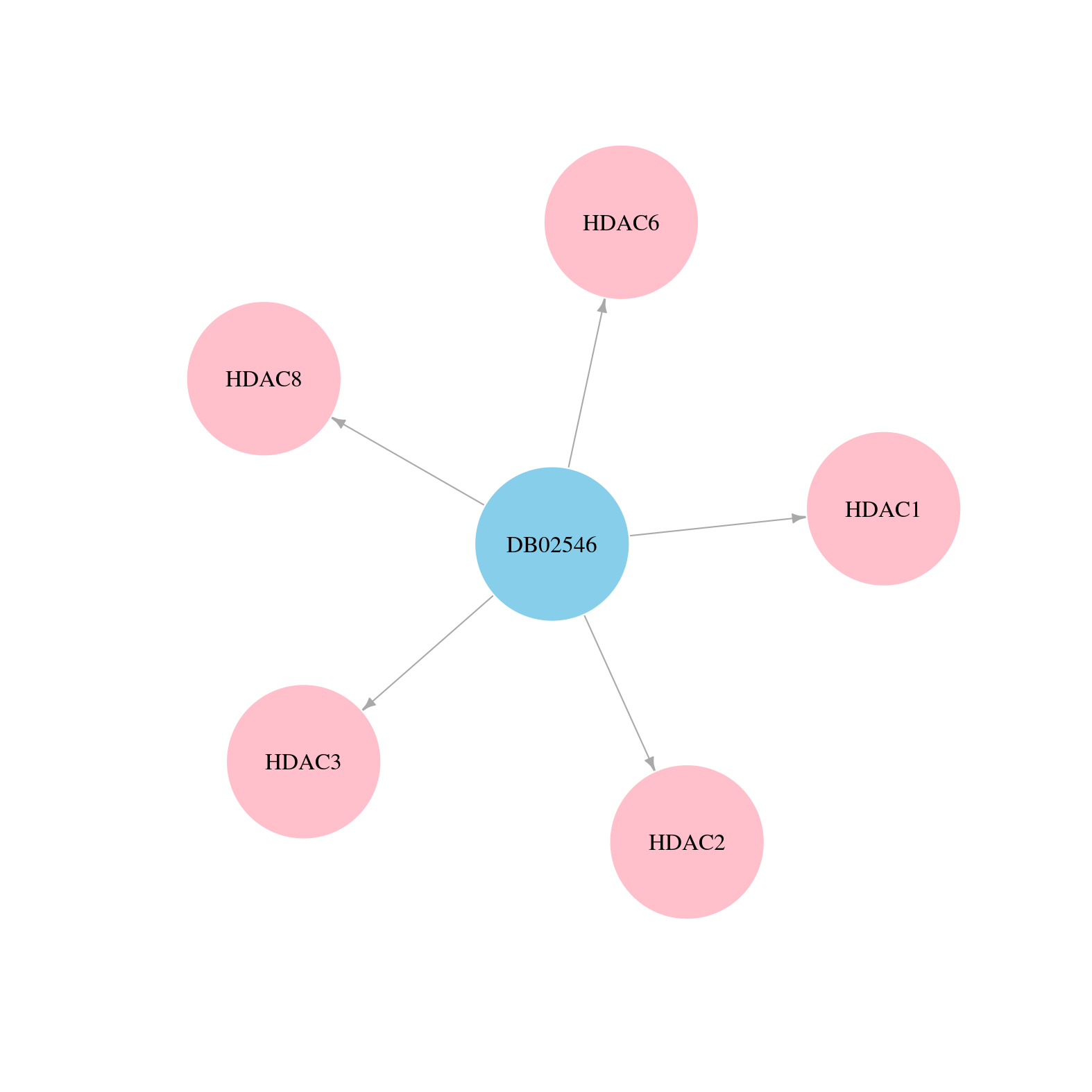
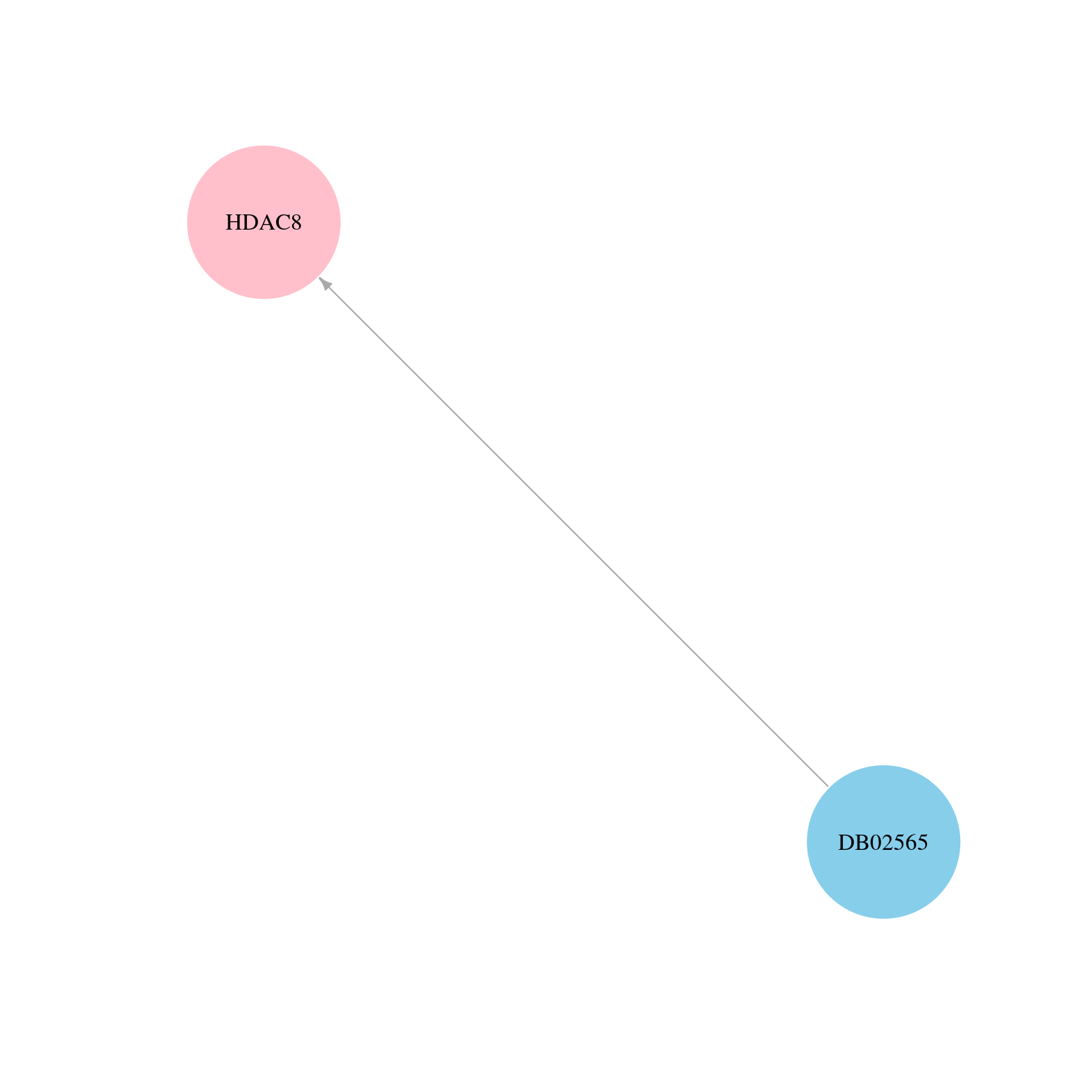
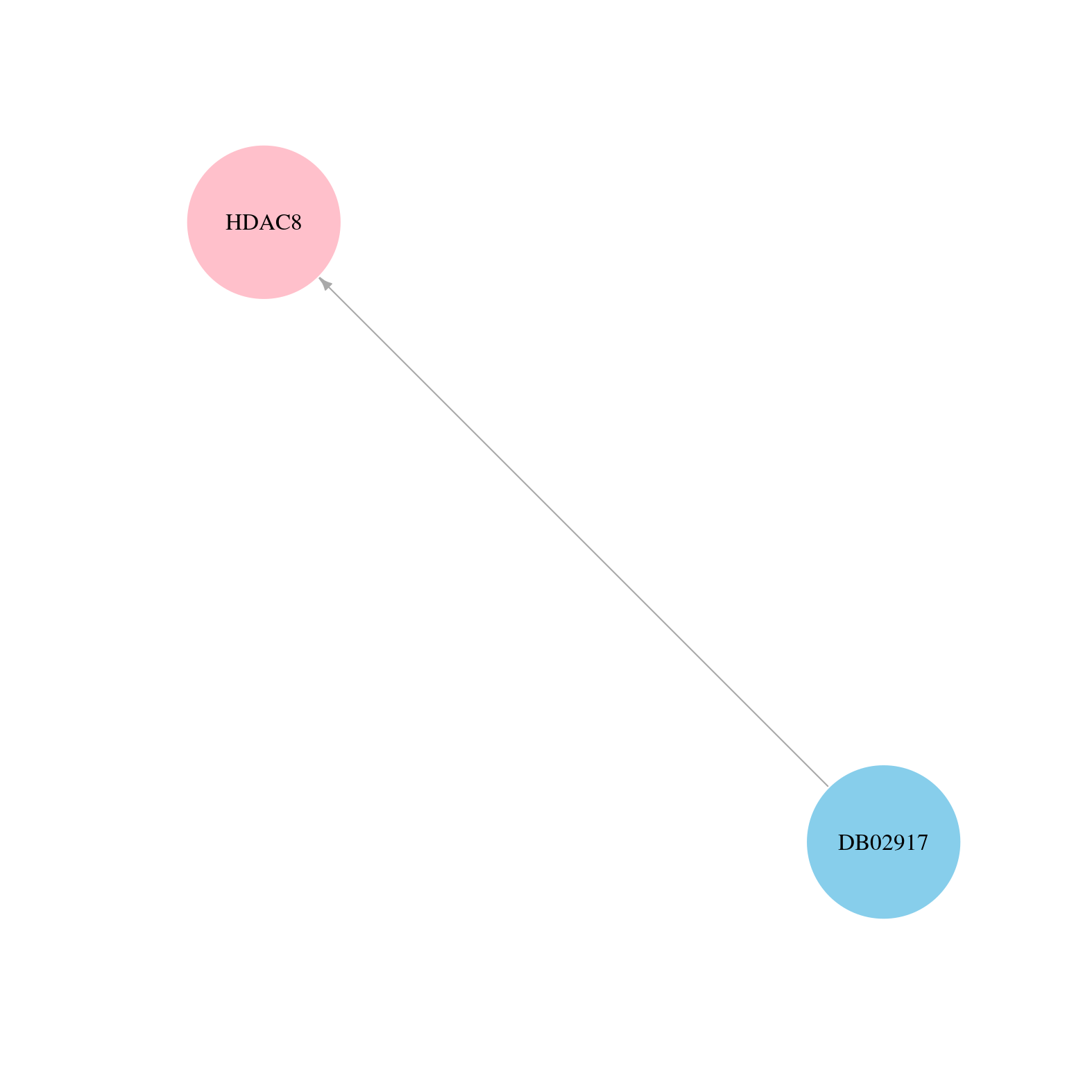
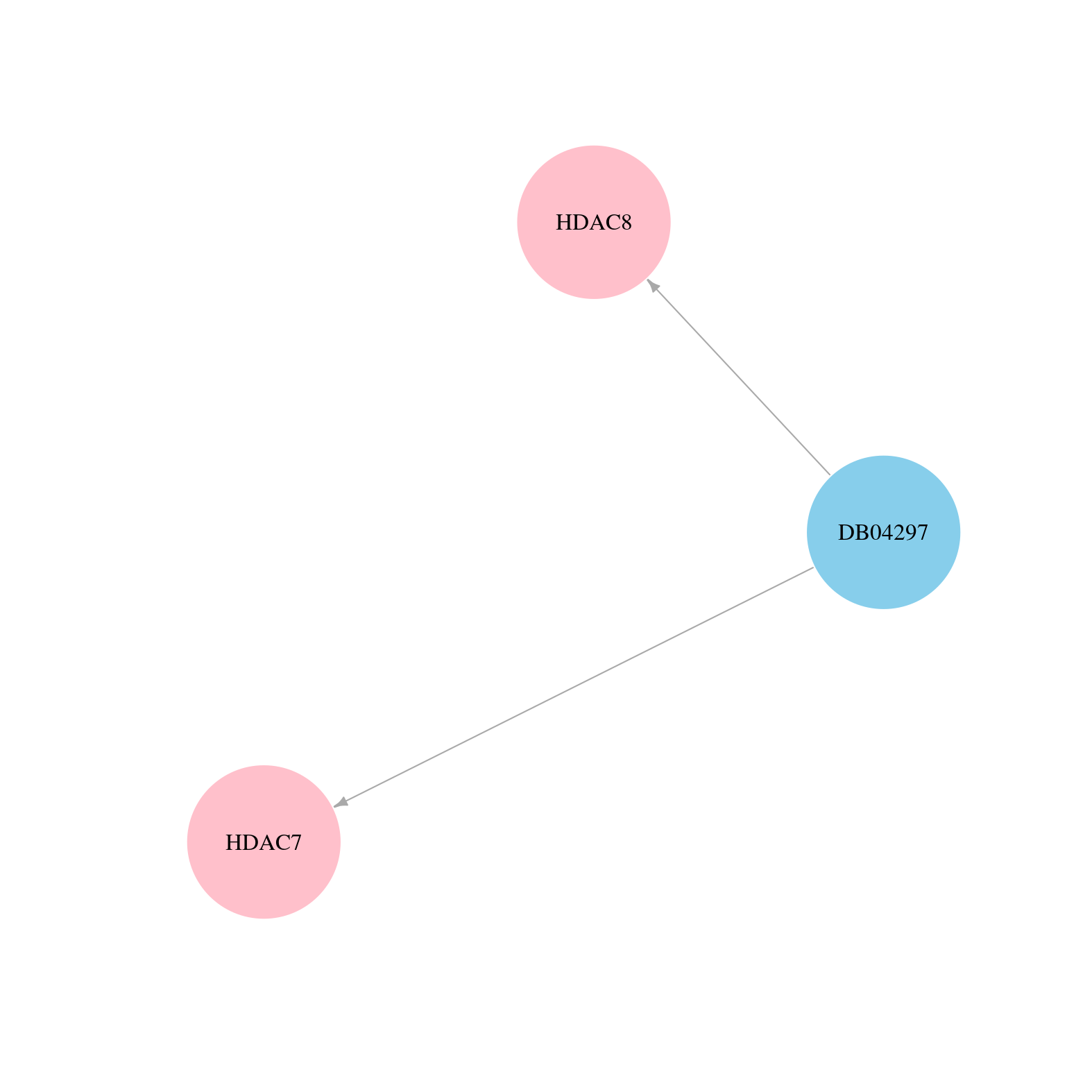
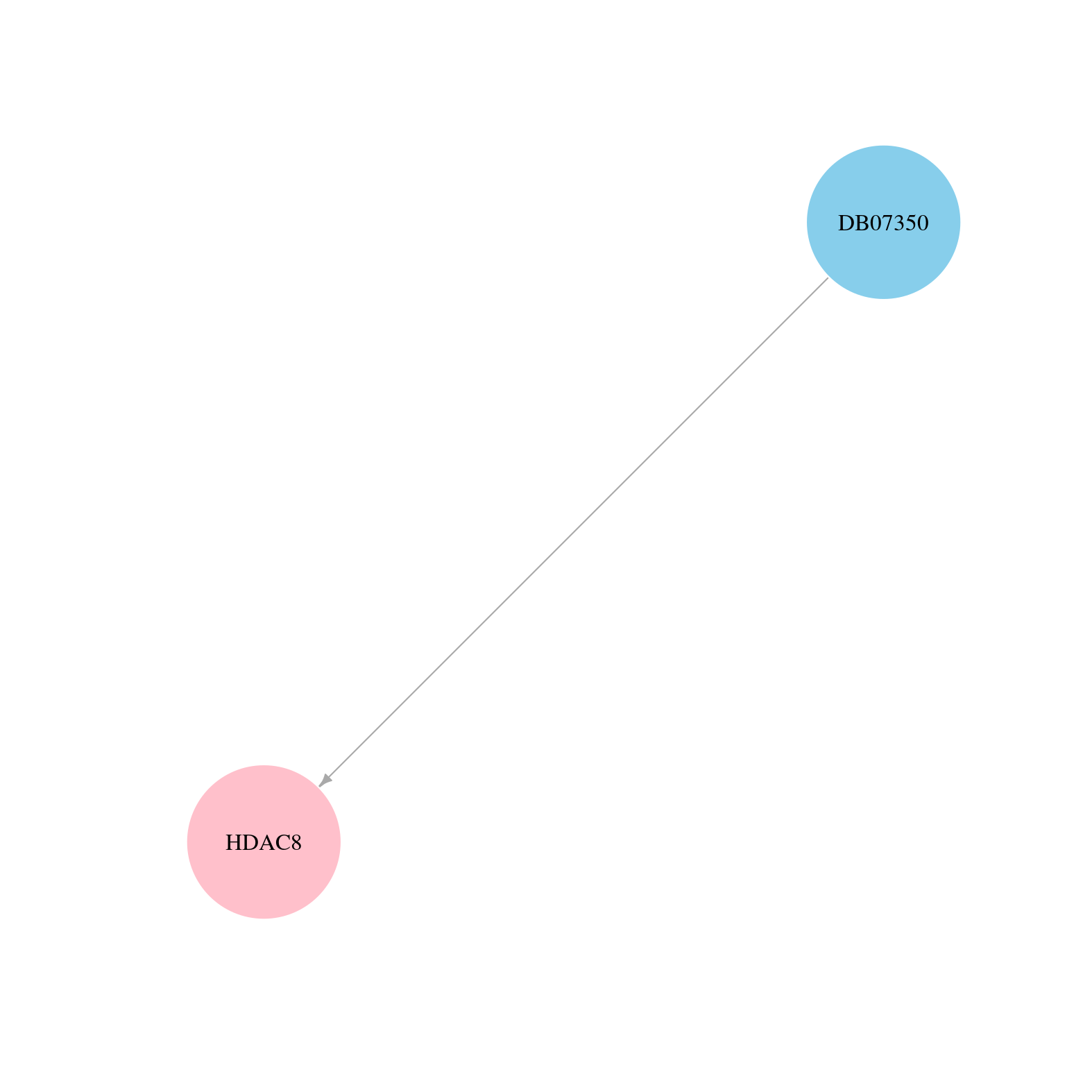
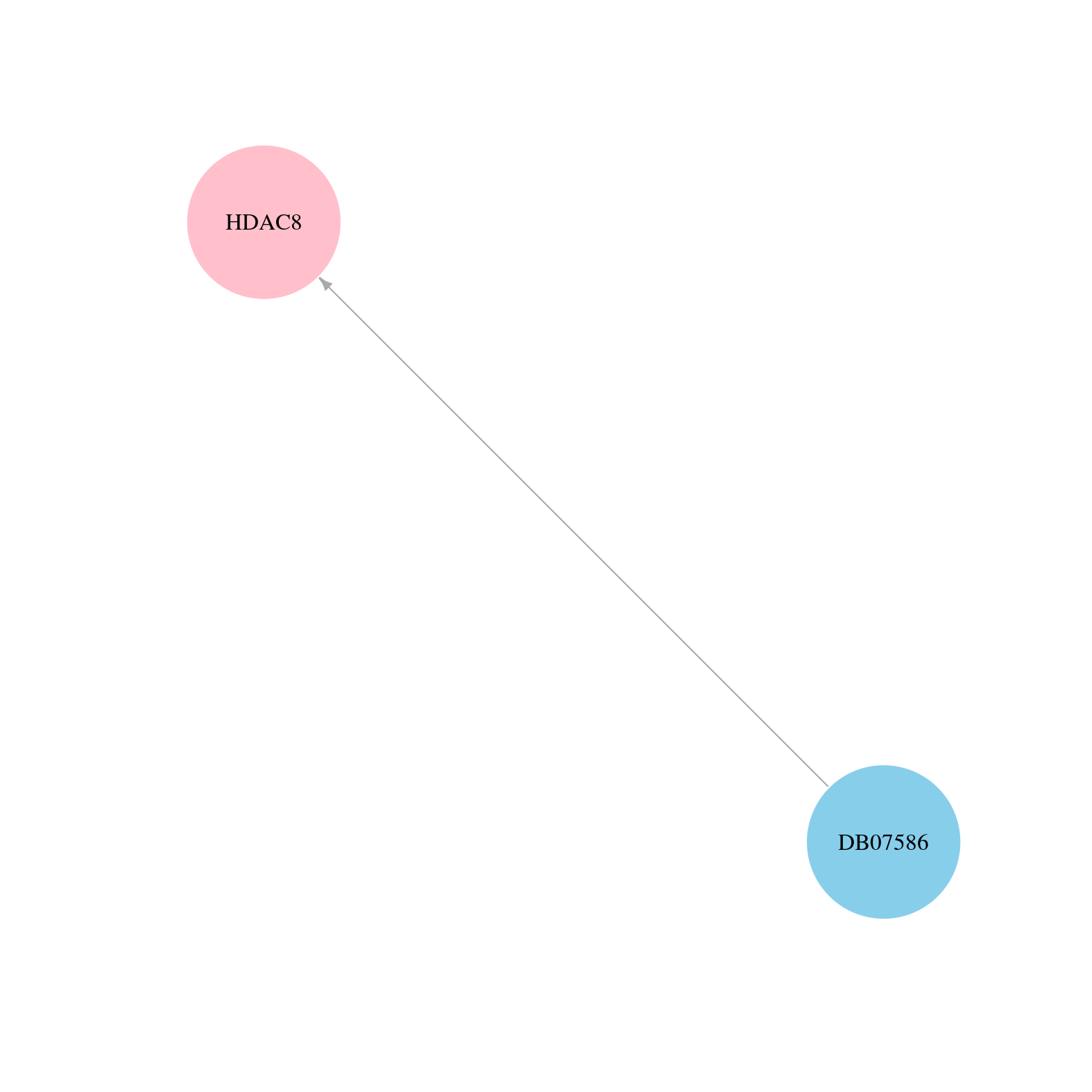
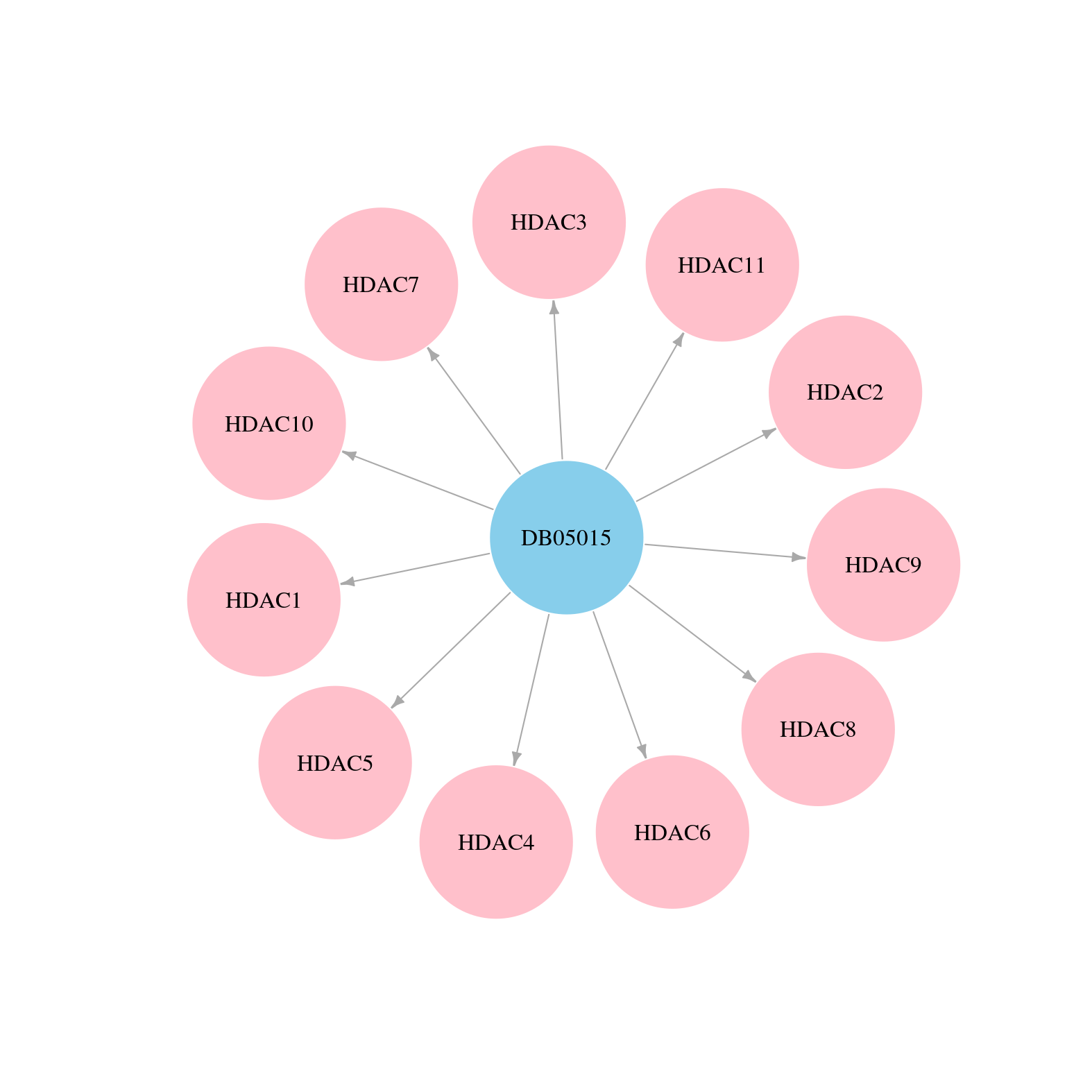
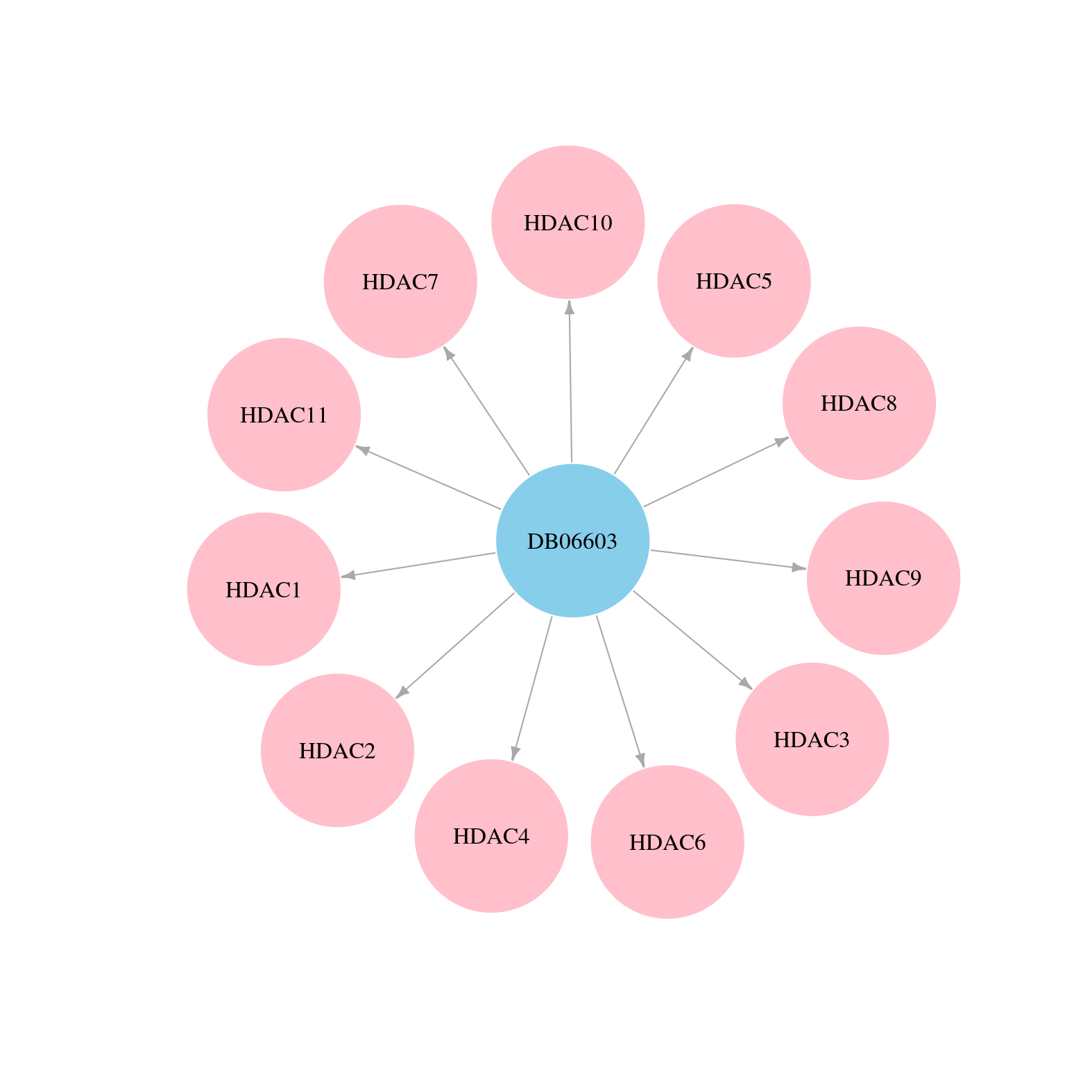
> Drugs from DrugBank database |
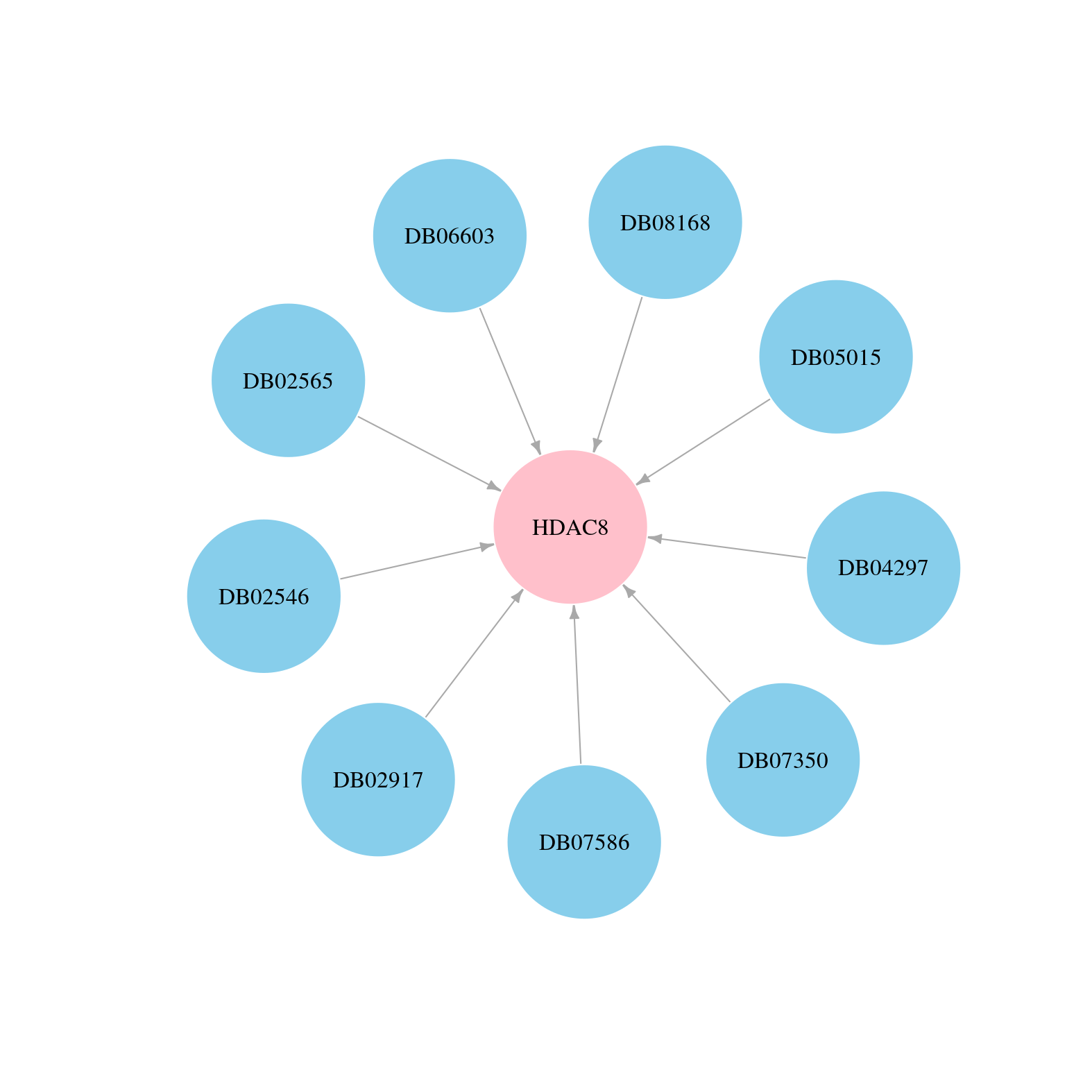
|
| Summary | |
|---|---|
| Symbol | HDAC8 |
| Name | histone deacetylase 8 |
| Aliases | HDACL1; MRXS6; histone deacetylase-like 1; Wilson-Turner X-linked mental retardation syndrome; CDA07; CDLS5; ...... |
| Location | Xq13.1 |
| External Links | HGNC, NCBI, Ensembl, Uniprot, GeneCards |
| Cancer Gene Databases | ONGene (Oncogene) , TSGene (Tumor suppressor gene) , NCG (Network of Cancer Genes) |
| Content |
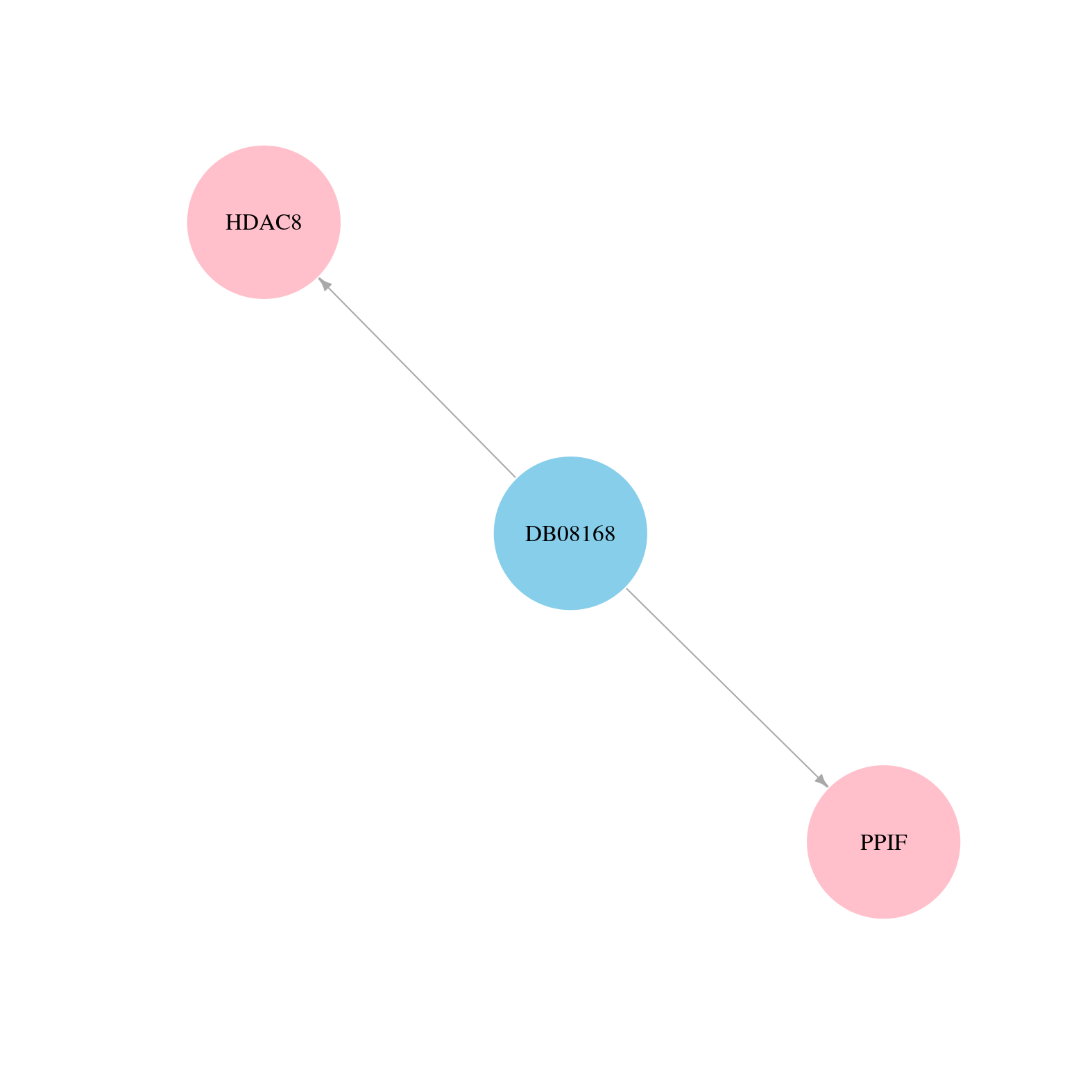
> Protein-Protein Interaction Network > miRNA Regulatory Relationship > Interactions from Text Mining |
|